Direct Regulation of DNA Repair by E2F and RB in Mammals and Plants: Core Function or Convergent Evolution?
Abstract
Simple Summary
Abstract
1. Introduction
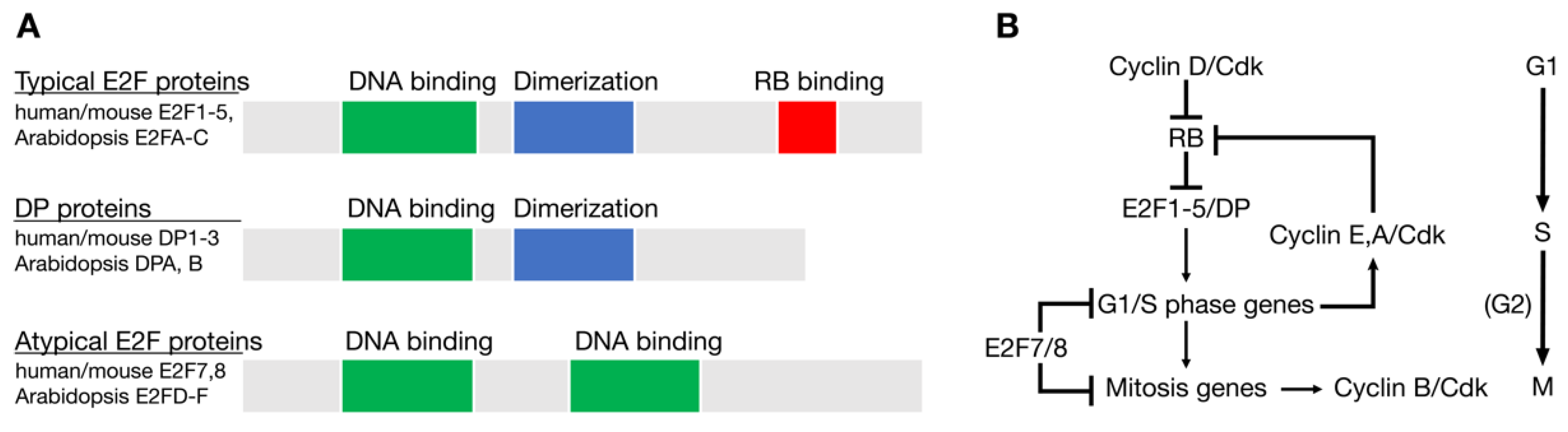
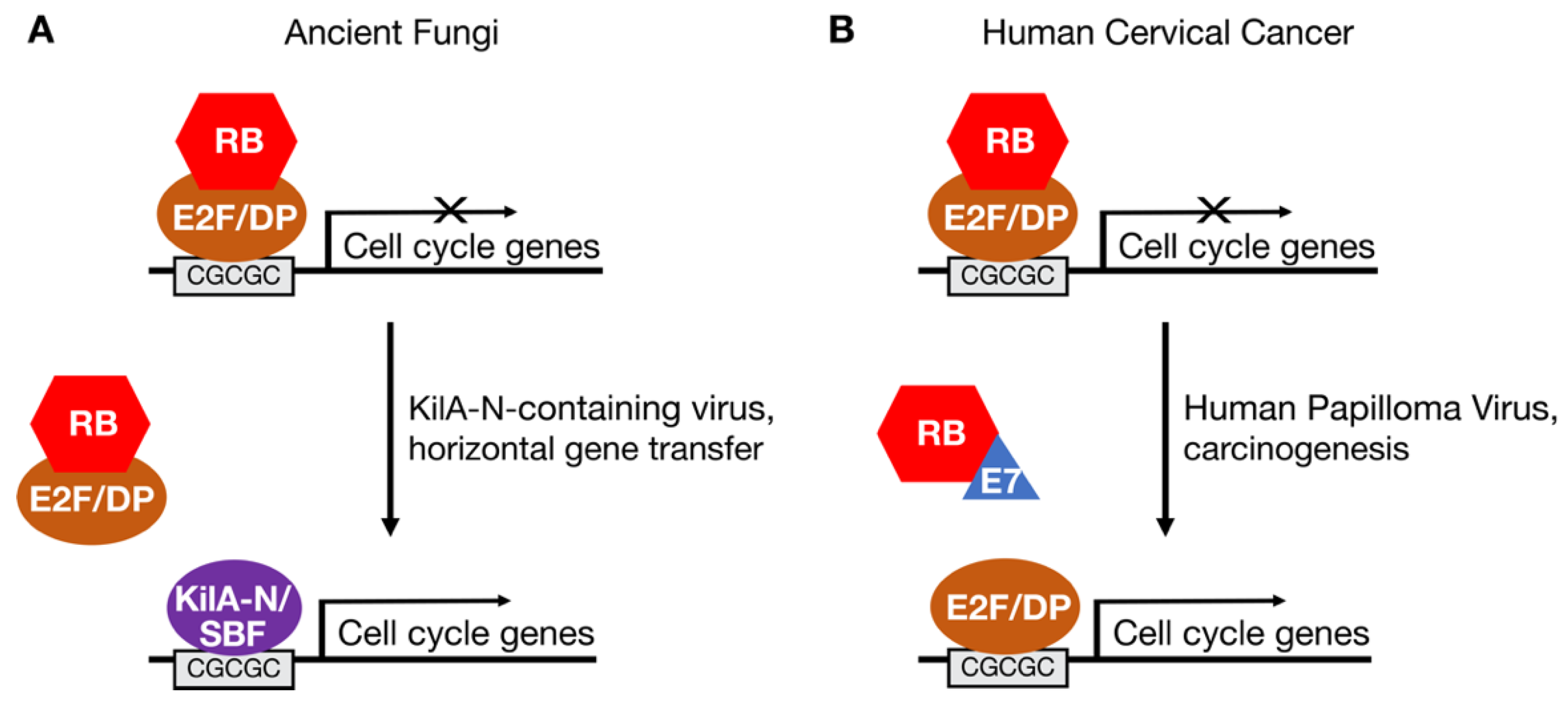
2. Evolutionary History of E2F and RB Homologs in Eukaryotes
3. Shared Functions of RB and E2F in Animals and Plants beyond the Mitotic Cell Cycle
3.1. Endoreplication
3.2. Differentiation
3.3. Repeat Silencing
3.4. Other Non-Canonical Functions
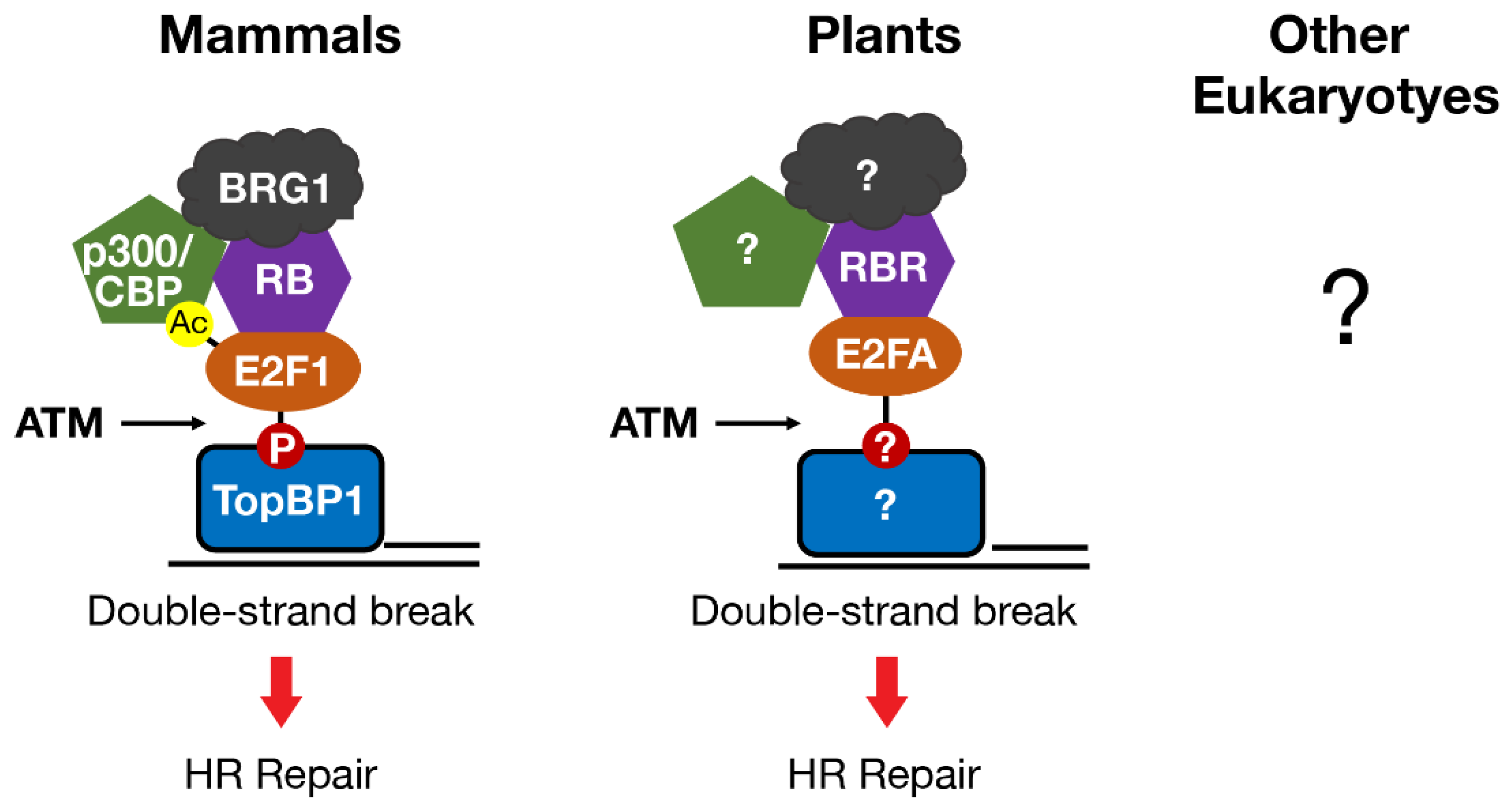
4. Homologs of E2F and RB in Mammals and Plants Directly Regulate DNA Double-Strand Break Repair
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Friend, S.H.; Bernards, R.; Rogelj, S.; Weinberg, R.A.; Rapaport, J.M.; Albert, D.M.; Dryja, T.P. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 1986, 323, 643–646. [Google Scholar] [CrossRef]
- Burkhart, D.L.; Sage, J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer 2008, 8, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Chellappan, S.P.; Hiebert, S.; Mudryj, M.; Horowitz, J.M.; Nevins, J.R. The E2F transcription factor is a cellular target for the RB protein. Cell 1991, 65, 1053–1061. [Google Scholar] [CrossRef]
- Nevins, J.R. E2F: A link between the Rb tumor suppressor protein and viral oncoproteins. Science 1992, 258, 424–429. [Google Scholar] [CrossRef]
- Helin, K.; Lees, J.A.; Vidal, M.; Dyson, N.; Harlow, E.; Fattaey, A. A cDNA encoding a pRB-binding protein with properties of the transcription factor E2F. Cell 1992, 70, 337–350. [Google Scholar] [CrossRef]
- Kaelin, W.G., Jr.; Krek, W.; Sellers, W.R.; DeCaprio, J.A.; Ajchenbaum, F.; Fuchs, C.S.; Chittenden, T.; Li, Y.; Farnham, P.J.; Blanar, M.A.; et al. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell 1992, 70, 351–364. [Google Scholar] [CrossRef]
- Grafi, G.; Burnett, R.J.; Helentjaris, T.; Larkins, B.A.; DeCaprio, J.A.; Sellers, W.R.; Kaelin, W.G., Jr. A maize cDNA encoding a member of the retinoblastoma protein family: Involvement in endoreduplication. Proc. Natl. Acad. Sci. USA 1996, 93, 8962–8967. [Google Scholar] [CrossRef] [PubMed]
- Lammens, T.; Li, J.; Leone, G.; De Veylder, L. Atypical E2Fs: New players in the E2F transcription factor family. Trends Cell Biol. 2009, 19, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Mariconti, L.; Pellegrini, B.; Cantoni, R.; Stevens, R.; Bergounioux, C.; Cella, R.; Albani, D. The E2F family of transcription factors from Arabidopsis thaliana. Novel and conserved components of the retinoblastoma/E2F pathway in plants. J. Biol. Chem. 2002, 277, 9911–9919. [Google Scholar] [CrossRef]
- Sekine, M.; Ito, M.; Uemukai, K.; Maeda, Y.; Nakagami, H.; Shinmyo, A. Isolation and characterization of the E2F-like gene in plants. FEBS Lett. 1999, 460, 117–122. [Google Scholar] [CrossRef]
- Trimarchi, J.M.; Lees, J.A. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 2002, 3, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Van den Heuvel, S.; Dyson, N.J. Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell Biol. 2008, 9, 713–724. [Google Scholar] [CrossRef]
- Du, W.; Vidal, M.; Xie, J.E.; Dyson, N. RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev. 1996, 10, 1206–1218. [Google Scholar] [CrossRef] [PubMed]
- De Jager, S.M.; Menges, M.; Bauer, U.M.; Murra, J.A. Arabidopsis E2F1 binds a sequence present in the promoter of S-phase-regulated gene AtCDC6 and is a member of a multigene family with differential activities. Plant Mol. Biol. 2001, 47, 555–568. [Google Scholar] [CrossRef]
- He, S.S.; Liu, J.; Xie, Z.; O’Neill, D.; Dotson, S. Arabidopsis E2Fa plays a bimodal role in regulating cell division and cell growth. Plant Mol. Biol. 2004, 56, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Zluhan-Martinez, E.; Perez-Koldenkova, V.; Ponce-Castaneda, M.V.; Sanchez, M.P.; Garcia-Ponce, B.; Miguel-Hernandez, S.; Alvarez-Buylla, E.R.; Garay-Arroyo, A. Beyond What Your Retina Can See: Similarities of Retinoblastoma Function between Plants and Animals, from Developmental Processes to Epigenetic Regulation. Int. J. Mol. Sci. 2020, 21, 4925. [Google Scholar] [CrossRef]
- Kent, L.N.; Leone, G. The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer 2019, 19, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, E.S.; Pruitt, S.C.; Hershberger, P.A.; Witkiewicz, A.K.; Goodrich, D.W. Cell Cycle and Beyond: Exploiting New RB1 Controlled Mechanisms for Cancer Therapy. Trends Cancer 2019, 5, 308–324. [Google Scholar] [CrossRef] [PubMed]
- Velez-Cruz, R.; Johnson, D.G. The Retinoblastoma (RB) Tumor Suppressor: Pushing Back against Genome Instability on Multiple Fronts. Int. J. Mol. Sci. 2017, 18, 1776. [Google Scholar] [CrossRef]
- Dick, F.A.; Goodrich, D.W.; Sage, J.; Dyson, N.J. Non-canonical functions of the RB protein in cancer. Nat. Rev. Cancer 2018, 18, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Peng, B.; Yao, L.; Zhang, X.; Sun, K.; Yang, X.; Yu, L. The ancient function of RB-E2F pathway: Insights from its evolutionary history. Biol. Direct 2010, 5, 55. [Google Scholar] [CrossRef]
- Cross, F.R.; Buchler, N.E.; Skotheim, J.M. Evolution of networks and sequences in eukaryotic cell cycle control. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 3532–3544. [Google Scholar] [CrossRef] [PubMed]
- Medina, E.M.; Turner, J.J.; Gordan, R.; Skotheim, J.M.; Buchler, N.E. Punctuated evolution and transitional hybrid network in an ancestral cell cycle of fungi. eLife 2016, 5, e09492. [Google Scholar] [CrossRef] [PubMed]
- Rauber, R.; Cabreira, C.; de Freitas, L.B.; Turchetto-Zolet, A.C.; Margis-Pinheiro, M. The evolutionary history of the E2F and DEL genes in Viridiplantae. Mol. Phylogenet. Evol. 2016, 99, 225–234. [Google Scholar] [CrossRef]
- Li, J.; Ran, C.; Li, E.; Gordon, F.; Comstock, G.; Siddiqui, H.; Cleghorn, W.; Chen, H.Z.; Kornacker, K.; Liu, C.G.; et al. Synergistic function of E2F7 and E2F8 is essential for cell survival and embryonic development. Dev. Cell 2008, 14, 62–75. [Google Scholar] [CrossRef]
- Desvoyes, B.; Gutierrez, C. Roles of plant retinoblastoma protein: Cell cycle and beyond. EMBO J. 2020, 39, e105802. [Google Scholar] [CrossRef]
- Costanzo, M.; Nishikawa, J.L.; Tang, X.; Millman, J.S.; Schub, O.; Breitkreuz, K.; Dewar, D.; Rupes, I.; Andrews, B.; Tyers, M. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 2004, 117, 899–913. [Google Scholar] [CrossRef] [PubMed]
- De Bruin, R.A.; McDonald, W.H.; Kalashnikova, T.I.; Yates, J., 3rd; Wittenberg, C. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 2004, 117, 887–898. [Google Scholar] [CrossRef]
- Taylor, I.A.; Treiber, M.K.; Olivi, L.; Smerdon, S.J. The X-ray structure of the DNA-binding domain from the Saccharomyces cerevisiae cell-cycle transcription factor Mbp1 at 2.1 A resolution. J. Mol. Biol. 1997, 272, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.M.; Koch, C.; Liu, Y.; Horton, J.R.; Knapp, D.; Nasmyth, K.; Cheng, X. Crystal structure of the DNA-binding domain of Mbp1, a transcription factor important in cell-cycle control of DNA synthesis. Structure 1997, 5, 349–358. [Google Scholar] [CrossRef]
- Zheng, N.; Fraenkel, E.; Pabo, C.O.; Pavletich, N.P. Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev. 1999, 13, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Koonin, E.V.; Aravind, L. Extensive domain shuffling in transcription regulators of DNA viruses and implications for the origin of fungal APSES transcription factors. Genome Biol. 2002, 3, RESEARCH0012. [Google Scholar] [CrossRef]
- Munger, K.; Howley, P.M. Human papillomavirus immortalization and transformation functions. Virus Res. 2002, 89, 213–228. [Google Scholar] [CrossRef]
- Guzman, F.; Fazeli, Y.; Khuu, M.; Salcido, K.; Singh, S.; Benavente, C.A. Retinoblastoma tumor suppressor protein roles in epigenetic regulation. Cancers 2020, 12, 2807. [Google Scholar] [CrossRef]
- Kuwabara, A.; Gruissem, W. Arabidopsis RETINOBLASTOMA-RELATED and Polycomb group proteins: Cooperation during plant cell differentiation and development. J. Exp. Bot. 2014, 65, 2667–2676. [Google Scholar] [CrossRef]
- Morris, E.J.; Dyson, N.J. Retinoblastoma protein partners. Adv. Cancer Res. 2001, 82, 1–54. [Google Scholar]
- Ach, R.A.; Durfee, T.; Miller, A.B.; Taranto, P.; Hanley-Bowdoin, L.; Zambryski, P.C.; Gruissem, W. RRB1 and RRB2 encode maize retinoblastoma-related proteins that interact with a plant D-type cyclin and geminivirus replication protein. Mol. Cell. Biol. 1997, 17, 5077–5086. [Google Scholar] [CrossRef] [PubMed]
- Nevins, J.R. Cell cycle targets of the DNA tumor viruses. Curr. Opin. Genet. Dev. 1994, 4, 130–134. [Google Scholar] [CrossRef]
- Lee, H.O.; Davidson, J.M.; Duronio, R.J. Endoreplication: Polyploidy with purpose. Genes Dev. 2009, 23, 2461–2477. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Z.; Ouseph, M.M.; Li, J.; Pecot, T.; Chokshi, V.; Kent, L.; Bae, S.; Byrne, M.; Duran, C.; Comstock, G.; et al. Canonical and atypical E2Fs regulate the mammalian endocycle. Nat. Cell Biol. 2012, 14, 1192–1202. [Google Scholar] [CrossRef] [PubMed]
- Kohn, M.J.; Bronson, R.T.; Harlow, E.; Dyson, N.J.; Yamasaki, L. Dp1 is required for extra-embryonic development. Development 2003, 130, 1295–1305. [Google Scholar] [CrossRef] [PubMed]
- Magyar, Z.; Horvath, B.; Khan, S.; Mohammed, B.; Henriques, R.; De Veylder, L.; Bako, L.; Scheres, B.; Bogre, L. Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO J. 2012, 31, 1480–1493. [Google Scholar] [CrossRef]
- Ouellet, J.; Roy, R. The lin-35/Rb and RNAi pathways cooperate to regulate a key cell cycle transition in C. elegans. BMC Dev. Biol. 2007, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- Park, J.A.; Ahn, J.W.; Kim, Y.K.; Kim, S.J.; Kim, J.K.; Kim, W.T.; Pai, H.S. Retinoblastoma protein regulates cell proliferation, differentiation, and endoreduplication in plants. Plant J. 2005, 42, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Sabelli, P.A.; Liu, Y.; Dante, R.A.; Lizarraga, L.E.; Nguyen, H.N.; Brown, S.W.; Klingler, J.P.; Yu, J.; LaBrant, E.; Layton, T.M.; et al. Control of cell proliferation, endoreduplication, cell size, and cell death by the retinoblastoma-related pathway in maize endosperm. Proc. Natl. Acad. Sci. USA 2013, 110, E1827–E1836. [Google Scholar] [CrossRef]
- Weng, L.; Zhu, C.; Xu, J.; Du, W. Critical role of active repression by E2F and Rb proteins in endoreplication during Drosophila development. EMBO J. 2003, 22, 3865–3875. [Google Scholar] [CrossRef]
- Desvoyes, B.; de Mendoza, A.; Ruiz-Trillo, I.; Gutierrez, C. Novel roles of plant RETINOBLASTOMA-RELATED (RBR) protein in cell proliferation and asymmetric cell division. J. Exp. Bot. 2014, 65, 2657–2666. [Google Scholar] [CrossRef]
- Ahlander, J.; Chen, X.B.; Bosco, G. The N-terminal domain of the Drosophila retinoblastoma protein Rbf1 interacts with ORC and associates with chromatin in an E2F independent manner. PLoS ONE 2008, 3, e2831. [Google Scholar] [CrossRef] [PubMed]
- Bosco, G.; Du, W.; Orr-Weaver, T.L. DNA replication control through interaction of E2F-RB and the origin recognition complex. Nat. Cell Biol. 2001, 3, 289–295. [Google Scholar] [CrossRef]
- Julian, L.M.; Vandenbosch, R.; Pakenham, C.A.; Andrusiak, M.G.; Nguyen, A.P.; McClellan, K.A.; Svoboda, D.S.; Lagace, D.C.; Park, D.S.; Leone, G.; et al. Opposing regulation of Sox2 by cell-cycle effectors E2f3a and E2f3b in neural stem cells. Cell Stem Cell 2013, 12, 440–452. [Google Scholar] [CrossRef]
- Kareta, M.S.; Gorges, L.L.; Hafeez, S.; Benayoun, B.A.; Marro, S.; Zmoos, A.F.; Cecchini, M.J.; Spacek, D.; Batista, L.F.; O’Brien, M.; et al. Inhibition of pluripotency networks by the Rb tumor suppressor restricts reprogramming and tumorigenesis. Cell Stem Cell 2015, 16, 39–50. [Google Scholar] [CrossRef]
- Wildwater, M.; Campilho, A.; Perez-Perez, J.M.; Heidstra, R.; Blilou, I.; Korthout, H.; Chatterjee, J.; Mariconti, L.; Gruissem, W.; Scheres, B. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 2005, 123, 1337–1349. [Google Scholar] [CrossRef] [PubMed]
- Gutzat, R.; Borghi, L.; Gruissem, W. Emerging roles of RETINOBLASTOMA-RELATED proteins in evolution and plant development. Trends Plant Sci. 2012, 17, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, P.L.; Wu, L.; de Bruin, A.; Chong, J.L.; Chen, W.Y.; Dureska, G.; Sites, E.; Pan, T.; Sharma, A.; Huang, K. Rb is critical in a mammalian tissue stem cell population. Genes Dev. 2007, 21, 85–97. [Google Scholar] [CrossRef]
- Chen, H.Z.; Tsai, S.Y.; Leone, G. Emerging roles of E2Fs in cancer: An exit from cell cycle control. Nat. Rev. Cancer 2009, 9, 785–797. [Google Scholar] [CrossRef]
- Classon, M.; Dyson, N. p107 and p130: Versatile proteins with interesting pockets. Exp. Cell Res. 2001, 264, 135–147. [Google Scholar] [CrossRef]
- Calo, E.; Quintero-Estades, J.A.; Danielian, P.S.; Nedelcu, S.; Berman, S.D.; Lees, J.A. Rb regulates fate choice and lineage commitment in vivo. Nature 2010, 466, 1110–1114. [Google Scholar] [CrossRef]
- Clarke, A.R.; Maandag, E.R.; van Roon, M.; van der Lugt, N.M.; van der Valk, M.; Hooper, M.L.; Berns, A.; te Riele, H. Requirement for a functional Rb-1 gene in murine development. Nature 1992, 359, 328–330. [Google Scholar] [CrossRef] [PubMed]
- Flowers, S.; Beck, G.R., Jr.; Moran, E. Transcriptional activation by pRB and its coordination with SWI/SNF recruitment. Cancer Res. 2010, 70, 8282–8287. [Google Scholar] [CrossRef]
- Flowers, S.; Xu, F.; Moran, E. Cooperative activation of tissue-specific genes by pRB and E2F1. Cancer Res. 2013, 73, 2150–2158. [Google Scholar] [CrossRef] [PubMed]
- Goupille, O.; Penglong, T.; Kadri, Z.; Granger-Locatelli, M.; Denis, R.; Luquet, S.; Badoual, C.; Fucharoen, S.; Maouche-Chretien, L.; Leboulch, P.; et al. The LXCXE Retinoblastoma Protein-Binding Motif of FOG-2 Regulates Adipogenesis. Cell Rep. 2017, 21, 3524–3535. [Google Scholar] [CrossRef]
- Jacks, T.; Fazeli, A.; Schmitt, E.M.; Bronson, R.T.; Goodell, M.A.; Weinberg, R.A. Effects of an Rb mutation in the mouse. Nature 1992, 359, 295–300. [Google Scholar] [CrossRef]
- Lee, E.Y.; Chang, C.Y.; Hu, N.; Wang, Y.C.; Lai, C.C.; Herrup, K.; Lee, W.H.; Bradley, A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 1992, 359, 288–294. [Google Scholar] [CrossRef]
- Julian, L.M.; Liu, Y.; Pakenham, C.A.; Dugal-Tessier, D.; Ruzhynsky, V.; Bae, S.; Tsai, S.Y.; Leone, G.; Slack, R.S.; Blais, A. Tissue-specific targeting of cell fate regulatory genes by E2f factors. Cell Death Differ. 2016, 23, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Leviczky, T.; Molnar, E.; Papdi, C.; Oszi, E.; Horvath, G.V.; Vizler, C.; Nagy, V.; Pauk, J.; Bogre, L.; Magyar, Z. E2FA and E2FB transcription factors coordinate cell proliferation with seed maturation. Development 2019, 146, dev179333. [Google Scholar] [CrossRef] [PubMed]
- Kadri, Z.; Shimizu, R.; Ohneda, O.; Maouche-Chretien, L.; Gisselbrecht, S.; Yamamoto, M.; Romeo, P.H.; Leboulch, P.; Chretien, S. Direct binding of pRb/E2F-2 to GATA-1 regulates maturation and terminal cell division during erythropoiesis. PLoS Biol. 2009, 7, e1000123. [Google Scholar] [CrossRef]
- Perilli, S.; Perez-Perez, J.M.; Di Mambro, R.; Llavata Peris, C.; Diaz-Trivino, S.; Del Bianco, M.; Pierdonati, E.; Moubayidin, L.; Cruz-Ramirez, A.; Costantino, P.; et al. RETINOBLASTOMA-RELATED protein stimulates cell differentiation in the Arabidopsis root meristem by interacting with cytokinin signaling. Plant Cell 2013, 25, 4469–4478. [Google Scholar] [CrossRef]
- Bouyer, D.; Heese, M.; Chen, P.; Harashima, H.; Roudier, F.; Gruttner, C.; Schnittger, A. Genome-wide identification of RETINOBLASTOMA RELATED 1 binding sites in Arabidopsis reveals novel DNA damage regulators. PLoS Genet. 2018, 14, e1007797. [Google Scholar] [CrossRef] [PubMed]
- Ishak, C.A.; Marshall, A.E.; Passos, D.T.; White, C.R.; Kim, S.J.; Cecchini, M.J.; Ferwati, S.; MacDonald, W.A.; Howlett, C.J.; Welch, I.D.; et al. An RB-EZH2 Complex Mediates Silencing of Repetitive DNA Sequences. Mol. Cell 2016, 64, 1074–1087. [Google Scholar] [CrossRef]
- Cecchini, M.J.; Dick, F.A. The biochemical basis of CDK phosphorylation-independent regulation of E2F1 by the retinoblastoma protein. Biochem. J. 2011, 434, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Dick, F.A.; Dyson, N. pRB contains an E2F1-specific binding domain that allows E2F1-induced apoptosis to be regulated separately from other E2F activities. Mol. Cell 2003, 12, 639–649. [Google Scholar] [CrossRef]
- Montoya-Durango, D.E.; Ramos, K.A.; Bojang, P.; Ruiz, L.; Ramos, I.N.; Ramos, K.S. LINE-1 silencing by retinoblastoma proteins is effected through the nucleosomal and remodeling deacetylase multiprotein complex. BMC Cancer 2016, 16, 38. [Google Scholar] [CrossRef]
- Henaff, E.; Vives, C.; Desvoyes, B.; Chaurasia, A.; Payet, J.; Gutierrez, C.; Casacuberta, J.M. Extensive amplification of the E2F transcription factor binding sites by transposons during evolution of Brassica species. Plant J. 2014, 77, 852–862. [Google Scholar] [CrossRef]
- Gu, X.; Jiang, D.; Yang, W.; Jacob, Y.; Michaels, S.D.; He, Y. Arabidopsis homologs of retinoblastoma-associated protein 46/48 associate with a histone deacetylase to act redundantly in chromatin silencing. PLoS Genet. 2011, 7, e1002366. [Google Scholar] [CrossRef] [PubMed]
- Feschotte, C.; Pritham, E.J. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 2007, 41, 331–368. [Google Scholar] [CrossRef]
- Wells, J.N.; Feschotte, C. A Field Guide to Eukaryotic Transposable Elements. Annu. Rev. Genet. 2020, 54, 539–561. [Google Scholar] [CrossRef] [PubMed]
- Gladden, A.B.; Diehl, J.A. The cyclin D1-dependent kinase associates with the pre-replication complex and modulates RB.MCM7 binding. J. Biol. Chem. 2003, 278, 9754–9760. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Maldonado, R.; Paolinelli, R.; Galbiati, L.; Giadrossi, S.; Giacca, M. Interaction of the retinoblastoma protein with Orc1 and its recruitment to human origins of DNA replication. PLoS ONE 2010, 5, e13720. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, P.; Winter, S.L.; Alexandrow, M.G. Cell cycle arrest by transforming growth factor beta1 near G1/S is mediated by acute abrogation of prereplication complex activation involving an Rb-MCM interaction. Mol. Cell. Biol. 2010, 30, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Avni, D.; Yang, H.; Martelli, F.; Hofmann, F.; ElShamy, W.M.; Ganesan, S.; Scully, R.; Livingston, D.M. Active localization of the retinoblastoma protein in chromatin and its response to S phase DNA damage. Mol. Cell 2003, 12, 735–746. [Google Scholar] [CrossRef]
- Hilgendorf, K.I.; Leshchiner, E.S.; Nedelcu, S.; Maynard, M.A.; Calo, E.; Ianari, A.; Walensky, L.D.; Lees, J.A. The retinoblastoma protein induces apoptosis directly at the mitochondria. Genes Dev. 2013, 27, 1003–1015. [Google Scholar] [CrossRef]
- Coschi, C.H.; Ishak, C.A.; Gallo, D.; Marshall, A.; Talluri, S.; Wang, J.; Cecchini, M.J.; Martens, A.L.; Percy, V.; Welch, I.; et al. Haploinsufficiency of an RB-E2F1-Condensin II complex leads to aberrant replication and aneuploidy. Cancer Discov. 2014, 4, 840–853. [Google Scholar] [CrossRef] [PubMed]
- Longworth, M.S.; Herr, A.; Ji, J.Y.; Dyson, N.J. RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dCAP-D3. Genes Dev. 2008, 22, 1011–1024. [Google Scholar] [CrossRef] [PubMed]
- Manning, A.L.; Longworth, M.S.; Dyson, N.J. Loss of pRB causes centromere dysfunction and chromosomal instability. Genes Dev. 2010, 24, 1364–1376. [Google Scholar] [CrossRef]
- Ishak, C.A.; Coschi, C.H.; Roes, M.V.; Dick, F.A. Disruption of CDK-resistant chromatin association by pRB causes DNA damage, mitotic errors, and reduces Condensin II recruitment. Cell Cycle 2017, 16, 1430–1439. [Google Scholar] [CrossRef][Green Version]
- Gonzalo, S.; Garcia-Cao, M.; Fraga, M.F.; Schotta, G.; Peters, A.H.; Cotter, S.E.; Eguia, R.; Dean, D.C.; Esteller, M.; Jenuwein, T.; et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat. Cell Biol. 2005, 7, 420–428. [Google Scholar] [CrossRef]
- Siddiqui, H.; Fox, S.R.; Gunawardena, R.W.; Knudsen, E.S. Loss of RB compromises specific heterochromatin modifications and modulates HP1alpha dynamics. J. Cell. Physiol. 2007, 211, 131–137. [Google Scholar] [CrossRef]
- Isaac, C.E.; Francis, S.M.; Martens, A.L.; Julian, L.M.; Seifried, L.A.; Erdmann, N.; Binne, U.K.; Harrington, L.; Sicinski, P.; Berube, N.G.; et al. The retinoblastoma protein regulates pericentric heterochromatin. Mol. Cell. Biol. 2006, 26, 3659–3671. [Google Scholar] [CrossRef] [PubMed]
- Biedermann, S.; Harashima, H.; Chen, P.; Heese, M.; Bouyer, D.; Sofroni, K.; Schnittger, A. The retinoblastoma homolog RBR1 mediates localization of the repair protein RAD51 to DNA lesions in Arabidopsis. EMBO J. 2017, 36, 1279–1297. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhu, F.; Weaks, R.L.; Biswas, A.K.; Guo, R.; Li, Y.; Johnson, D.G. E2F1 promotes the recruitment of DNA repair factors to sites of DNA double-strand breaks. Cell Cycle 2011, 10, 1287–1294. [Google Scholar] [CrossRef]
- Horvath, B.M.; Kourova, H.; Nagy, S.; Nemeth, E.; Magyar, Z.; Papdi, C.; Ahmad, Z.; Sanchez-Perez, G.F.; Perilli, S.; Blilou, I.; et al. Arabidopsis RETINOBLASTOMA RELATED directly regulates DNA damage responses through functions beyond cell cycle control. EMBO J. 2017, 36, 1261–1278. [Google Scholar] [CrossRef]
- Lang, J.; Smetana, O.; Sanchez-Calderon, L.; Lincker, F.; Genestier, J.; Schmit, A.C.; Houlne, G.; Chaboute, M.E. Plant gammaH2AX foci are required for proper DNA DSB repair responses and colocalize with E2F factors. New Phytol. 2012, 194, 353–363. [Google Scholar] [CrossRef]
- Liu, K.; Lin, F.T.; Ruppert, J.M.; Lin, W.C. Regulation of E2F1 by BRCT domain-containing protein TopBP1. Mol. Cell. Biol. 2003, 23, 3287–3304. [Google Scholar] [CrossRef] [PubMed]
- Velez-Cruz, R.; Manickavinayaham, S.; Biswas, A.K.; Clary, R.W.; Premkumar, T.; Cole, F.; Johnson, D.G. RB localizes to DNA double-strand breaks and promotes DNA end resection and homologous recombination through the recruitment of BRG1. Genes Dev. 2016, 30, 2500–2512. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.H.; Kim, K.P. E2F1 facilitates DNA break repair by localizing to break sites and enhancing the expression of homologous recombination factors. Exp. Mol. Med. 2019, 51, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gentric, N.; Masoud, K.; Journot, R.P.; Cognat, V.; Chaboute, M.E.; Noir, S.; Genschik, P. The F-Box-Like Protein FBL17 Is a Regulator of DNA-Damage Response and Colocalizes with RETINOBLASTOMA RELATED1 at DNA Lesion Sites. Plant Physiol. 2020, 183, 1295–1305. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Yam, J.C.; Tham, C.C.; Pang, C.P.; Chu, W.K. RB Regulates DNA Double Strand Break Repair Pathway Choice by Mediating CtIP Dependent End Resection. Int. J. Mol. Sci. 2020, 21, 9176. [Google Scholar] [CrossRef]
- Manickavinayaham, S.; Velez-Cruz, R.; Biswas, A.K.; Bedford, E.; Klein, B.J.; Kutateladze, T.G.; Liu, B.; Bedford, M.T.; Johnson, D.G. E2F1 acetylation directs p300/CBP-mediated histone acetylation at DNA double-strand breaks to facilitate repair. Nat. Commun. 2019, 10, 4951. [Google Scholar] [CrossRef]
- Xiao, H.; Goodrich, D.W. The retinoblastoma tumor suppressor protein is required for efficient processing and repair of trapped topoisomerase II-DNA-cleavable complexes. Oncogene 2005, 24, 8105–8113. [Google Scholar] [CrossRef]
- Chen, Z.; Higgins, J.D.; Hui, J.T.; Li, J.; Franklin, F.C.; Berger, F. Retinoblastoma protein is essential for early meiotic events in Arabidopsis. EMBO J. 2011, 30, 744–755. [Google Scholar] [CrossRef] [PubMed]
- Dunaief, J.L.; Strober, B.E.; Guha, S.; Khavari, P.A.; Alin, K.; Luban, J.; Begemann, M.; Crabtree, G.R.; Goff, S.P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 1994, 79, 119–130. [Google Scholar] [CrossRef]
- Fry, C.J.; Pearson, A.; Malinowski, E.; Bartley, S.M.; Greenblatt, J.; Farnham, P.J. Activation of the murine dihydrofolate reductase promoter by E2F1. A requirement for CBP recruitment. J. Biol. Chem. 1999, 274, 15883–15891. [Google Scholar] [CrossRef]
- Ogiwara, H.; Kohno, T. CBP and p300 histone acetyltransferases contribute to homologous recombination by transcriptionally activating the BRCA1 and RAD51 genes. PLoS ONE 2012, 7, e52810. [Google Scholar] [CrossRef] [PubMed]
- Pietrzak, J.; Ploszaj, T.; Pulaski, L.; Robaszkiewicz, A. EP300-HDAC1-SWI/SNF functional unit defines transcription of some DNA repair enzymes during differentiation of human macrophages. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 198–208. [Google Scholar] [CrossRef] [PubMed]
- Strobeck, M.W.; Knudsen, K.E.; Fribourg, A.F.; DeCristofaro, M.F.; Weissman, B.E.; Imbalzano, A.N.; Knudsen, E.S. BRG-1 is required for RB-mediated cell cycle arrest. Proc. Natl. Acad. Sci. USA 2000, 97, 7748–7753. [Google Scholar] [CrossRef] [PubMed]
- Trouche, D.; Cook, A.; Kouzarides, T. The CBP co-activator stimulates E2F1/DP1 activity. Nucleic Acids Res. 1996, 24, 4139–4145. [Google Scholar] [CrossRef] [PubMed]
- Chiu, L.Y.; Gong, F.; Miller, K.M. Bromodomain proteins: Repairing DNA damage within chromatin. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160286. [Google Scholar] [CrossRef]
- Harrod, A.; Lane, K.A.; Downs, J.A. The role of the SWI/SNF chromatin remodelling complex in the response to DNA double strand breaks. DNA Repair 2020, 93, 102919. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Lee, S.Y.; Miller, K.M. Preserving genome integrity and function: The DNA damage response and histone modifications. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 208–241. [Google Scholar] [CrossRef]
- Murr, R.; Loizou, J.I.; Yang, Y.G.; Cuenin, C.; Li, H.; Wang, Z.Q.; Herceg, Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 2006, 8, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H. Chromatin Remodeling and Epigenetic Regulation in Plant DNA Damage Repair. Int. J. Mol. Sci. 2019, 20, 4093. [Google Scholar] [CrossRef]
- Biswas, A.K.; Mitchell, D.L.; Johnson, D.G. E2F1 Responds to Ultraviolet Radiation by Directly Stimulating DNA Repair and Suppressing Carcinogenesis. Cancer Res. 2014, 74, 3369–3377. [Google Scholar] [CrossRef] [PubMed]
- Marshall, A.E.; Roes, M.V.; Passos, D.T.; DeWeerd, M.C.; Chaikovsky, A.C.; Sage, J.; Howlett, C.J.; Dick, F.A. RB1 deletion in retinoblastoma protein pathway-disrupted cells results in DNA damage and cancer progression. Mol. Cell. Biol. 2019, 39, e00105-19. [Google Scholar] [CrossRef] [PubMed]
- Lavin, M.F.; Yeo, A.J. Clinical potential of ATM inhibitors. Mutat. Res. 2020, 821, 111695. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manickavinayaham, S.; Dennehey, B.K.; Johnson, D.G. Direct Regulation of DNA Repair by E2F and RB in Mammals and Plants: Core Function or Convergent Evolution? Cancers 2021, 13, 934. https://doi.org/10.3390/cancers13050934
Manickavinayaham S, Dennehey BK, Johnson DG. Direct Regulation of DNA Repair by E2F and RB in Mammals and Plants: Core Function or Convergent Evolution? Cancers. 2021; 13(5):934. https://doi.org/10.3390/cancers13050934
Chicago/Turabian StyleManickavinayaham, Swarnalatha, Briana K. Dennehey, and David G. Johnson. 2021. "Direct Regulation of DNA Repair by E2F and RB in Mammals and Plants: Core Function or Convergent Evolution?" Cancers 13, no. 5: 934. https://doi.org/10.3390/cancers13050934
APA StyleManickavinayaham, S., Dennehey, B. K., & Johnson, D. G. (2021). Direct Regulation of DNA Repair by E2F and RB in Mammals and Plants: Core Function or Convergent Evolution? Cancers, 13(5), 934. https://doi.org/10.3390/cancers13050934





