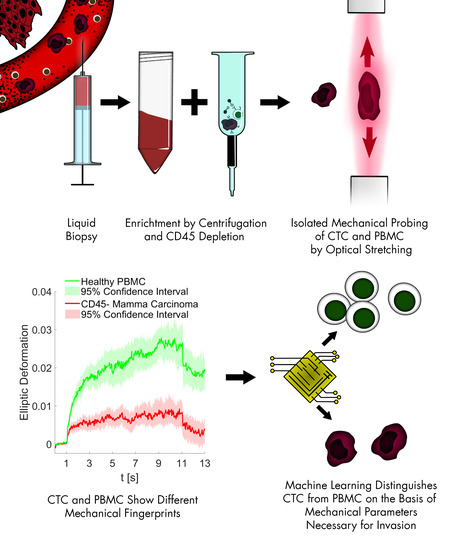
The Mechanical Fingerprint of Circulating Tumor Cells (CTCs) in Breast Cancer Patients
Abstract
Simple Summary
Abstract
1. Introduction
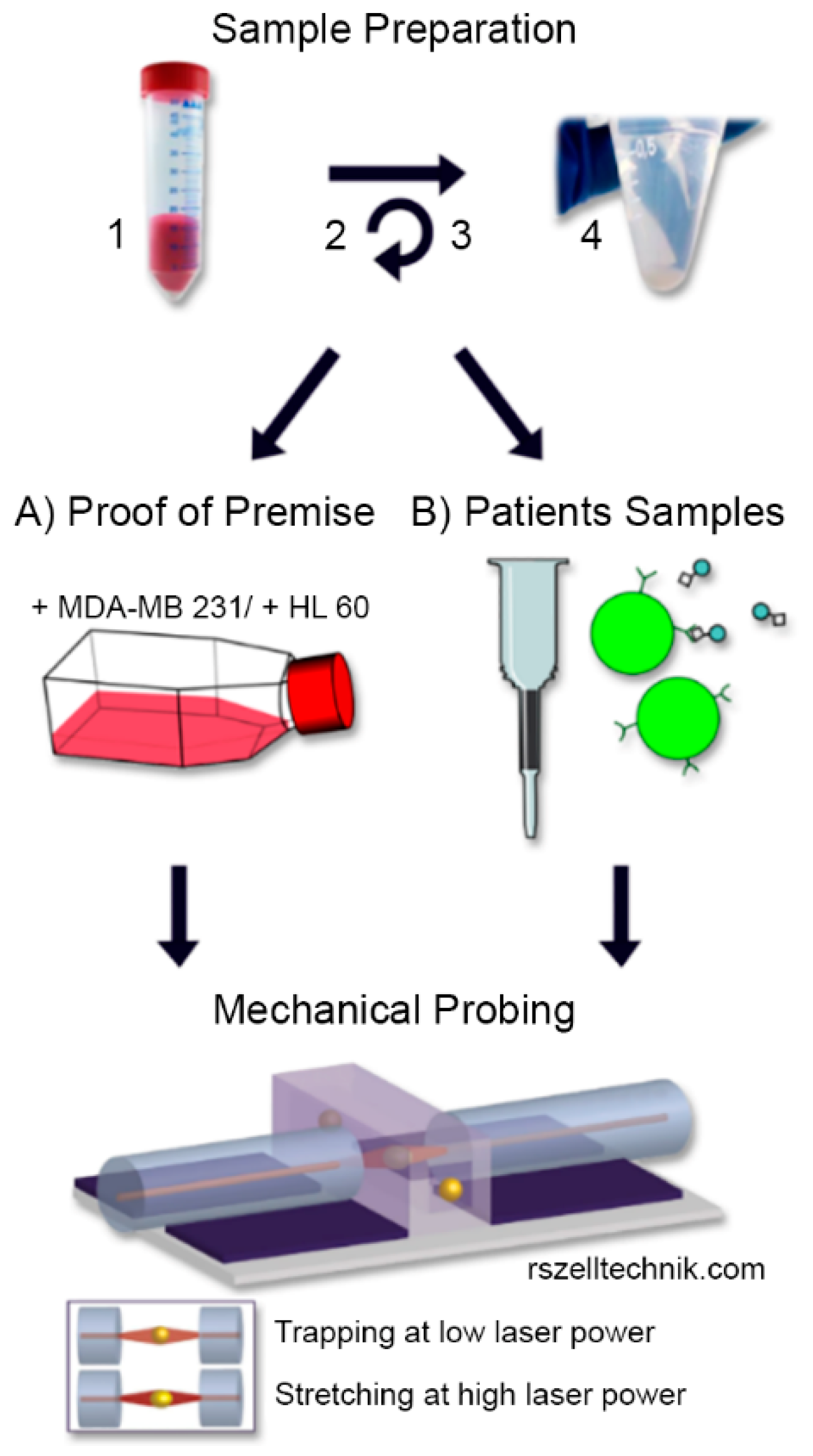
2. Materials and Methods
2.1. Study Population, Blood Samples and Informed Consent
2.2. Cell Culture
2.3. Sample Preparation and CTC Enrichment
2.4. Cell Rheological Measurements
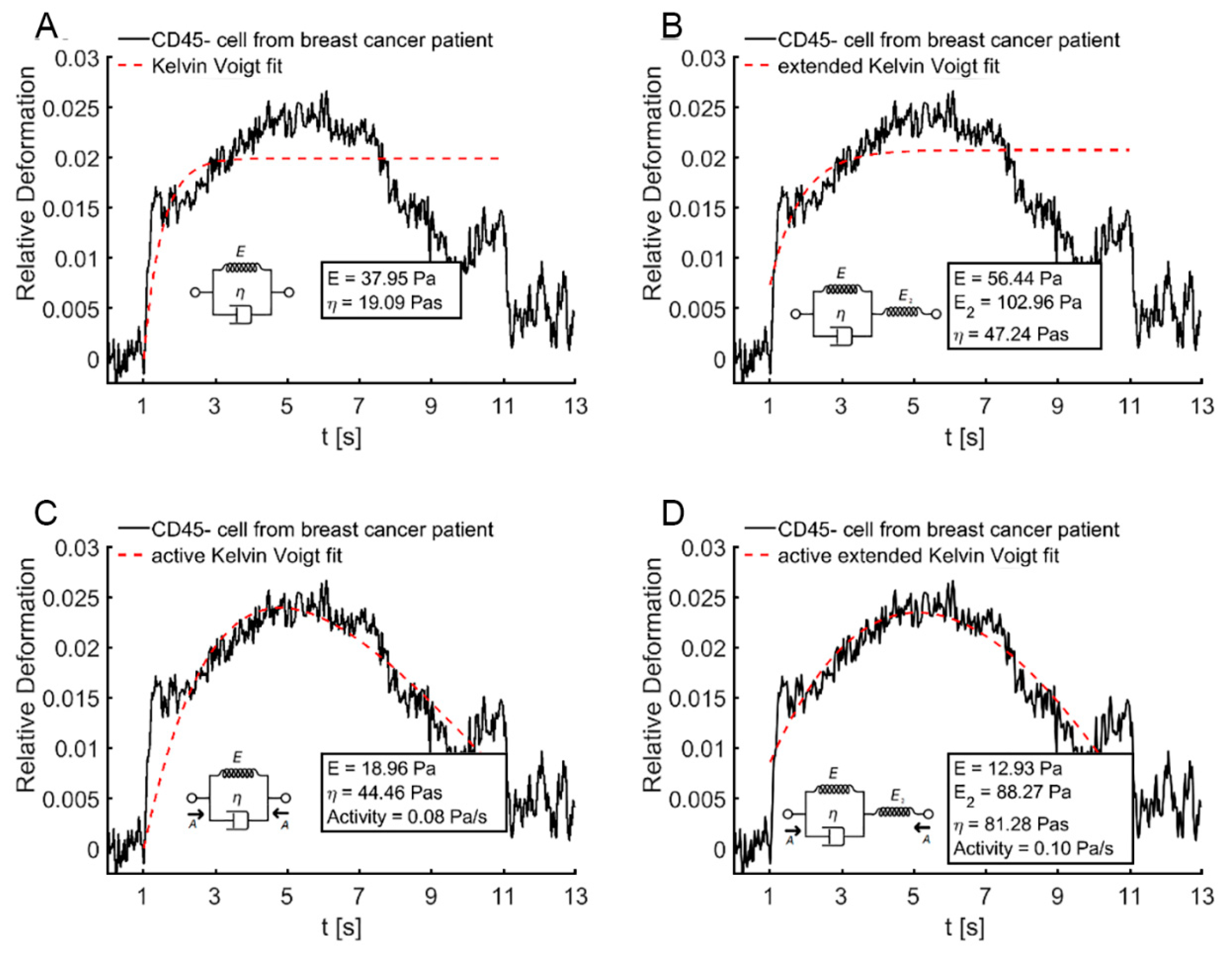
2.5. Kelvin–Voigt Fitting
2.6. Machine Learning
2.7. Statistical Analysis
3. Results
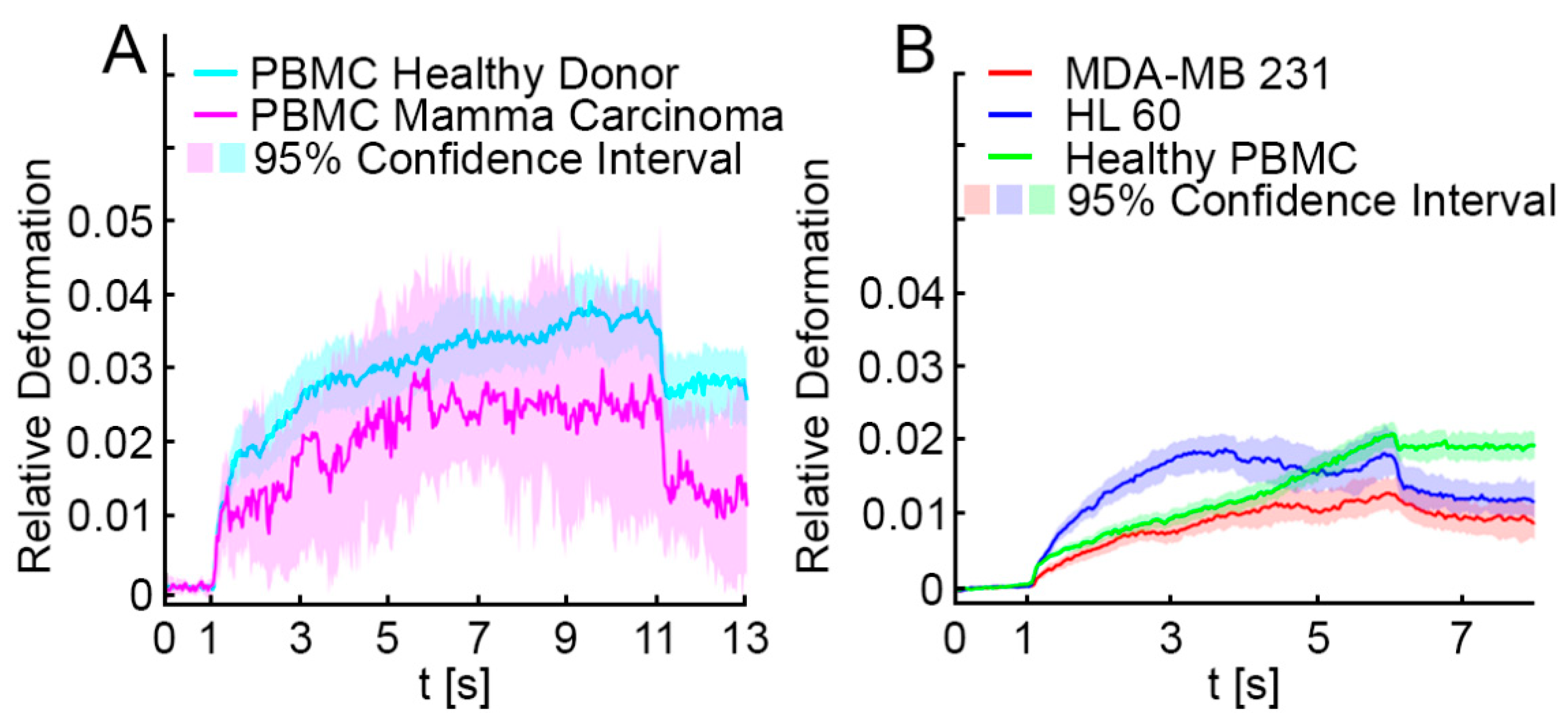
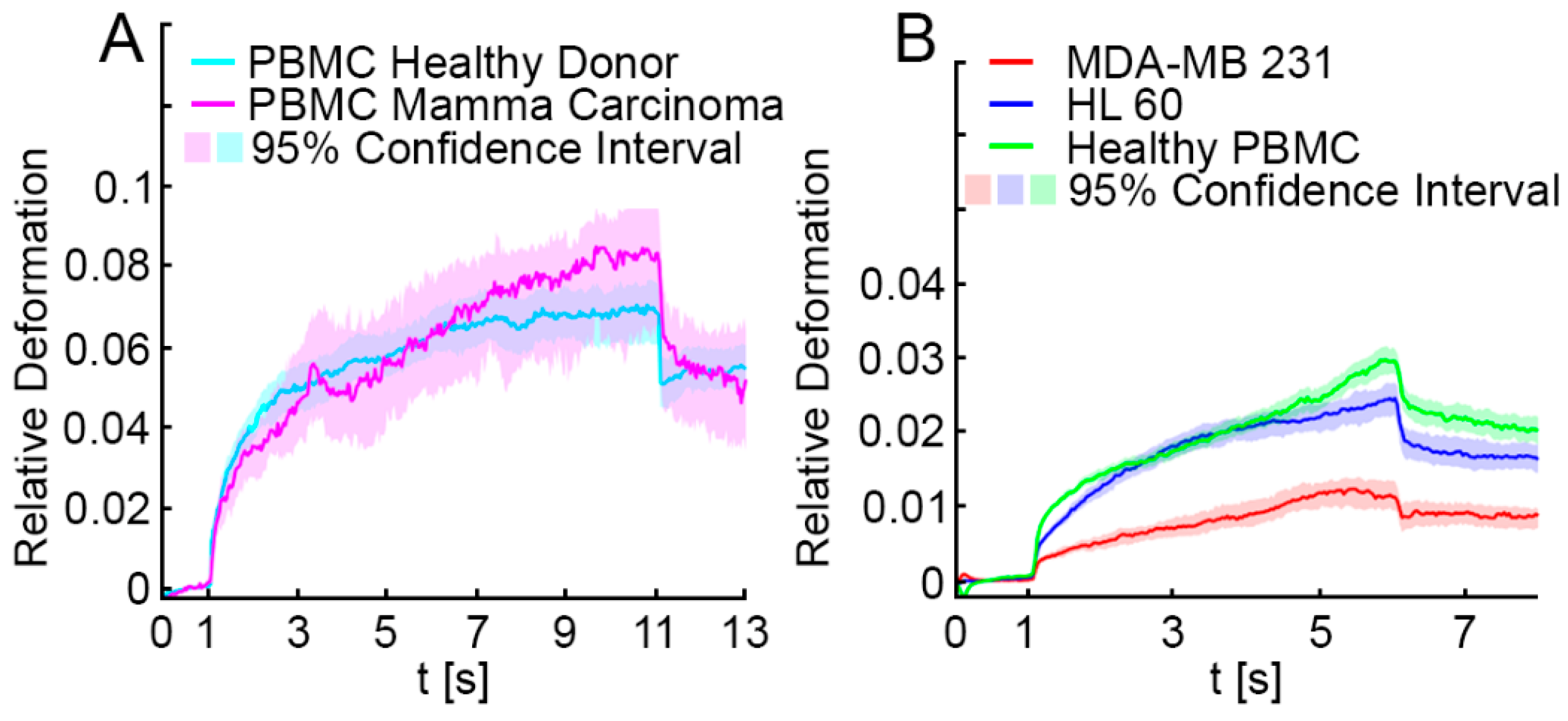
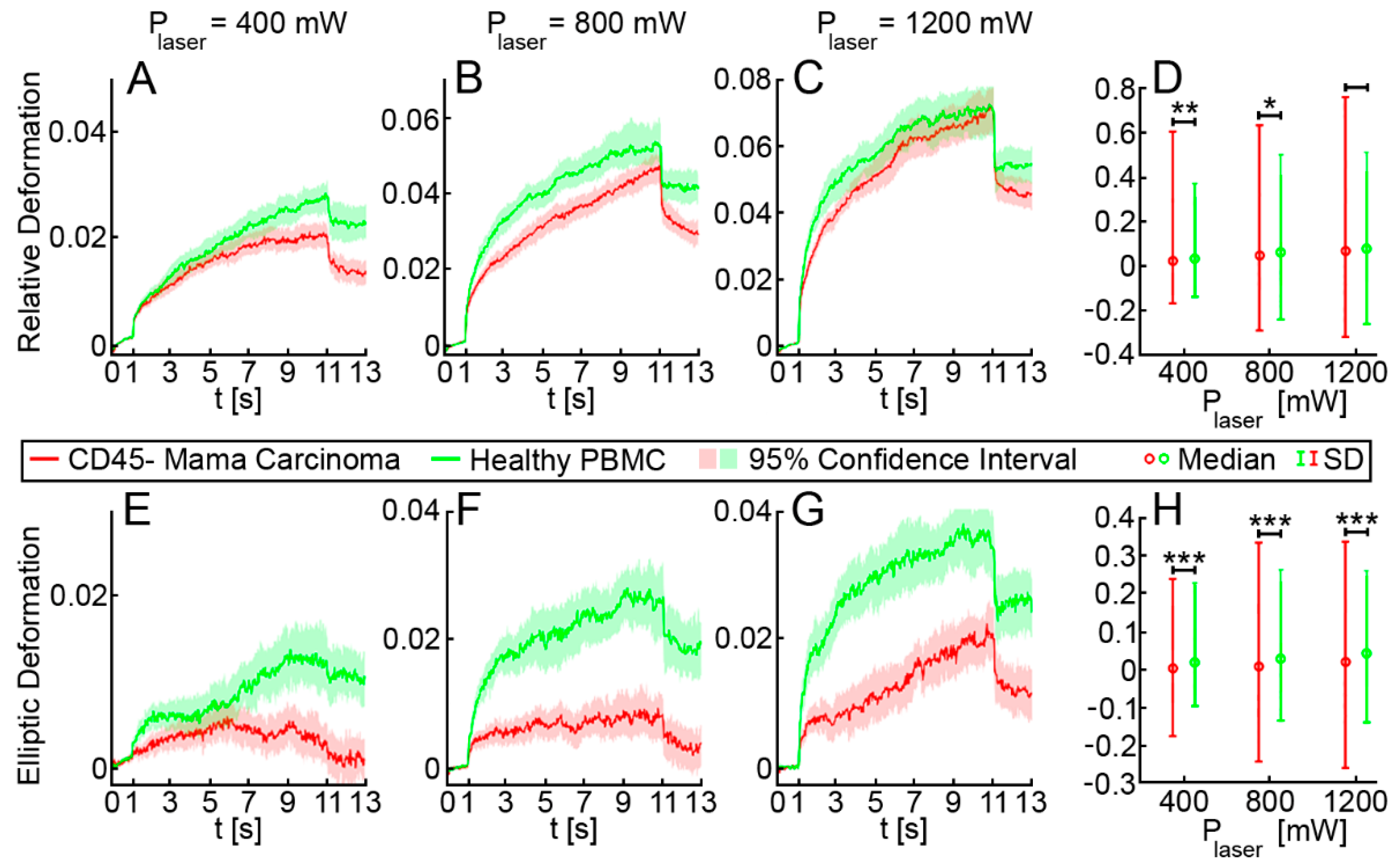
3.1. Optical Stretching Reveals Significant Differences in the Resistance between White Blood Cells, MDA-MB 231 and HL-60
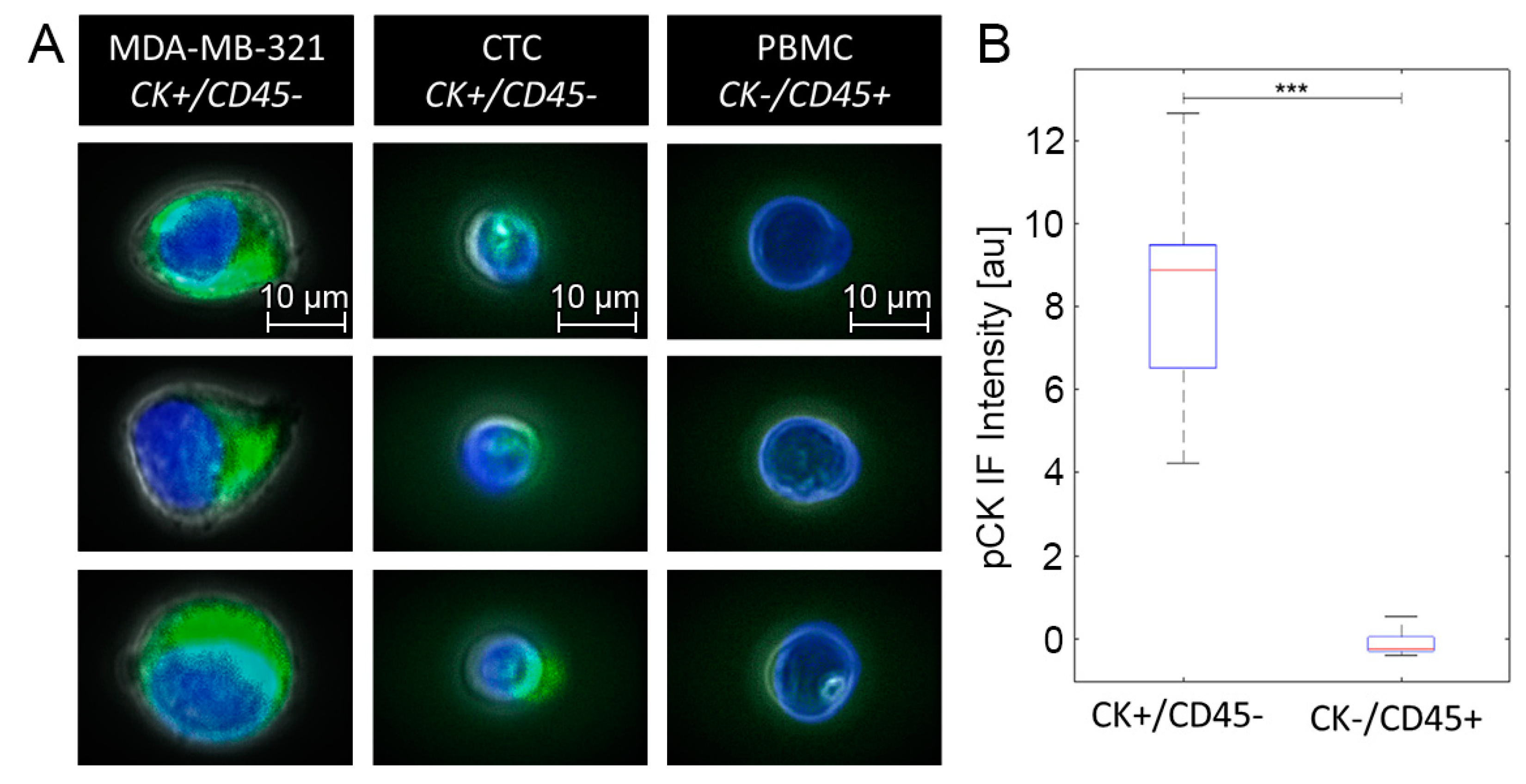
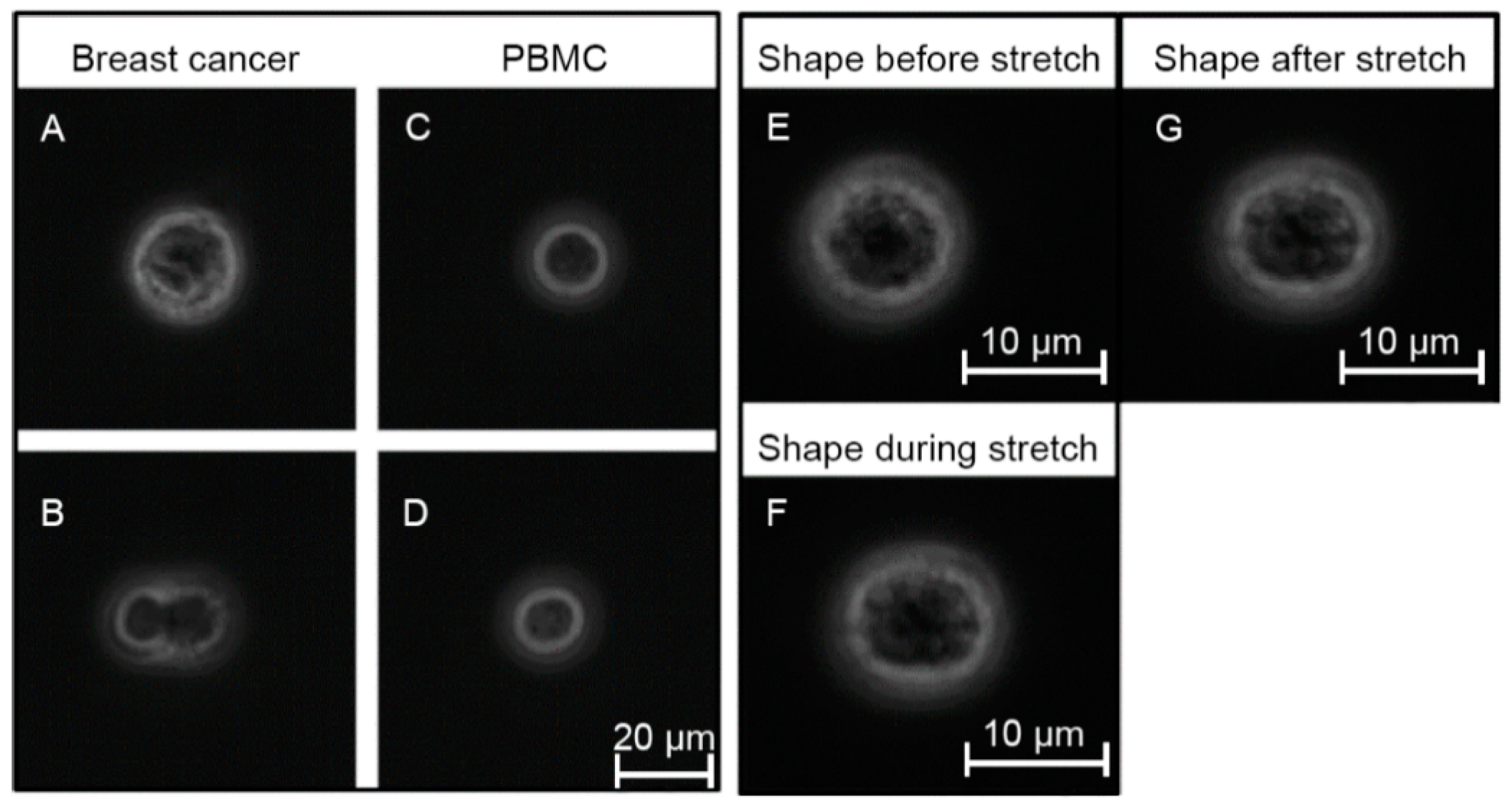
3.2. Elliptic Deformation and Shape Restoration Discriminate Blood Cells and CTC Candidates from Mamma Carcinoma
3.3. CTC Candidates Can Be Distinguished from Blood Cells by Machine Learning
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Breast Cancer Patients | Healthy Donors | ||||
|---|---|---|---|---|---|
| CD45+ | CD45− | Ratio of CD45− to CD45+ | CD45+ | CD45− | Ratio of CD45− to CD45+ |
| 3,000,000 | 10,000 | 0.33% | 10,000,000 | 10,000 | 0.10% |
| 1,300,000 | 10,000 | 0.77% | 3,750,000 | 12,500 | 0.33% |
| 7500,000 | 10,000 | 0.13% | 6,500,000 | 25,000 | 0.38% |
| 10,000,000 | 100,000 | 1.00% | 10,200,000 | 10,000 | 0.10% |
| 12,000,000 | 20,000 | 0.17% | 25,000,000 | 100,000 | 0.40% |
| 8,000,000 | 10,000 | 0.13% | |||
| 10,000,000 | 70,000 | 0.70% | |||
| 8,000,000 | 200,000 | 2.50% | |||
| 12,000,000 | 200,000 | 1.67% | |||
| 11,500,000 | 300,000 | 2.61% | |||
| 7,440,000 | 10,000 | 0.13% | |||
| 6,580,000 | 10,000 | 0.15% | |||
| 10,400,000 | 1,000,000 | 9.62% | |||
| 12,000,000 | 300,000 | 2.50% | |||
| 8,600,000 | 10,000 | 0.12% | |||
| 15,800,000 | 1,500,000 | 9.49% | |||
| 13,500,000 | 400,000 | 2.96% | |||
| 9,500,000 | 80,000 | 0.84% | |||
| 9,500,000 | 3,000,000 | 31.58% | |||
| 20,500,000 | 10,000 | 0.05% | |||
| 11,250,000 | 135,000 | 1.20% | |||
| 10,750,000 | 350,000 | 3.26% | |||
| Mean: 3.26% | Mean: 0.25% | ||||
| Median: 0.92% | Median: 0.33% | ||||
| Range: 0.05–31.58 | Range: 0.1–0.4 | ||||
Appendix B
| Excluding Shape Restoration | Including Shape Restoration | |||||||
|---|---|---|---|---|---|---|---|---|
| Parameter | 400 mW | 800 mW | 1200 mW | Pooled | 400 mW | 800 mW | 1200 mW | Pooled |
| 1. | CR (0.017) | CA (0.016) | CA (0.030) | CR (0.015) | CR (0.016) | CR (0.017) | CR (0.032) | CA (0.018) |
| 2. | CA (0.015) | eRD (0.007) | CR (0.030) | CA (0.012) | RD (0.009) | CA (0.015) | Restoration (0.012) | CR (0.013) |
| 3. | eRD (0.010) | RD (0.007) | Rotation (0.026) | Rotation (0.008) | RD (0.006) | ERD (0.008) | CA (0.011) | Restoration (0.011) |
| 4. | RD (0.009) | Rotation (0.007) | eRD (0.010) | eRD (0.004) | eRD (0.005) | Restoration (0.006) | Rotation (0.008) | Rotation (0.008) |
| 5. | RD (0.009) | CR (0.006) | eRD (0.009) | eRD (0.004) | RD (0.004) | RD (0.006) | RD (0.005) | eRD (0.006) |
References
- Klein, C.A. Parallel progression of primary tumours and metastases. Nat. Rev. Cancer 2009, 9, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabières, C.; Pantel, K. Challenges in circulating tumour cell research. Nat. Rev. Cancer 2014, 14, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Patriarca, C.; Macchi, R.M.; Marschner, A.K.; Mellstedt, H. Epithelial cell adhesion molecule expression (CD326) in cancer: A short review. Cancer Treat. Rev. 2012, 38, 68–75. [Google Scholar] [CrossRef]
- Hyun, K.-A.; Koo, G.-B.; Han, H.; Sohn, J.; Choi, W.; Kim, S.-I.; Jung, H.-I.; Kim, Y.-S. Epithelial-to-mesenchymal transition leads to loss of EpCAM and different physical properties in circulating tumor cells from metastatic breast cancer. Oncotarget 2016, 7, 24677–24687. [Google Scholar] [CrossRef]
- Grover, P.K.; Cummins, A.G.; Price, T.J.; Roberts-Thomson, I.C.; Hardingham, J.E. Circulating tumour cells: The evolving concept and the inadequacy of their enrichment by EpCAM-based methodology for basic and clinical cancer research. Ann. Oncol. 2014, 25, 1506–1516. [Google Scholar] [CrossRef]
- Aktas, B.; Tewes, M.; Fehm, T.; Hauch, S.; Kimmig, R.; Kasimir-Bauer, S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009, 11, R46. [Google Scholar] [CrossRef]
- Gertler, R.; Rosenberg, R.; Fuehrer, K.; Dahm, M.; Nekarda, H.; Siewert, J.R. Detection of circulating tumor cells in blood using an optimized density gradient centrifugation. Recent results in cancer research. Fortschritte der Krebsforschung. Progres dans les recherches sur le cancer. Methods Mol. Biol. 2003, 162, 149–155. [Google Scholar] [CrossRef]
- Guo, T.; Stankiewicz, E.; Mao, X.; Lu, Y.-J. The Isolation and Analysis of Circulating Tumor Cells. In Methods in Molecular Biology; Clifton, N.J., Ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2019; Volume 2054, pp. 115–128. [Google Scholar]
- Zheng, S.; Lin, H.K.; Lu, B.; Williams, A.; Datar, R.; Cote, R.J.; Tai, Y.-C. 3D microfilter device for viable circulating tumor cell (CTC) enrichment from blood. Biomed. Microdevices 2011, 13, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Desitter, I.; Guerrouahen, B.S.; Benali-Furet, N.; Wechsler, J.; Jänne, P.A.; Kuang, Y.; Yanagita, M.; Wang, L.; Berkowitz, J.A.; Distel, R.J.; et al. A new device for rapid isolation by size and characterization of rare circulating tumor cells. Anticancer Res. 2011, 31, 427–441. [Google Scholar]
- Gagnon, Z.R. Cellular dielectrophoresis: Applications to the characterization, manipulation, separation and patterning of cells. Electrophoresis 2011, 32, 2466–2487. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Jafferji, I.; Garza, M.; Melnikova, V.O.; Hasegawa, D.K.; Pethig, R.; Davis, D.W. ApoStream™, a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics 2012, 6, 024133. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.J.; Yobas, L.; Lee, G.Y.H.; Ong, C.N.; Lim, C.T. Microdevice for the isolation and enumeration of cancer cells from blood. Biomed. Microdevices 2009, 11, 883–892. [Google Scholar] [CrossRef]
- Mohamed, H.; McCurdy, L.D.; Szarowski, D.H.; Duva, S.; Turner, J.N.; Caggana, M. Development of a Rare Cell Fractionation Device: Application for Cancer Detection. IEEE Trans. NanoBiosci. 2004, 3, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Koch, C.; Joosse, S.A.; Schneegans, S.; Wilken, O.J.W.; Janning, M.; Loreth, D.; Müller, V.; Prieske, K.; Banys-Paluchowski, M.; Horst, L.J.; et al. Pre-Analytical and Analytical Variables of Label-Independent Enrichment and Automated Detection of Circulating Tumor Cells in Cancer Patients. Cancers 2020, 12, 442. [Google Scholar] [CrossRef]
- Lianidou, E.S.; Markou, A. Circulating Tumor Cells in Breast Cancer: Detection Systems, Molecular Characterization, and Future Challenges. Clin. Chem. 2011, 57, 1242–1255. [Google Scholar] [CrossRef]
- Gabriel, M.T.; Calleja, L.R.; Chalopin, A.; Ory, B.; Heymann, D. Circulating Tumor Cells: A Review of Non–EpCAM-Based Approaches for Cell Enrichment and Isolation. Clin. Chem. 2016, 62, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Zhang, H.; Zhu, Y.; Liang, Y.; Yuan, Z.; Li, J.; Li, J.; Li, X.; Jia, Y.; He, T.; et al. Circulating tumor cells in cancer patients: Developments and clinical applications for immunotherapy. Mol. Cancer 2020, 19, 1–12. [Google Scholar] [CrossRef]
- Nel, I.; Baba, H.A.; Ertle, J.; Weber, F.; Sitek, B.; Eisenacher, M.; Meyer, H.E.; Schlaak, J.F.; Hoffmann, A.-C. Individual Profiling of Circulating Tumor Cell Composition and Therapeutic Outcome in Patients with Hepatocellular Carcinoma. Transl. Oncol. 2013, 6, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Nel, I.; Gauler, T.; Hoffmann, A.-C. Circulating tumor cell composition and outcome in patients with solid tumors. Int. J. Clin. Pharmacol. Ther. 2014, 52, 74–75. [Google Scholar] [CrossRef]
- Marrinucci, D.; Bethel, K.; Bruce, R.H.; Curry, D.N.; Hsieh, B.; Humphrey, M.; Krivacic, R.T.; Kroener, J.; Kroener, L.; Ladanyi, A.; et al. Case study of the morphologic variation of circulating tumor cells. Hum. Pathol. 2007, 38, 514–519. [Google Scholar] [CrossRef]
- Islam, M.; Raj, A.; McFarland, B.; Brink, H.M.; Ciciliano, J.; Fay, M.; Myers, D.R.; Flowers, C.; Waller, E.K.; Lam, W.; et al. Stiffness based enrichment of leukemia cells using microfluidics. APL Bioeng. 2020, 4, 036101. [Google Scholar] [CrossRef]
- Vigmostad, S.C.; Krog, B.L.; Nauseef, J.T.; Henry, M.D.; Keshav, V. Alterations in cancer cell mechanical properties after fluid shear stress exposure: A micropipette aspiration study. Cell Heal. Cytoskelet. 2015, 7, 25–35. [Google Scholar] [CrossRef][Green Version]
- Ghassemi, P.; Ren, X.; Foster, B.M.; Kerr, B.A.; Agah, M. Post-enrichment circulating tumor cell detection and enumeration via deformability impedance cytometry. Biosens. Bioelectron. 2020, 150, 111868. [Google Scholar] [CrossRef]
- Seltmann, K.; Fritsch, A.W.; Käs, J.A.; Magin, T.M. Keratins significantly contribute to cell stiffness and impact invasive behavior. Proc. Natl. Acad. Sci. USA 2013, 110, 18507–18512. [Google Scholar] [CrossRef] [PubMed]
- Guck, J.; Ananthakrishnan, R.; Mahmood, H.; Moon, T.J.; Cunningham, C.C.; Käs, J. The Optical Stretcher: A Novel Laser Tool to Micromanipulate Cells. Biophys. J. 2001, 81, 767–784. [Google Scholar] [CrossRef]
- Fletcher, D.A.; Mullins, R.D. Cell mechanics and the cytoskeleton. Nature 2010, 463, 485–492. [Google Scholar] [CrossRef]
- Grosser, S.; Fritsch, A.W.; Kießling, T.R.; Stange, R.; Käs, J.A. The lensing effect of trapped particles in a dual-beam optical trap. Opt. Express 2015, 23, 5221. [Google Scholar] [CrossRef] [PubMed]
- Mierke, C.T. The Role of the Optical Stretcher Is Crucial in the Investigation of Cell Mechanics Regulating Cell Adhesion and Motility. Front. Cell Dev. Biol. 2019, 7, 184. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Bragheri, F.; Minzioni, P. A Comprehensive Review of Optical Stretcher for Cell Mechanical Characterization at Single-Cell Level. Micromachines 2016, 7, 90. [Google Scholar] [CrossRef] [PubMed]
- Schneck, H.; Gierke, B.; Uppenkamp, F.; Behrens, B.; Niederacher, D.; Stoecklein, N.H.; Templin, M.F.; Pawlak, M.; Fehm, T.; Neubauer, H.; et al. EpCAM-Independent Enrichment of Circulating Tumor Cells in Metastatic Breast Cancer. PLoS ONE 2015, 10, e0144535. [Google Scholar] [CrossRef]
- Greaves, G.N.; Greer, A.L.; Lakes, R.S.; Rouxel, T. Poisson’s ratio and modern materials. Nat. Mater. 2011, 10, 823–837. [Google Scholar] [CrossRef]
- Guck, J.; Schinkinger, S.; Lincoln, B.; Wottawah, F.; Ebert, S.; Romeyke, M.; Lenz, D.; Erickson, H.M.; Ananthakrishnan, R.; Mitchell, D.; et al. Optical Deformability as an Inherent Cell Marker for Testing Malignant Transformation and Metastatic Competence. Biophys. J. 2005, 88, 3689–3698. [Google Scholar] [CrossRef]
- Gyger, M.; Stange, R.; Kießling, T.R.; Fritsch, A.; Kostelnik, K.B.; Beck-Sickinger, A.G.; Zink, M.; Käs, J.A. Active contractions in single suspended epithelial cells. Eur. Biophys. J. 2014, 43, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Reduzzi, C.; Vismara, M.; Gerratana, L.; Silvestri, M.; De Braud, F.; Raspagliesi, F.; Verzoni, E.; Di Cosimo, S.; Locati, L.D.; Cristofanilli, M.; et al. The curious phenomenon of dual-positive circulating cells: Longtime overlooked tumor cells. Semin. Cancer Biol. 2020, 60, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Nel, I.; Jehn, U.; Gauler, T.; Hoffmann, A.-C. Individual profiling of circulating tumor cell composition in patients with non-small cell lung cancer receiving platinum based treatment. Transl. Lung Cancer Res. 2014, 3, 100–106. [Google Scholar] [PubMed]
- Fedosov, D.A.; Gompper, G. White blood cell margination in microcirculation. Soft Matter 2014, 10, 2961–2970. [Google Scholar] [CrossRef]
- Munn, L.L.; Dupin, M.M. Blood Cell Interactions and Segregation in Flow. Ann. Biomed. Eng. 2008, 36, 534–544. [Google Scholar] [CrossRef] [PubMed]
- Rowan, B.G.; Gimble, J.M.; Sheng, M.; Anbalagan, M.; Jones, R.K.; Frazier, T.P.; Asher, M.; Lacayo, E.A.; Friedlander, P.L.; Kutner, R.; et al. Human Adipose Tissue-Derived Stromal/Stem Cells Promote Migration and Early Metastasis of Triple Negative Breast Cancer Xenografts. PLoS ONE 2014, 9, e89595. [Google Scholar] [CrossRef]
- Shen, Y.; Schmidt, B.U.S.; Kubitschke, H.; Morawetz, E.W.; Wolf, B.; Käs, J.A.; Losert, W. Detecting heterogeneity in and between breast cancer cell lines. Cancer Converg. 2020, 4, 1–11. [Google Scholar] [CrossRef]
- Lincoln, B.; Wottawah, F.; Schinkinger, S.; Ebert, S.; Guck, J. High-Throughput Rheological Measurements with an Optical Stretcher. In Cell Mechanics; Elsevier: Amsterdam, The Netherlands, 2007; Volume 83, pp. 397–432. [Google Scholar] [CrossRef]
- Van Liedekerke, P.; Neitsch, J.; Johann, T.; Warmt, E.; Gonzàlez-Valverde, I.; Hoehme, S.; Grosser, S.; Kaes, J.; Drasdo, D. A quantitative high-resolution computational mechanics cell model for growing and regenerating tissues. Biomech. Model. Mechanobiol. 2019, 19, 189–220. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Ward, K.A.; Li, W.-I.; Zimmer, S.; Davis, T. Viscoelastic properties of transformed cells: Role in tumor cell progression and metastasis formation. Biorheology 1991, 28, 301–313. [Google Scholar] [CrossRef]
- Remmerbach, T.W.; Wottawah, F.; Dietrich, J.; Lincoln, B.; Wittekind, C.; Guck, J. Oral Cancer Diagnosis by Mechanical Phenotyping. Cancer Res. 2009, 69, 1728–1732. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, A.; Höckel, M.; Kiessling, T.; Nnetu, K.D.; Wetzel, F.; Zink, M.; Käs, J.A. Are biomechanical changes necessary for tumour progression? Nat. Phys. 2010, 6, 730–732. [Google Scholar] [CrossRef]
- Panhwar, M.H.; Czerwinski, F.; Dabbiru, V.A.S.; Komaragiri, Y.; Fregin, B.; Biedenweg, D.; Nestler, P.; Pires, R.H.; Otto, O. High-throughput cell and spheroid mechanics in virtual fluidic channels. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Altmann, A.; Toloşi, L.; Sander, O.; Lengauer, T. Permutation importance: A corrected feature importance measure. Bioinformatics 2010, 26, 1340–1347. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]

| Characteristic | Mamma Carcinoma |
|---|---|
| (n = 14) * | |
| Age (years) | 26–85 |
| Median | 63 |
| Histology | |
| ductal | 10 |
| lobular | 2 |
| mixed | 2 |
| Stage | |
| T0 | 1 |
| Tis | 3 |
| T1 | 7 |
| T2 | 3 |
| Lymphnode | |
| N0 | 10 |
| N1 | 2 |
| Nx | 2 |
| Grade | |
| G1 | 5 |
| G2 | 4 |
| G3 | 5 |
| Subtype | |
| luminal A | 4 |
| luminal B | 4 |
| HER2 enriched | 1 |
| triple negative | 3 |
| DCIS | 2 |
| Setting | |
| primary | 12 |
| recurrent | 2 |
| Therapy | |
| neoadjuvant CT | 3 |
| neoadjuvant endocrine | 3 |
| adjuvant CT | 1 |
| adjuvant endocrine | 9 |
| adjuvant RT | 1 |
| adjuvant Trastuzumab | 1 |
| none | 1 |
| Laser Power (mW) | Excluding Shape Restoration | Including Shape Restoration | ||||
|---|---|---|---|---|---|---|
| Accuracy | Sensitivity | Specificity | Accuracy | Sensitivity | Specificity | |
| 400 | 0.63 | 0.72 | 0.52 | 0.64 | 0.73 | 0.52 |
| 800 | 0.66 | 0.76 | 0.55 | 0.65 | 0.76 | 0.51 |
| 1200 | 0.66 | 0.72 | 0.60 | 0.69 | 0.74 | 0.63 |
| averaged | 0.65 | 0.73 | 0.56 | 0.66 | 0.74 | 0.55 |
| pooled | 0.65 | 0.72 | 0.56 | 0.66 | 0.77 | 0.53 |
| Stepwise Data Input | Accuracy | Sensitivity | Specificity |
|---|---|---|---|
| Cell area, cell radius | 0.55 | 0.61 | 0.48 |
| +Relative deformation | 0.57 | 0.61 | 0.53 |
| +Elliptic deformation | 0.59 | 0.65 | 0.52 |
| +Shape restoration | 0.62 | 0.70 | 0.55 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nel, I.; Morawetz, E.W.; Tschodu, D.; Käs, J.A.; Aktas, B. The Mechanical Fingerprint of Circulating Tumor Cells (CTCs) in Breast Cancer Patients. Cancers 2021, 13, 1119. https://doi.org/10.3390/cancers13051119
Nel I, Morawetz EW, Tschodu D, Käs JA, Aktas B. The Mechanical Fingerprint of Circulating Tumor Cells (CTCs) in Breast Cancer Patients. Cancers. 2021; 13(5):1119. https://doi.org/10.3390/cancers13051119
Chicago/Turabian StyleNel, Ivonne, Erik W. Morawetz, Dimitrij Tschodu, Josef A. Käs, and Bahriye Aktas. 2021. "The Mechanical Fingerprint of Circulating Tumor Cells (CTCs) in Breast Cancer Patients" Cancers 13, no. 5: 1119. https://doi.org/10.3390/cancers13051119
APA StyleNel, I., Morawetz, E. W., Tschodu, D., Käs, J. A., & Aktas, B. (2021). The Mechanical Fingerprint of Circulating Tumor Cells (CTCs) in Breast Cancer Patients. Cancers, 13(5), 1119. https://doi.org/10.3390/cancers13051119