Functional States in Tumor-Initiating Cell Differentiation in Human Colorectal Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
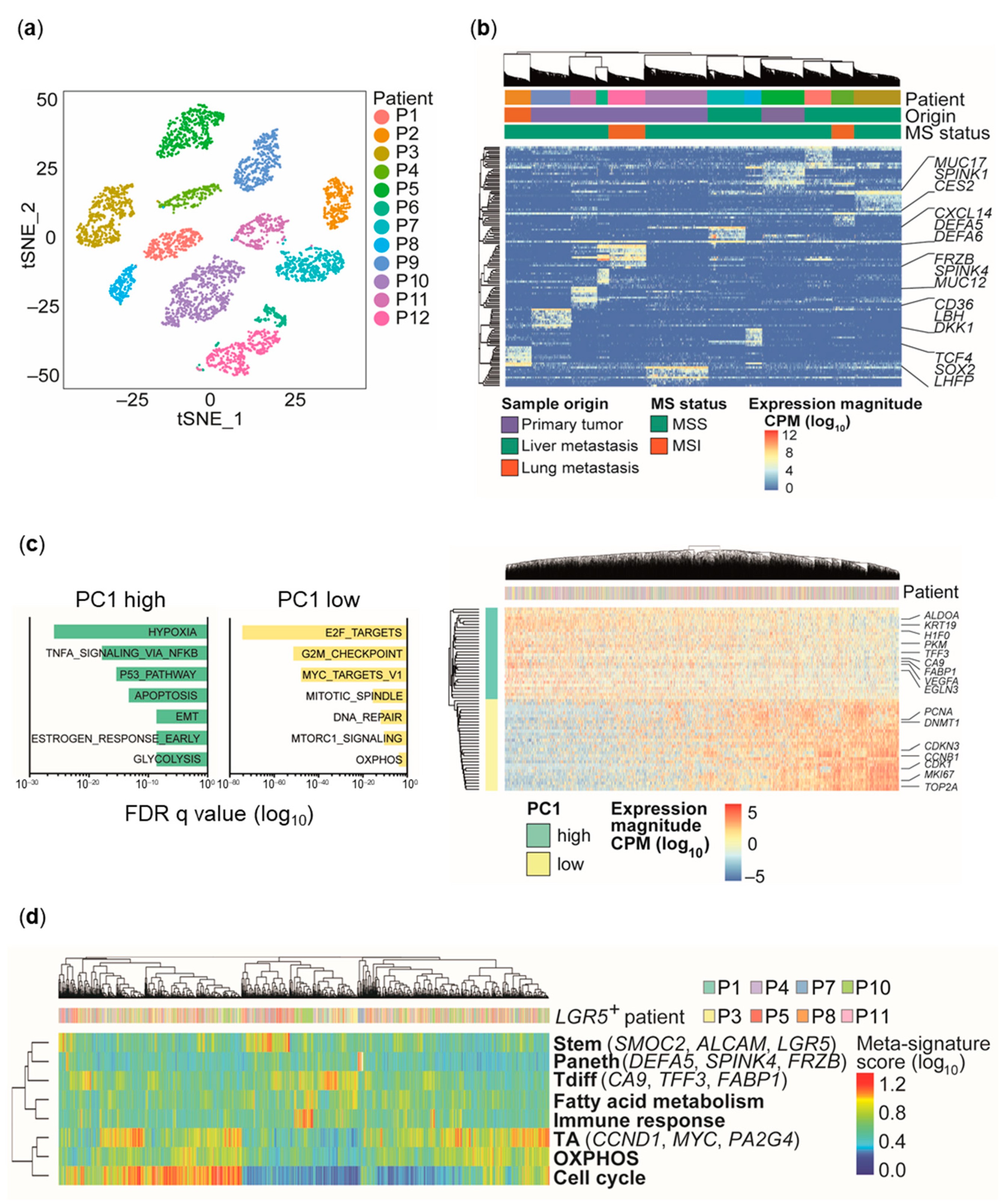
2.1. Transcriptional Heterogeneity of Patient-Derived CRC Spheroid Cultures
2.2. Distinct Cell Types and Cell States in Individual CRC Spheroids
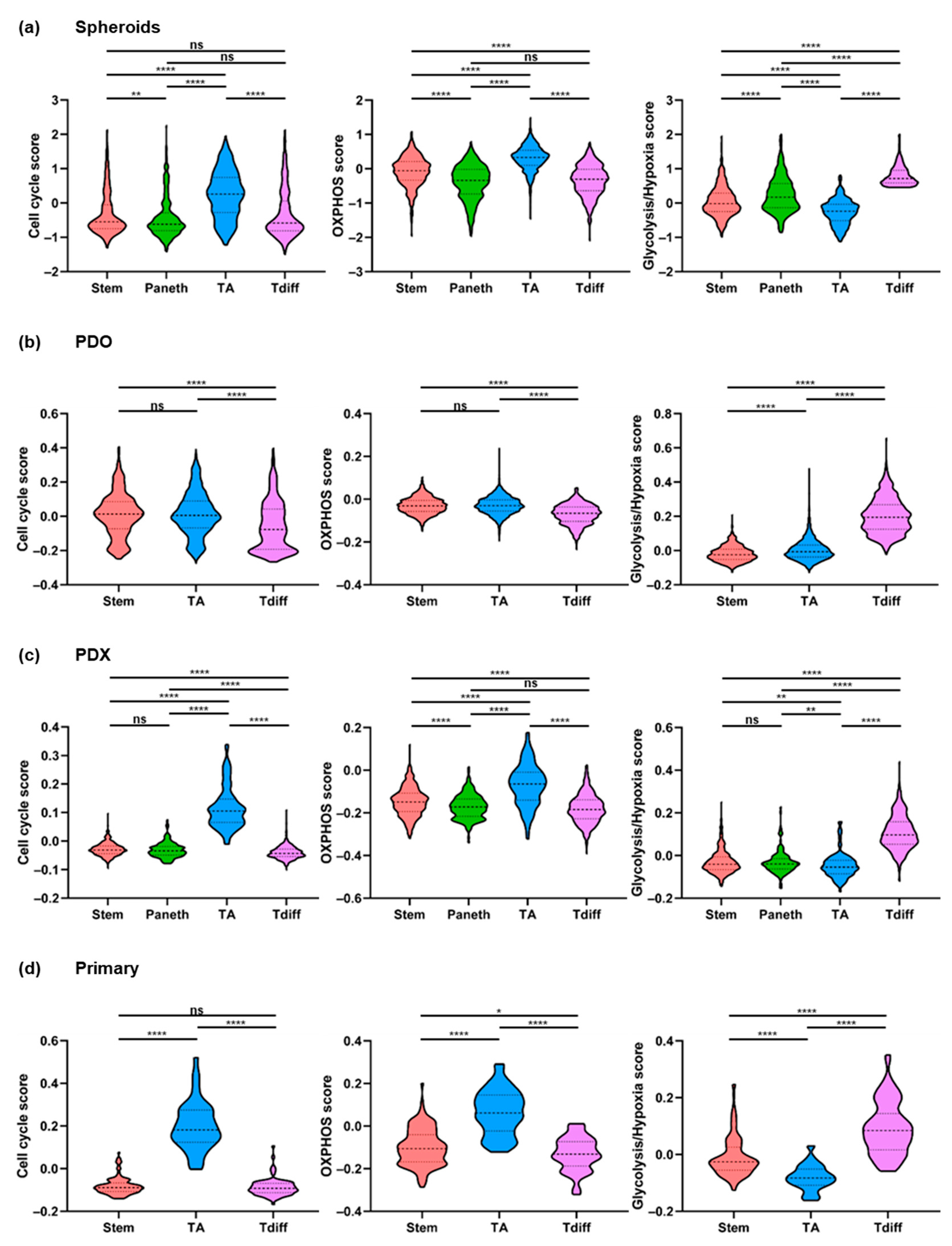
2.3. Cell Cycle and Proliferative Activity of Human CRC Cells
2.4. Divergent Cell Type-Associated Energy Metabolic Preferences
2.5. Cell States in Patient-Derived CRC Organoids, Xenografts, and Primary Tumors
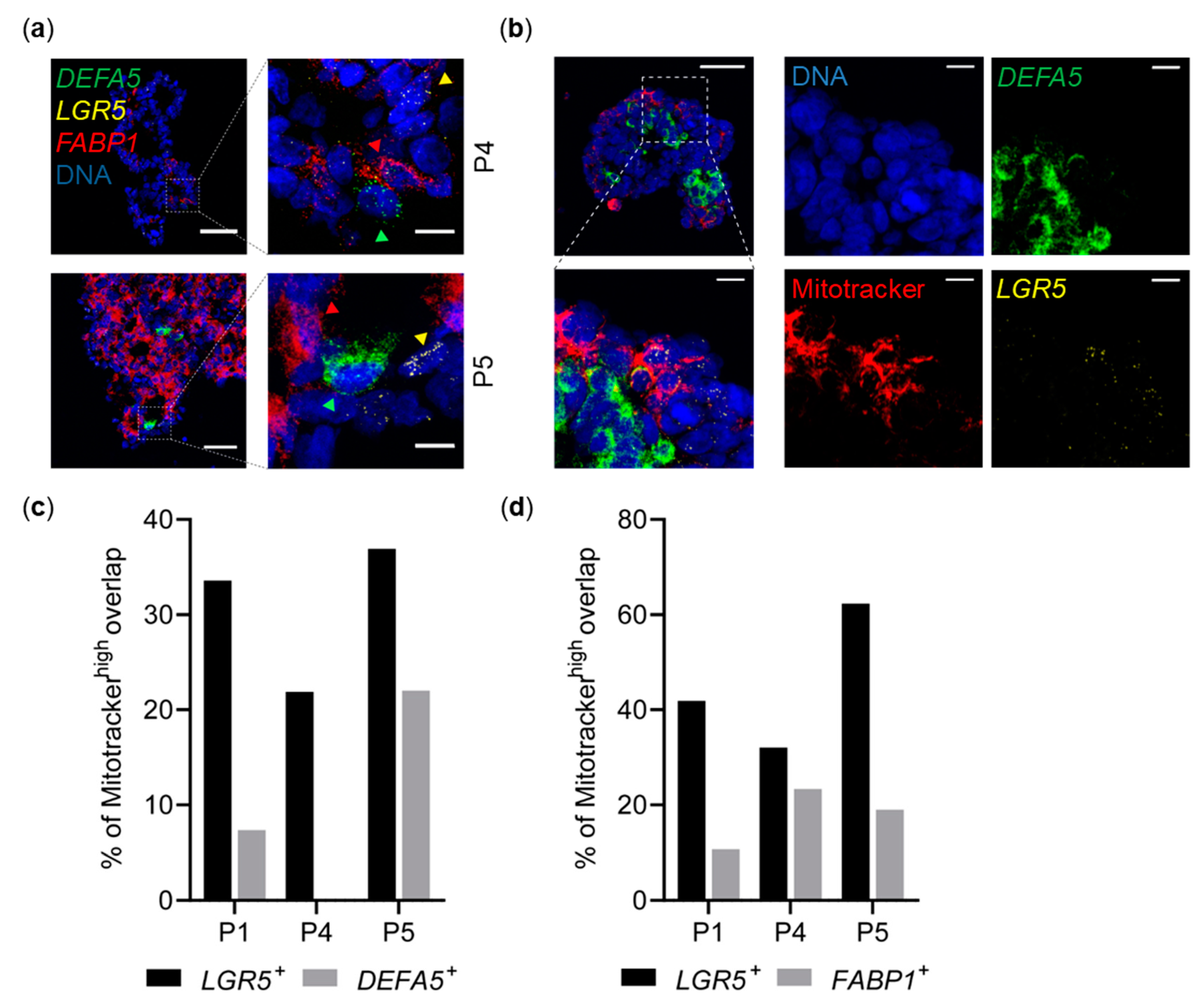
2.6. Spatial Distribution of OXPHOS and Distinct Cell Types in CRC Spheroids
2.7. Heterogeneous Energy Metabolism in Patient Tumors
2.8. Heterogeneous Energy Metabolism in Patient-Derived Models
2.9. Increased Spheroid and Tumor Formation Capacity in OXPHOSHigh Cells
3. Discussion
4. Materials and Methods
4.1. Primary CRC Spheroids and Organoids
4.2. Laboratory Animals
4.3. scRNA-seq of Spheroids
4.4. scRNA-seq of Tumors, PDXs, PDOs
4.5. Preprocessing and Analysis of iCELL8 Data
4.6. Preprocessing and Analysis of 10x Data
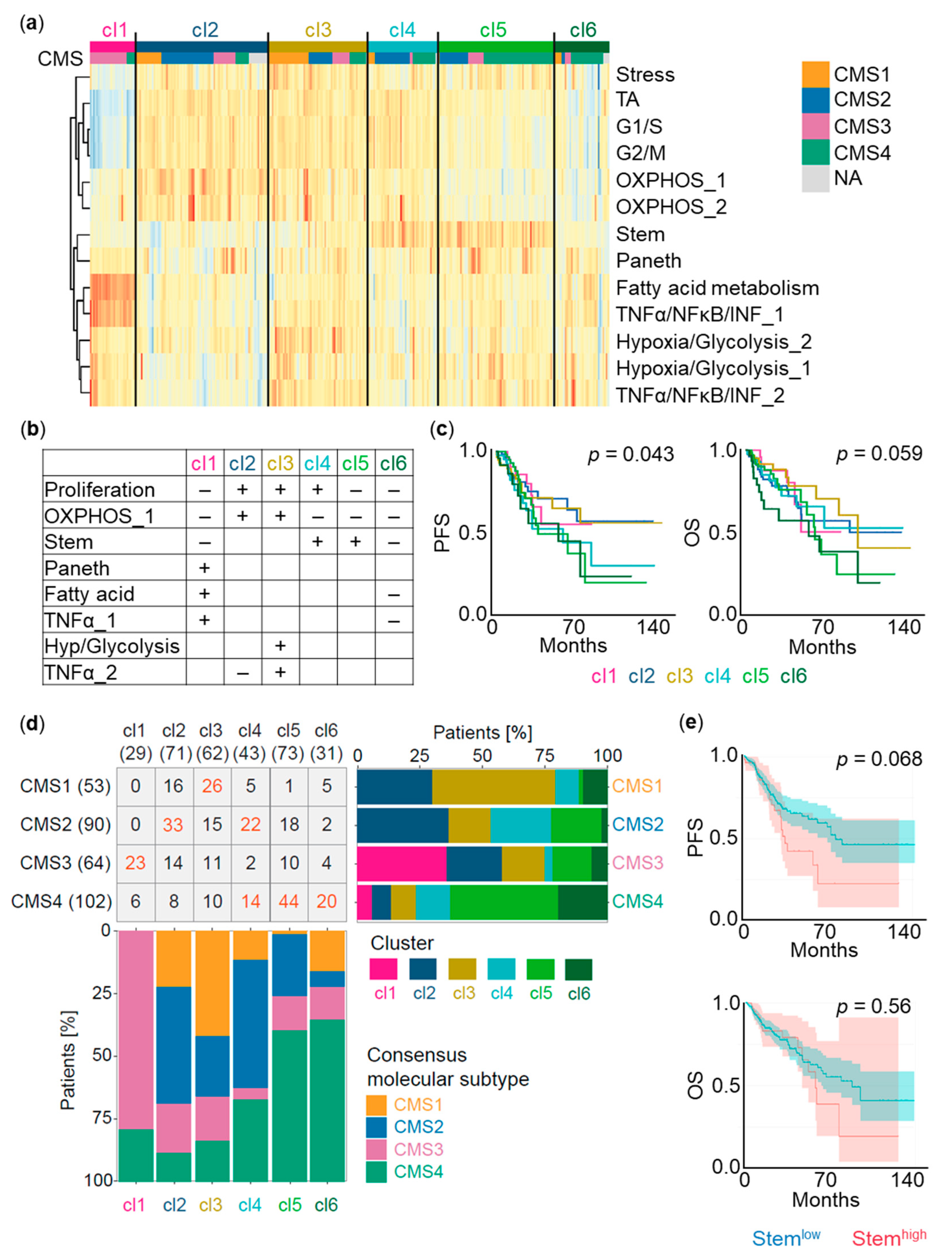
4.7. Patient Clustering and Survival Analysis
4.8. Genetic Labelling of Spheroids
4.9. RNA-FISH
4.10. Flow Cytometry and Sorting of Metabolic Subpopulations
4.11. Assessment of SFC Frequency
4.12. Assessment of TIC Frequency
4.13. Co-Cultivation Experiments
4.14. Inhibitor Treatments
4.15. Immunohistochemistry
4.16. Mass Spectrometry
4.17. Quantification and Statistical Analysis
4.18. Data and Code Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jordan, C.T.; Guzman, M.L.; Noble, M. Cancer stem cells. N. Engl. J. Med. 2006, 355, 1253–1261. [Google Scholar] [CrossRef]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells: Current status and evolving complexities. Cell Stem Cell 2012, 10, 717–728. [Google Scholar] [CrossRef]
- Dieter, S.M.; Ball, C.R.; Hoffmann, C.M.; Nowrouzi, A.; Herbst, F.; Zavidij, O.; Abel, U.; Arens, A.; Weichert, W.; Brand, K.; et al. Distinct types of tumor-initiating cells form human colon cancer tumors and metastases. Cell Stem Cell 2011, 9, 357–365. [Google Scholar] [CrossRef]
- Giessler, K.M.; Kleinheinz, K.; Huebschmann, D.; Balasubramanian, G.P.; Dubash, T.D.; Dieter, S.M.; Siegl, C.; Herbst, F.; Weber, S.; Hoffmann, C.M.; et al. Genetic subclone architecture of tumor clone-initiating cells in colorectal cancer. J. Exp. Med. 2017, 214, 2073–2088. [Google Scholar] [CrossRef]
- Cortina, C.; Turon, G.; Stork, D.; Hernando-Momblona, X.; Sevillano, M.; Aguilera, M.; Tosi, S.; Merlos-Suárez, A.; Stephan-Otto Attolini, C.; Sancho, E.; et al. A genome editing approach to study cancer stem cells in human tumors. EMBO Mol. Med. 2017, 9, 869–879. [Google Scholar] [CrossRef]
- Barker, N. Adult intestinal stem cells: Critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol. 2014, 15, 19–33. [Google Scholar] [CrossRef]
- Merlos-Suárez, A.; Barriga, F.M.; Jung, P.; Iglesias, M.; Céspedes, M.V.; Rossell, D.; Sevillano, M.; Hernando-Momblona, X.; da Silva-Diz, V.; Muñoz, P.; et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 2011, 8, 511–524. [Google Scholar] [CrossRef]
- Bormann, F.; Rodríguez-Paredes, M.; Lasitschka, F.; Edelmann, D.; Musch, T.; Benner, A.; Bergman, Y.; Dieter, S.M.; Ball, C.R.; Glimm, H.; et al. Cell-of-Origin DNA Methylation Signatures Are Maintained during Colorectal Carcinogenesis. Cell Rep. 2018, 23, 3407–3418. [Google Scholar] [CrossRef]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef]
- Blanpain, C.; Fuchs, E. Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science 2014, 344, 1242281. [Google Scholar] [CrossRef]
- Varga, J.; Greten, F.R. Cell plasticity in epithelial homeostasis and tumorigenesis. Nat. Cell Biol. 2017, 19, 1133–1141. [Google Scholar] [CrossRef]
- De Sousa e Melo, F.; Kurtova, A.V.; Harnoss, J.M.; Kljavin, N.; Hoeck, J.D.; Hung, J.; Anderson, J.E.; Storm, E.E.; Modrusan, Z.; Koeppen, H.; et al. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature 2017, 543, 676–680. [Google Scholar] [CrossRef]
- Shimokawa, M.; Ohta, Y.; Nishikori, S.; Matano, M.; Takano, A.; Fujii, M.; Date, S.; Sugimoto, S.; Kanai, T.; Sato, T. Visualization and targeting of LGR5+ human colon cancer stem cells. Nature 2017, 545, 187–192. [Google Scholar] [CrossRef]
- Ball, C.R.; Oppel, F.; Ehrenberg, K.R.; Dubash, T.D.; Dieter, S.M.; Hoffmann, C.M.; Abel, U.; Herbst, F.; Koch, M.; Werner, J.; et al. Succession of transiently active tumor-initiating cell clones in human pancreatic cancer xenografts. EMBO Mol. Med. 2017, 9, 918–932. [Google Scholar] [CrossRef]
- Dalerba, P.; Dylla, S.J.; Park, I.K.; Liu, R.; Wang, X.; Cho, R.W.; Hoey, T.; Gurney, A.; Huang, E.H.; Simeone, D.M.; et al. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 10158–10163. [Google Scholar] [CrossRef]
- Kreso, A.; O’Brien, C.A.; van Galen, P.; Gan, O.I.; Notta, F.; Brown, A.M.; Ng, K.; Ma, J.; Wienholds, E.; Dunant, C.; et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science 2013, 339, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Ricci-Vitiani, L.; Lombardi, D.G.; Pilozzi, E.; Biffoni, M.; Todaro, M.; Peschle, C.; De Maria, R. Identification and expansion of human colon-cancer-initiating cells. Nature 2007, 445, 111–115. [Google Scholar] [CrossRef]
- Goldstein, L.D.; Chen, Y.J.; Dunne, J.; Mir, A.; Hubschle, H.; Guillory, J.; Yuan, W.; Zhang, J.; Stinson, J.; Jaiswal, B.; et al. Massively parallel nanowell-based single-cell gene expression profiling. BMC Genom. 2017, 18, 519. [Google Scholar] [CrossRef]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H., 2nd; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef]
- Tirosh, I.; Venteicher, A.S.; Hebert, C.; Escalante, L.E.; Patel, A.P.; Yizhak, K.; Fisher, J.M.; Rodman, C.; Mount, C.; Filbin, M.G.; et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 2016, 539, 309–313. [Google Scholar] [CrossRef]
- Rodríguez-Colman, M.J.; Schewe, M.; Meerlo, M.; Stigter, E.; Gerrits, J.; Pras-Raves, M.; Sacchetti, A.; Hornsveld, M.; Oost, K.C.; Snippert, H.J.; et al. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature 2017, 543, 424–427. [Google Scholar] [CrossRef]
- Lee, D.D.; Seung, H.S. Learning the parts of objects by non-negative matrix factorization. Nature 1999, 401, 788–791. [Google Scholar] [CrossRef] [PubMed]
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017, 171, 1611–1624.e1624. [Google Scholar] [CrossRef] [PubMed]
- Barker, N.; Ridgway, R.A.; van Es, J.H.; van de Wetering, M.; Begthel, H.; van den Born, M.; Danenberg, E.; Clarke, A.R.; Sansom, O.J.; Clevers, H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 2009, 457, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Barker, N.; van Es, J.H.; Kuipers, J.; Kujala, P.; van den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef]
- Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef]
- Eide, P.W.; Bruun, J.; Lothe, R.A.; Sveen, A. CMScaller: An R package for consensus molecular subtyping of colorectal cancer pre-clinical models. Sci. Rep. 2017, 7, 16618. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Li, H.; Courtois, E.T.; Sengupta, D.; Tan, Y.; Chen, K.H.; Goh, J.J.L.; Kong, S.L.; Chua, C.; Hon, L.K.; Tan, W.S.; et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat. Genet. 2017, 49, 708–718. [Google Scholar] [CrossRef]
- Falkowska-Hansen, B.; Kollar, J.; Grüner, B.M.; Schanz, M.; Boukamp, P.; Siveke, J.; Rethwilm, A.; Kirschner, M. An inducible Tet-Off-H2B-GFP lentiviral reporter vector for detection and in vivo isolation of label-retaining cells. Exp. Cell Res. 2010, 316, 1885–1895. [Google Scholar] [CrossRef]
- Haegebarth, A.; Clevers, H. Wnt signaling, lgr5, and stem cells in the intestine and skin. Am. J. Pathol. 2009, 174, 715–721. [Google Scholar] [CrossRef]
- Oliver, P.L.; Finelli, M.J.; Edwards, B.; Bitoun, E.; Butts, D.L.; Becker, E.B.; Cheeseman, M.T.; Davies, B.; Davies, K.E. Oxr1 is essential for protection against oxidative stress-induced neurodegeneration. PLoS Genet. 2011, 7, e1002338. [Google Scholar] [CrossRef]
- Li, W.; Kennedy, D.; Shao, Z.; Wang, X.; Kamdar, A.K.; Weber, M.; Mislick, K.; Kiefer, K.; Morales, R.; Agatisa-Boyle, B.; et al. Paraoxonase 2 prevents the development of heart failure. Free Radic. Biol. Med. 2018, 121, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Liu, X.; Gjoerup, O.; Zhang, A.; Yuan, X.; Balk, S.P.; Schneider, M.C.; Lu, M.L. Activation of p21-activated kinase 6 by MAP kinase kinase 6 and p38 MAP kinase. J. Biol. Chem. 2005, 280, 3323–3330. [Google Scholar] [CrossRef]
- Klein, A.M.; Mazutis, L.; Akartuna, I.; Tallapragada, N.; Veres, A.; Li, V.; Peshkin, L.; Weitz, D.A.; Kirschner, M.W. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015, 161, 1187–1201. [Google Scholar] [CrossRef]
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M.; et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015, 161, 1202–1214. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, N.; Sachs, N.; Wiebrands, K.; Ellenbroek, S.I.; Fumagalli, A.; Lyubimova, A.; Begthel, H.; van den Born, M.; van Es, J.H.; Karthaus, W.R.; et al. Reg4+ deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon. Proc. Natl. Acad. Sci. USA 2016, 113, E5399–E5407. [Google Scholar] [CrossRef]
- Baker, A.M.; Graham, T.A.; Elia, G.; Wright, N.A.; Rodriguez-Justo, M. Characterization of LGR5 stem cells in colorectal adenomas and carcinomas. Sci. Rep. 2015, 5, 8654. [Google Scholar] [CrossRef]
- Sato, T.; van Es, J.H.; Snippert, H.J.; Stange, D.E.; Vries, R.G.; van den Born, M.; Barker, N.; Shroyer, N.F.; van de Wetering, M.; Clevers, H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011, 469, 415–418. [Google Scholar] [CrossRef]
- Dalerba, P.; Kalisky, T.; Sahoo, D.; Rajendran, P.S.; Rothenberg, M.E.; Leyrat, A.A.; Sim, S.; Okamoto, J.; Johnston, D.M.; Qian, D.; et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat. Biotechnol. 2011, 29, 1120–1127. [Google Scholar] [CrossRef]
- Tirier, S.M.; Park, J.; Preußer, F.; Amrhein, L.; Gu, Z.; Steiger, S.; Mallm, J.P.; Krieger, T.; Waschow, M.; Eismann, B.; et al. Pheno-seq-linking visual features and gene expression in 3D cell culture systems. Sci. Rep. 2019, 9, 12367. [Google Scholar] [CrossRef]
- Heytler, P.G.; Prichard, W.W. A new class of uncoupling agents–carbonyl cyanide phenylhydrazones. Biochem. Biophys. Res. Commun. 1962, 7, 272–275. [Google Scholar] [CrossRef]
- Joung, J.G.; Oh, B.Y.; Hong, H.K.; Al-Khalidi, H.; Al-Alem, F.; Lee, H.O.; Bae, J.S.; Kim, J.; Cha, H.U.; Alotaibi, M.; et al. Tumor Heterogeneity Predicts Metastatic Potential in Colorectal Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 7209–7216. [Google Scholar] [CrossRef]
- Yan, K.S.; Janda, C.Y.; Chang, J.; Zheng, G.X.Y.; Larkin, K.A.; Luca, V.C.; Chia, L.A.; Mah, A.T.; Han, A.; Terry, J.M.; et al. Non-equivalence of Wnt and R-spondin ligands during Lgr5+ intestinal stem-cell self-renewal. Nature 2017, 545, 238–242. [Google Scholar] [CrossRef]
- Dieter, S.M.; Glimm, H.; Ball, C.R. Colorectal cancer-initiating cells caught in the act. EMBO Mol. Med. 2017, 9, 856–858. [Google Scholar] [CrossRef]
- Puig, I.; Tenbaum, S.P.; Chicote, I.; Arqués, O.; Martínez-Quintanilla, J.; Cuesta-Borrás, E.; Ramírez, L.; Gonzalo, P.; Soto, A.; Aguilar, S.; et al. TET2 controls chemoresistant slow-cycling cancer cell survival and tumor recurrence. J. Clin. Investig. 2018, 128, 3887–3905. [Google Scholar] [CrossRef] [PubMed]
- Ayyaz, A.; Kumar, S.; Sangiorgi, B.; Ghoshal, B.; Gosio, J.; Ouladan, S.; Fink, M.; Barutcu, S.; Trcka, D.; Shen, J.; et al. Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature 2019, 569, 121–125. [Google Scholar] [CrossRef]
- Grün, D.; Lyubimova, A.; Kester, L.; Wiebrands, K.; Basak, O.; Sasaki, N.; Clevers, H.; van Oudenaarden, A. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 2015, 525, 251–255. [Google Scholar] [CrossRef]
- Yan, K.S.; Gevaert, O.; Zheng, G.X.Y.; Anchang, B.; Probert, C.S.; Larkin, K.A.; Davies, P.S.; Cheng, Z.F.; Kaddis, J.S.; Han, A.; et al. Intestinal Enteroendocrine Lineage Cells Possess Homeostatic and Injury-Inducible Stem Cell Activity. Cell Stem Cell 2017, 21, 78–90.e76. [Google Scholar] [CrossRef]
- Haber, A.L.; Biton, M.; Rogel, N.; Herbst, R.H.; Shekhar, K.; Smillie, C.; Burgin, G.; Delorey, T.M.; Howitt, M.R.; Katz, Y.; et al. A single-cell survey of the small intestinal epithelium. Nature 2017, 551, 333–339. [Google Scholar] [CrossRef]
- Chen, E.C.; Karl, T.A.; Kalisky, T.; Gupta, S.K.; O’Brien, C.A.; Longacre, T.A.; van de Rijn, M.; Quake, S.R.; Clarke, M.F.; Rothenberg, M.E. KIT Signaling Promotes Growth of Colon Xenograft Tumors in Mice and Is Up-Regulated in a Subset of Human Colon Cancers. Gastroenterology 2015, 149, 705–717.e702. [Google Scholar] [CrossRef]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef]
- Biton, M.; Haber, A.L.; Rogel, N.; Burgin, G.; Beyaz, S.; Schnell, A.; Ashenberg, O.; Su, C.W.; Smillie, C.; Shekhar, K.; et al. T Helper Cell Cytokines Modulate Intestinal Stem Cell Renewal and Differentiation. Cell 2018, 175, 1307–1320.e1322. [Google Scholar] [CrossRef]
- Velten, L.; Haas, S.F.; Raffel, S.; Blaszkiewicz, S.; Islam, S.; Hennig, B.P.; Hirche, C.; Lutz, C.; Buss, E.C.; Nowak, D.; et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat. Cell Biol. 2017, 19, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, A.; Oost, K.C.; Kester, L.; Morgner, J.; Bornes, L.; Bruens, L.; Spaargaren, L.; Azkanaz, M.; Schelfhorst, T.; Beerling, E.; et al. Plasticity of Lgr5-Negative Cancer Cells Drives Metastasis in Colorectal Cancer. Cell Stem Cell 2020, 26, 569–578.e567. [Google Scholar] [CrossRef] [PubMed]
- Morral, C.; Stanisavljevic, J.; Hernando-Momblona, X.; Mereu, E.; Álvarez-Varela, A.; Cortina, C.; Stork, D.; Slebe, F.; Turon, G.; Whissell, G.; et al. Zonation of Ribosomal DNA Transcription Defines a Stem Cell Hierarchy in Colorectal Cancer. Cell Stem Cell 2020, 26, 845–861 e812. [Google Scholar] [CrossRef]
- Zheng, J. Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation (Review). Oncol. Lett. 2012, 4, 1151–1157. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Kwon, H.; Han, C.; Zhang, J.; Dash, S.; Lim, K.; Wu, T. Active glycolytic metabolism in CD133(+) hepatocellular cancer stem cells: Regulation by MIR-122. Oncotarget 2015, 6, 40822–40835. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Gentles, A.; Nair, R.V.; Huang, M.; Lin, Y.; Lee, C.Y.; Cai, S.; Scheeren, F.A.; Kuo, A.H.; Diehn, M. Targeting unique metabolic properties of breast tumor initiating cells. Stem Cells 2014, 32, 1734–1745. [Google Scholar] [CrossRef]
- Palorini, R.; Votta, G.; Balestrieri, C.; Monestiroli, A.; Olivieri, S.; Vento, R.; Chiaradonna, F. Energy metabolism characterization of a novel cancer stem cell-like line 3AB-OS. J. Cell. Biochem. 2014, 115, 368–379. [Google Scholar] [CrossRef]
- Shen, Y.A.; Lin, C.H.; Chi, W.H.; Wang, C.Y.; Hsieh, Y.T.; Wei, Y.H.; Chen, Y.J. Resveratrol Impedes the Stemness, Epithelial-Mesenchymal Transition, and Metabolic Reprogramming of Cancer Stem Cells in Nasopharyngeal Carcinoma through p53 Activation. Evid.-Based Complementary Altern. Med. ECAM 2013, 2013, 590393. [Google Scholar] [CrossRef]
- Viale, A.; Pettazzoni, P.; Lyssiotis, C.A.; Ying, H.; Sánchez, N.; Marchesini, M.; Carugo, A.; Green, T.; Seth, S.; Giuliani, V.; et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 2014, 514, 628–632. [Google Scholar] [CrossRef]
- Janiszewska, M.; Suvà, M.L.; Riggi, N.; Houtkooper, R.H.; Auwerx, J.; Clément-Schatlo, V.; Radovanovic, I.; Rheinbay, E.; Provero, P.; Stamenkovic, I. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev. 2012, 26, 1926–1944. [Google Scholar] [CrossRef]
- Lagadinou, E.D.; Sach, A.; Callahan, K.; Rossi, R.M.; Neering, S.J.; Minhajuddin, M.; Ashton, J.M.; Pei, S.; Grose, V.; O’Dwyer, K.M.; et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell 2013, 12, 329–341. [Google Scholar] [CrossRef]
- Elgendy, M.; Cirò, M.; Hosseini, A.; Weiszmann, J.; Mazzarella, L.; Ferrari, E.; Cazzoli, R.; Curigliano, G.; DeCensi, A.; Bonanni, B.; et al. Combination of Hypoglycemia and Metformin Impairs Tumor Metabolic Plasticity and Growth by Modulating the PP2A-GSK3β-MCL-1 Axis. Cancer Cell 2019, 35, 798–815.e795. [Google Scholar] [CrossRef]
- Wiener, Z.; Högström, J.; Hyvönen, V.; Band, A.M.; Kallio, P.; Holopainen, T.; Dufva, O.; Haglund, C.; Kruuna, O.; Oliver, G.; et al. Prox1 promotes expansion of the colorectal cancer stem cell population to fuel tumor growth and ischemia resistance. Cell Rep. 2014, 8, 1943–1956. [Google Scholar] [CrossRef] [PubMed]
- Ragusa, S.; Cheng, J.; Ivanov, K.I.; Zangger, N.; Ceteci, F.; Bernier-Latmani, J.; Milatos, S.; Joseph, J.M.; Tercier, S.; Bouzourene, H.; et al. PROX1 promotes metabolic adaptation and fuels outgrowth of Wnt(high) metastatic colon cancer cells. Cell Rep. 2014, 8, 1957–1973. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.; Kim, H.; Kim, D.; Ban, S.; Oh, S.; Ji, S.; Kang, D.; Lee, H.; Ahn, T.S.; Kim, H.J.; et al. Defensin alpha 6 (DEFA6) is a prognostic marker in colorectal cancer. Cancer Biomark. Sect. A Dis. Markers 2019, 24, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Rothenberg, M.E.; Nusse, Y.; Kalisky, T.; Lee, J.J.; Dalerba, P.; Scheeren, F.; Lobo, N.; Kulkarni, S.; Sim, S.; Qian, D.; et al. Identification of a cKit+ colonic crypt base secretory cell that supports Lgr5+ stem cells in mice. Gastroenterology 2012, 142, 1195–1205 e1196. [Google Scholar] [CrossRef] [PubMed]
- Altmann, G.G. Morphological observations on mucus-secreting nongoblet cells in the deep crypts of the rat ascending colon. Am. J. Anat. 1983, 167, 95–117. [Google Scholar] [CrossRef]
- Dieter, S.M.; Giessler, K.M.; Kriegsmann, M.; Dubash, T.D.; Möhrmann, L.; Schulz, E.R.; Siegl, C.; Weber, S.; Strakerjahn, H.; Oberlack, A.; et al. Patient-derived xenografts of gastrointestinal cancers are susceptible to rapid and delayed B-lymphoproliferation. Int. J. Cancer 2017, 140, 1356–1363. [Google Scholar] [CrossRef]
- Dubash, T.D.; Hoffmann, C.M.; Oppel, F.; Giessler, K.M.; Weber, S.; Dieter, S.M.; Hüllein, J.; Zenz, T.; Herbst, F.; Scholl, C.; et al. Phenotypic differentiation does not affect tumorigenicity of primary human colon cancer initiating cells. Cancer Lett. 2016, 371, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Riffle, S.; Hegde, R.S. Modeling tumor cell adaptations to hypoxia in multicellular tumor spheroids. J. Exp. Clin. Cancer Res. 2017, 36, 102. [Google Scholar] [CrossRef] [PubMed]
- Fujii, M.; Shimokawa, M.; Date, S.; Takano, A.; Matano, M.; Nanki, K.; Ohta, Y.; Toshimitsu, K.; Nakazato, Y.; Kawasaki, K.; et al. A Colorectal Tumor Organoid Library Demonstrates Progressive Loss of Niche Factor Requirements during Tumorigenesis. Cell Stem Cell 2016, 18, 827–838. [Google Scholar] [CrossRef]
- Van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef]
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.; Van Es, J.H.; Van den Brink, S.; Van Houdt, W.J.; Pronk, A.; Van Gorp, J.; Siersema, P.D.; et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Liu, B.; Wang, J.; Wei, S.; Qi, Z.; Wang, S.; Fu, W.; Chen, Y.G. A growth factor-free culture system underscores the coordination between Wnt and BMP signaling in Lgr5+ intestinal stem cell maintenance. Cell Discov. 2018, 4, 49. [Google Scholar] [CrossRef] [PubMed]
- Castro, F.; Dirks, W.G.; Fähnrich, S.; Hotz-Wagenblatt, A.; Pawlita, M.; Schmitt, M. High-throughput SNP-based authentication of human cell lines. Int. J. Cancer 2013, 132, 308–314. [Google Scholar] [CrossRef]
- Schmitt, M.; Pawlita, M. High-throughput detection and multiplex identification of cell contaminations. Nucleic Acids Res. 2009, 37, e119. [Google Scholar] [CrossRef]
- Girardot, C.; Scholtalbers, J.; Sauer, S.; Su, S.Y.; Furlong, E.E. Je, a versatile suite to handle multiplexed NGS libraries with unique molecular identifiers. BMC Bioinform. 2016, 17, 419. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Wu, Z.; Wu, Z. Exploration, visualization, and preprocessing of high-dimensional data. Methods Mol. Biol. 2010, 620, 267–284. [Google Scholar] [CrossRef] [PubMed]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., 3rd; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902 e1821. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- Lin, T.-Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft coco: Common objects in context. In Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014; pp. 740–755. [Google Scholar]
- Baretton, G. Pathology of the lower gastrointestinal tract. Der Pathol. 2011, 32, 273–274. [Google Scholar] [CrossRef][Green Version]
- Kriegsmann, K.; Cremer, M.; Zgorzelski, C.; Harms, A.; Muley, T.; Winter, H.; Kazdal, D.; Warth, A.; Kriegsmann, M. Agreement of CK5/6, p40, and p63 immunoreactivity in non-small cell lung cancer. Pathology 2019, 51, 240–245. [Google Scholar] [CrossRef]
- Erich, K.; Reinle, K.; Müller, T.; Munteanu, B.; Sammour, D.A.; Hinsenkamp, I.; Gutting, T.; Burgermeister, E.; Findeisen, P.; Ebert, M.P.; et al. Spatial Distribution of Endogenous Tissue Protease Activity in Gastric Carcinoma Mapped by MALDI Mass Spectrometry Imaging. Mol. Cell. Proteom. MCP 2019, 18, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]

| Patient | Sex | Origin | Site | Stage (UICC) | MS status | TP53 | APC | KRAS |
|---|---|---|---|---|---|---|---|---|
| P1 | m | liver met | Rectum | IV | MSS | X | X | X |
| P2 | m | lung met | Caecum | IV | MSS | X | X | X |
| P3 | f | liver met | Rectum | IV | MSS | X | X | X |
| P4 | f | liver met | Ascending colon | IV | MSI | X | X | X |
| P5 | f | primary | Transverse colon | IV | MSS | - | - | - |
| P6 | f | primary | Caecum | IV | MSS | X | - | - |
| P7 | m | liver met | Sigmoid | IV | MSS | - | X | X |
| P8 | m | liver met | Caecum | IV | MSS | X | - | - |
| P9 | m | primary | Rectum | IV | MSS | X | - | X |
| P10 | m | primary | Sigmoid | IIIB | MSS | N/A | N/A | N/A |
| P11 | m | primary | Rectum and caecum | IIIB | MSS | - | - | - |
| P12 | m | primary | Rectum and transverse colon | II | MSI | X | X | X |
| O1 | f | liver met | Sigmoid | IV | MSS | X | X | X |
| O2 | f | liver met | Caecum | IV | MSS | X | X | X |
| O3 | f | liver met | Ascending colon | IV | MSI | - | X | - |
| X1 | m | primary | Rectum | I | N/A | N/A | N/A | N/A |
| X2 | m | primary | Ascending colon | II | MSI | N/A | N/A | - |
| T1 | m | primary | Sigmoid | III | N/A | N/A | N/A | N/A |
| T2 | m | primary | Ascending colon | IV | N/A | N/A | N/A | N/A |
| T3 | m | primary | Ascending colon | IVa | MSS | N/A | N/A | X |
| CRC Spheroids | ||||
| Patient | Mean Reads Per Cell | Cell Number after QC | Mean Detected Genes Per Cell | LGR5 Score |
| P1 | 348,016 | 325 | 3535 | 12.85 |
| P2 | 261,595 | 309 | 4072 | 0.23 |
| P3 | 460,471 | 551 | 4537 | 6.43 |
| P4 | 1,061,813 | 263 | 4186 | 87.72 |
| P5 | 334,099 | 502 | 3943 | 4.61 |
| P6 | 1,276,856 | 141 | 5116 | 0.03 |
| P7 | 359,362 | 434 | 4335 | 10.18 |
| P8 | 190,170 | 197 | 4174 | 3.38 |
| P9 | 527,407 | 464 | 4354 | 0.00 |
| P10 | 391,680 | 736 | 3418 | 3.35 |
| P11 | 505,439 | 308 | 4036 | 1.43 |
| P12 | 454,258 | 433 | 3977 | 0.00 |
| CRC PDOs, PDXs, Tumors | ||||
| Sample | Mean Reads Per Cell | Cell Number after QC | Mean Detected Genes Per Cell (Hs) | Mean Detected Genes Per Cell (Mm) |
| O1 | 120,218 | 5550 | 5542 | - |
| O2 | 169,086 | 3003 | 5425 | - |
| O3 | 73,415 | 8785 | 4176 | - |
| X1 | 238,836 | 1475 | 1841 | 2281 |
| X2 | 237,891 | 1070 | 4598 | 2415 |
| T1 | 1,333,884 | 362 (136) | 3646 | - |
| T2 | 847,472 | 538 (77) | 4090 | - |
| T3 | 623,942 | 724 (40) | 2474 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zowada, M.K.; Tirier, S.M.; Dieter, S.M.; Krieger, T.G.; Oberlack, A.; Chua, R.L.; Huerta, M.; Ten, F.W.; Laaber, K.; Park, J.; et al. Functional States in Tumor-Initiating Cell Differentiation in Human Colorectal Cancer. Cancers 2021, 13, 1097. https://doi.org/10.3390/cancers13051097
Zowada MK, Tirier SM, Dieter SM, Krieger TG, Oberlack A, Chua RL, Huerta M, Ten FW, Laaber K, Park J, et al. Functional States in Tumor-Initiating Cell Differentiation in Human Colorectal Cancer. Cancers. 2021; 13(5):1097. https://doi.org/10.3390/cancers13051097
Chicago/Turabian StyleZowada, Martina K., Stephan M. Tirier, Sebastian M. Dieter, Teresa G. Krieger, Ava Oberlack, Robert Lorenz Chua, Mario Huerta, Foo Wei Ten, Karin Laaber, Jeongbin Park, and et al. 2021. "Functional States in Tumor-Initiating Cell Differentiation in Human Colorectal Cancer" Cancers 13, no. 5: 1097. https://doi.org/10.3390/cancers13051097
APA StyleZowada, M. K., Tirier, S. M., Dieter, S. M., Krieger, T. G., Oberlack, A., Chua, R. L., Huerta, M., Ten, F. W., Laaber, K., Park, J., Jechow, K., Müller, T., Kalxdorf, M., Kriegsmann, M., Kriegsmann, K., Herbst, F., Krijgsveld, J., Schneider, M., Eils, R., ... Ball, C. R. (2021). Functional States in Tumor-Initiating Cell Differentiation in Human Colorectal Cancer. Cancers, 13(5), 1097. https://doi.org/10.3390/cancers13051097








