Perspective of Immune Checkpoint Inhibitors in Thymic Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. The TCGA Genomic and Clinical Data
2.2. Transcriptomic Data
2.3. The Human Protein Atlas
2.4. Stemness-Associated Scores
2.5. The Gene Set Enrichment Analysis
2.6. Validation Sets
2.7. Statistical Analyses
3. Results
3.1. TIF1 Family Members Are Differentially Expressed across Solid Tumors
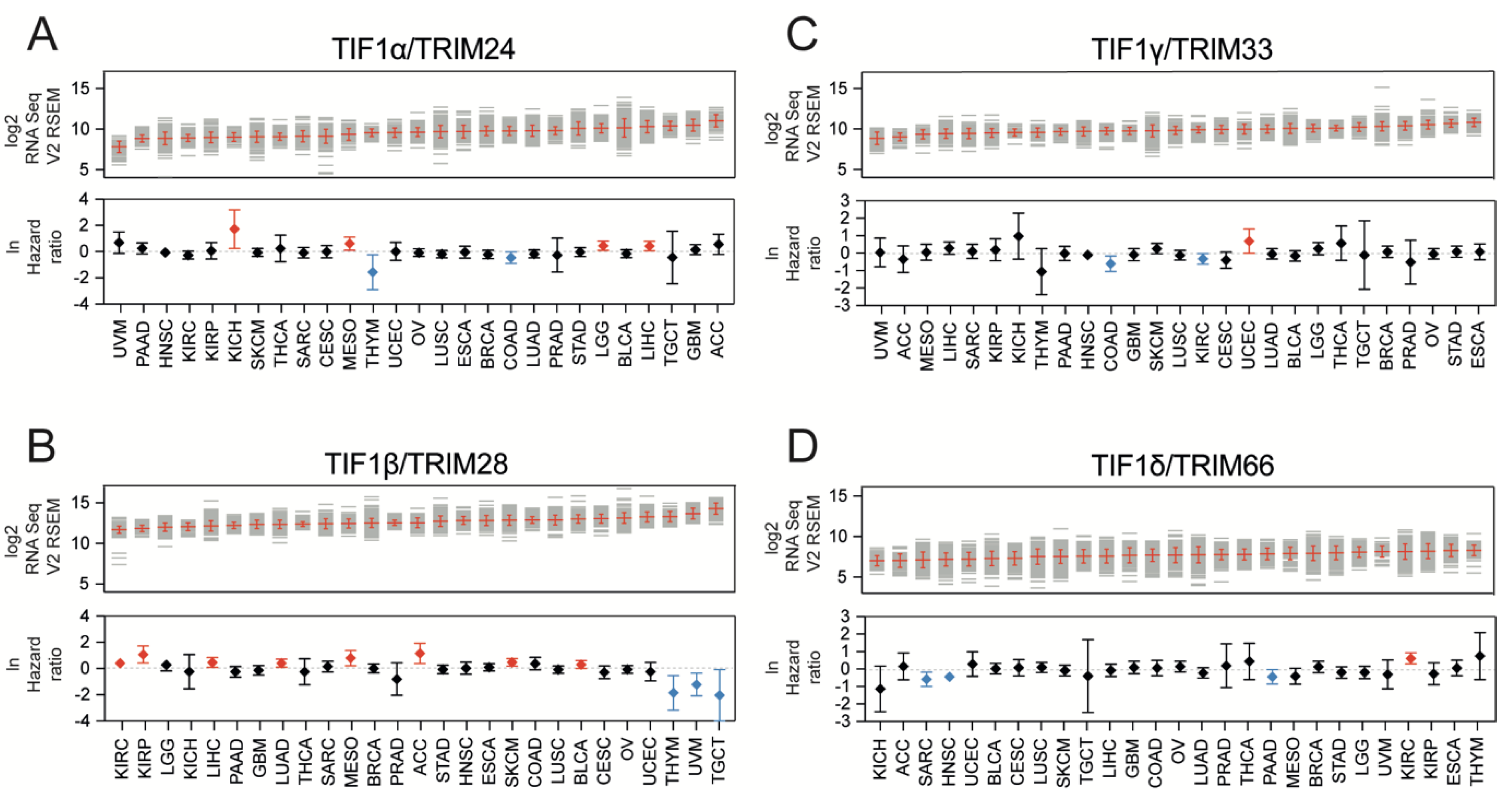
3.2. TIF1α/TRIM24 and TIF1β/TRIM28 Are Positively Associated While TIF1γ/TRIM33 and TIF1δ/TRIM66 Are Negatively Associated with Tumor Stemness
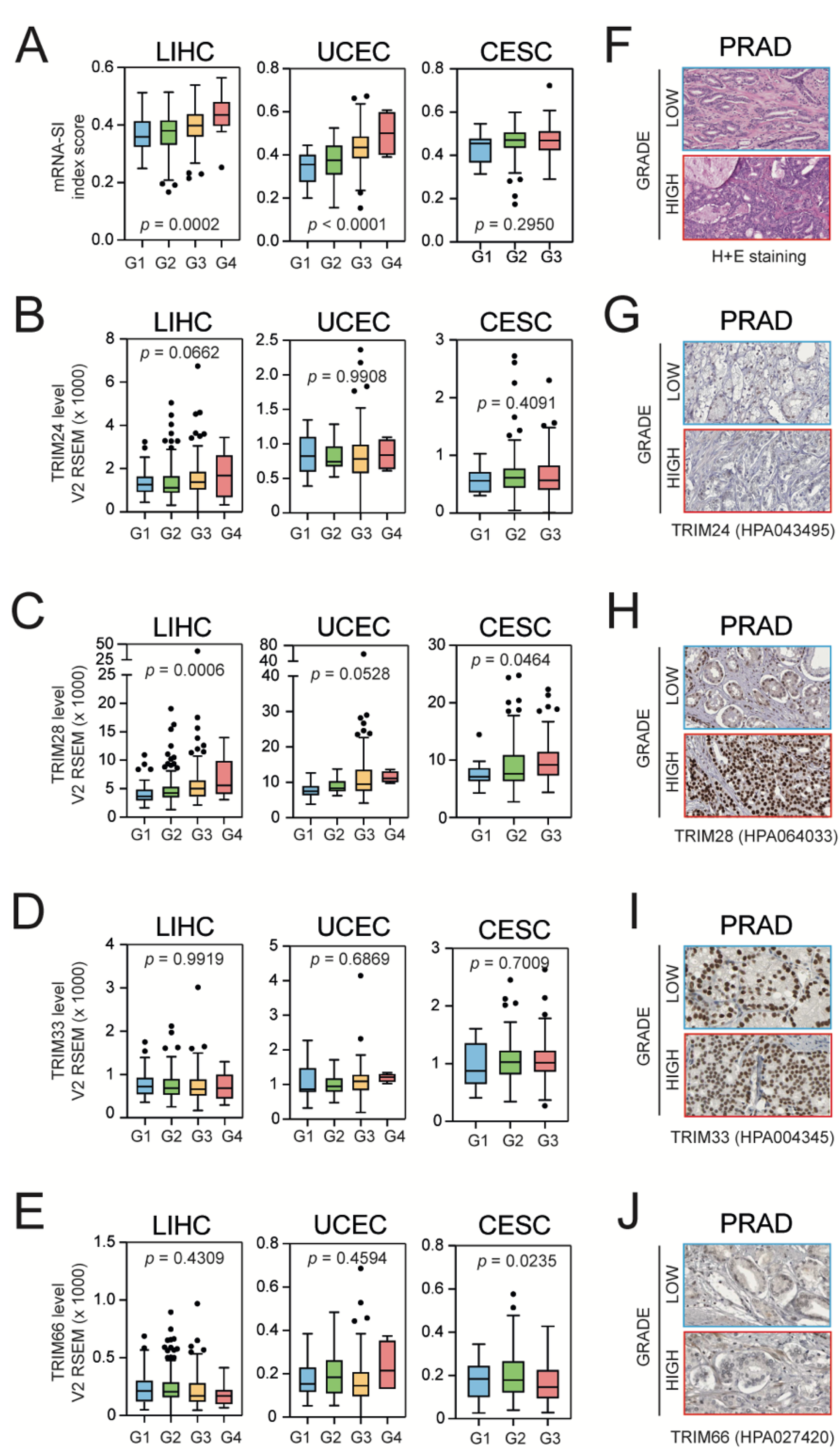
3.3. Higher Grade Tumors Exhibit a Higher Level of mRNA-SI and Elevated TIF1β/TRIM28 Expression
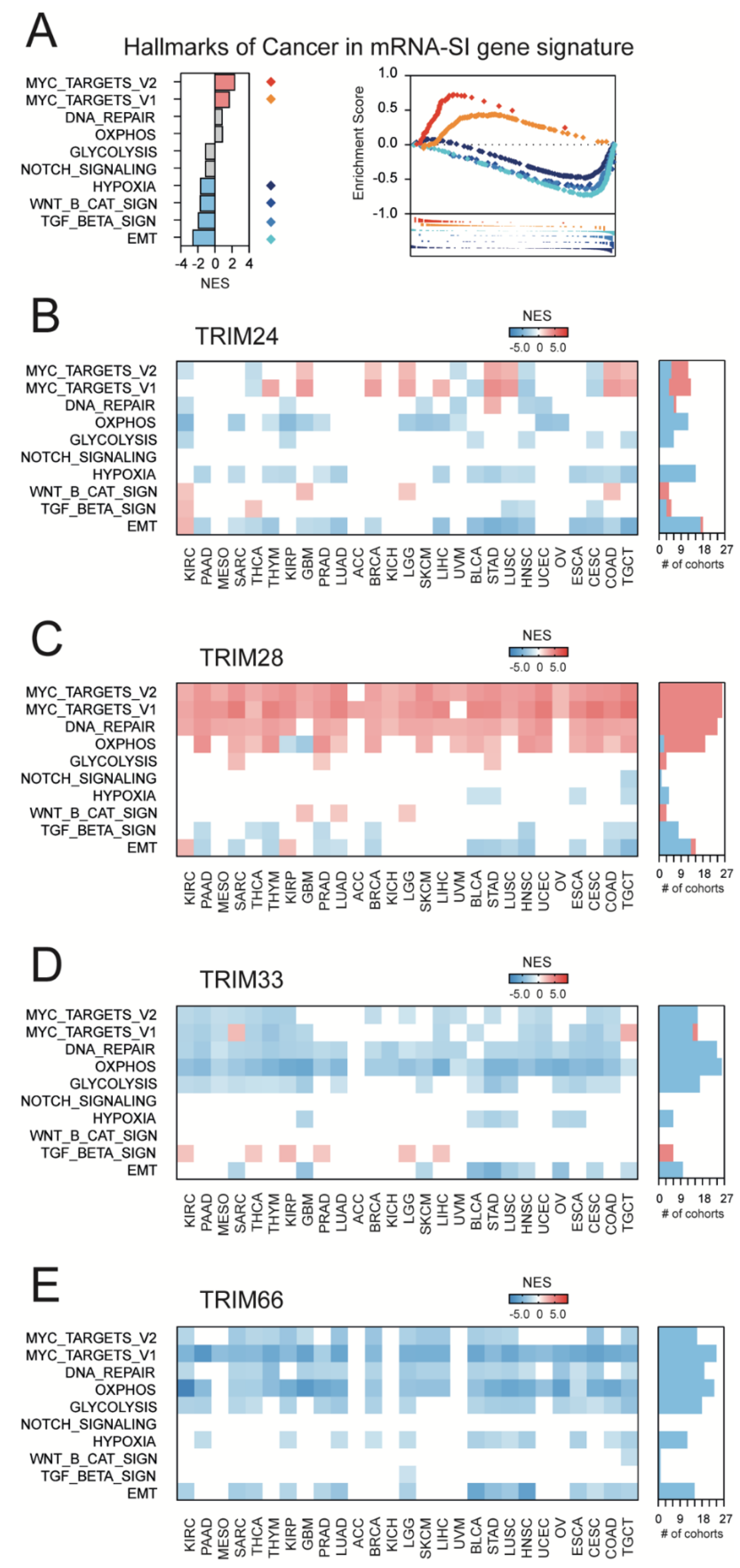
3.4. TRIM28-Associated Gene Expression Profiles Are Significantly Enriched with the Targets for the c-Myc Transcription Factor That Mirrors the Enrichment of mRNA-SI Gene Signature with the “Hallmarks Of Cancer” Terms
3.5. TRIM28-Associated Transcriptome Profiles Are Significantly Enriched, while TRIM66-Associated Transcriptome Profiles Are Significantly Depleted with Stemness-Associated Gene Signatures across Solid TCGA Tumor Types
3.6. Stemness-High Tumors Are Significantly Associated with a Worse Prognosis, and TRIM28 High Expression May Predict a “Stemness-High” Phenotype
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Friedmann-Morvinski, D.; Verma, I.M. Dedifferentiation and reprogramming: Origins of cancer stem cells. EMBO Rep. 2014, 15, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Czerwinska, P.; Kaminska, B. Regulation of breast cancer stem cell features. Contemp. Oncol. 2015, 19, A7–A15. [Google Scholar] [CrossRef] [PubMed]
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells: Current status and evolving complexities. Cell Stem Cell 2012, 10, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Ben-Porath, I.; Thomson, M.W.; Carey, V.J.; Ge, R.; Bell, G.W.; Regev, A.; Weinberg, R.A. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008, 40, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.J.; Liu, H.; Ridky, T.W.; Cassarino, D.; Segal, E.; Chang, H.Y. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell 2008, 2, 333–344. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, B.; Miura, T.; Brandenberger, R.; Mejido, J.; Luo, Y.; Yang, A.X.; Joshi, B.H.; Ginis, I.; Thies, R.S.; Amit, M.; et al. Gene expression in human embryonic stem cell lines: Unique molecular signature. Blood 2012, 103, 2956–2964. [Google Scholar] [CrossRef]
- Malta, T.M.; Sokolov, A.; Gentles, A.J.; Burzykowski, T.; Poisson, L.; Weinstein, J.N.; Kaminska, B.; Huelsken, J.; Omberg, L.; Gevaert, O.; et al. Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell 2018, 173, 338–354. [Google Scholar] [CrossRef]
- Wainwright, E.N.; Scaffidi, P. Epigenetics and Cancer Stem Cells: Unleashing, Hijacking, and Restricting Cellular Plasticity. Trends Cancer 2017, 3, 372–386. [Google Scholar] [CrossRef]
- McAvera, R.M.; Crawford, L.J. TIF1 Proteins in Genome Stability and Cancer. Cancers 2020, 12, 2094. [Google Scholar] [CrossRef]
- Jaworska, A.M.; Wlodarczyk, N.A.; Mackiewicz, A.; Czerwinska, P. The role of TRIM family proteins in the regulation of cancer stem cell self-renewal. Stem Cells 2020, 38, 165–173. [Google Scholar]
- Li, H.; Sun, L.; Tang, Z.; Fu, L.; Xu, Y.; Li, Z.; Luo, W.; Qiu, X.; Wang, E. Overexpression of TRIM24 Correlates with Tumor Progression in Non-Small Cell Lung Cancer. PLoS ONE 2012, 7, e37657. [Google Scholar] [CrossRef]
- Cui, Z.; Cao, W.; Li, J.; Song, X.; Mao, L.; Chen, W. TRIM24 Overexpression Is Common in Locally Advanced Head and Neck Squamous Cell Carcinoma and Correlates with Aggressive Malignant Phenotypes. PLoS ONE 2013, 8, e63887. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-H.; Yin, A.-A.; Cheng, J.-X.; Huang, H.-Y.; Li, X.-M.; Zhang, Y.-Q.; Han, N.; Zhang, X. TRIM24 promotes glioma progression and enhances chemoresistance through activation of the PI3K/Akt signaling pathway. Oncogene 2015, 34, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Lv, D.; Li, Y.; Zhang, W.; Alvarez, A.A.; Song, L.; Tang, J.; Gao, W.-Q.; Hu, B.; Cheng, S.-Y.; Feng, H. TRIM24 is an oncogenic transcriptional co-activator of STAT3 in glioblastoma. Nat. Commun. 2017, 8, 1454. [Google Scholar] [CrossRef]
- Zhang, L.; Yin, Y.; Chen, H.; Feng, S.; Liu, J.; Chen, L.; Fu, W.-L.; Sun, G.-C.; Yu, X.-G.; Xu, D.-G. TRIM24 promotes stemness and invasiveness of glioblastoma cells via activating Sox2 expression. Neuro Oncol. 2020, 22, 1797–1808. [Google Scholar] [CrossRef] [PubMed]
- Allton, K.; Jain, A.K.; Herz, H.-M.; Tsai, W.-W.; Jung, S.Y.; Qin, J.; Bergmann, A.; Johnson, R.L.; Barton, M.C. Trim24 targets endogenous p53 for degradation. Proc. Natl. Acad. Sci. USA 2009, 106, 11612–11616. [Google Scholar] [CrossRef]
- Jain, A.K.; Allton, K.; Iacovino, M.; Mahen, E.; Milczarek, R.J.; Zwaka, T.P.; Kyba, M.; Barton, M.C. p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS Biol. 2012, 10, e1001268. [Google Scholar] [CrossRef]
- Rafiee, M.-R.; Girardot, C.; Sigismondo, G.; Krijgsveld, J. Expanding the Circuitry of Pluripotency by Selective Isolation of Chromatin-Associated Proteins. Mol. Cell. 2016, 64, 624–635. [Google Scholar] [CrossRef]
- Addison, J.B.; Koontz, C.; Fugett, J.H.; Creighton, C.J.; Chen, D.; Farrugia, M.K.; Padon, R.R.; Voronkova, M.A.; McLaughlin, S.L.; Livengood, R.H.; et al. KAP1 Promotes Proliferation and Metastatic Progression of Breast Cancer Cells. Cancer Res. 2014, 75, 344–355. [Google Scholar] [CrossRef]
- Czerwinska, P.; Shah, P.K.; Tomczak, K.; Klimczak, M.; Mazurek, S.; Sozanska, B.; Biecek, P.; Korski, K.; Filas, V.; Mackiewicz, A.; et al. TRIM28 multi-domain protein regulates cancer stem cell population in breast tumor development. Oncotarget 2017, 8, 863–882. [Google Scholar] [CrossRef] [PubMed]
- Yokoe, T.; Toiyama, Y.; Okugawa, Y.; Tanaka, K.; Ohi, M.; Inoue, Y.; Mohri, Y.; Miki, C.; Kusunoki, M. KAP1 is associated with peritoneal carcinomatosis in gastric cancer. Ann. Surg. Oncol. 2010, 17, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Yang, S.; Fu, X.; Feng, J.; Xu, S.; Ying, G. High Levels of KAP1 Expression Are Associated with Aggressive Clinical Features in Ovarian Cancer. Int. J. Mol. Sci. 2015, 16, 363–377. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.-X.; Cai, J.-J.; Chen, L.-C.; Yue, Q.; Gong, Y.; Yao, Y.; Mao, Y. TRIM28 as an independent prognostic marker plays critical roles in glioma progression. J. Neurooncol. 2016, 126, 19–26. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, J.; Li, Q.; Ma, H.; Xu, Z.; Gao, Y. KAP1 is overexpressed in hepatocellular carcinoma and its clinical significance. Int. J. Clin. Oncol. 2016, 21, 927–933. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Zhan, L.; Jiang, J.; Pan, Y.; Zhang, H.; Li, X.; Pen, F.; Wang, M.; Qin, R.; Sun, C. KAP-1 is overexpressed and correlates with increased metastatic ability and tumorigenicity in pancreatic cancer. Med. Oncol. 2014, 31, 25. [Google Scholar] [CrossRef] [PubMed]
- Oleksiewicz, U.; Gładych, M.; Raman, A.T.; Heyn, H.; Mereu, E.; Chlebanowska, P.; Andrzejewska, A.; Sozanska, B.; Samant, N.; Fak, K.; et al. TRIM28 and Interacting KRAB-ZNFs Control Self-Renewal of Human Pluripotent Stem Cells through Epigenetic Repression of Pro-differentiation Genes. Stem Cell Rep. 2017, 9, 2065–2080. [Google Scholar] [CrossRef]
- Klimczak, M.; Czerwinska, P.; Mazurek, S.; Sozanska, B.; Biecek, P.; Mackiewicz, A.; Wiznerowicz, M. TRIM28 epigenetic corepressor is indispensable for stable induced pluripotent stem cell formation. Stem Cell Res. 2017, 23, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Czerwinska, P.; Jaworska, A.M.; Wlodarczyk, N.A.; Mackiewicz, A.A. Melanoma Stem Cell-Like Phenotype and Significant Suppression of Immune Response within a Tumor Are Regulated by TRIM28 Protein. Cancers 2020, 12, 2998. [Google Scholar] [CrossRef]
- Ding, Z.; Jin, G.; Wang, W.; Chen, W.; Wu, Y.; Ai, X.; Chen, L.; Zhang, W.-G.; Liang, H.-F.; Laurence, A.; et al. Reduced expression of transcriptional intermediary factor 1 gamma promotes metastasis and indicates poor prognosis of hepatocellular carcinoma. Hepatology 2014, 60, 1620–1636. [Google Scholar] [CrossRef]
- Pommier, R.M.; Gout, J.; Vincent, D.F.; Alcaraz, L.B.; Chuvin, N.; Arfi, V.; Martel, S.; Kaniewski, B.; Devailly, G.; Fourel, G.; et al. TIF1γ Suppresses Tumor Progression by Regulating Mitotic Checkpoints and Chromosomal Stability. Cancer Res. 2015, 75, 4335–4350. [Google Scholar] [CrossRef]
- Xue, J.; Chen, Y.; Wu, Y.; Wang, Z.; Zhou, A.; Zhang, S.; Lin, L.; Aldape, K.; Majumder, S.; Lu, Z.; et al. Tumour suppressor TRIM33 targets nuclear β-catenin degradation. Nat. Commun. 2015, 6, 6156. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Ding, Z.; Liang, H.; Zhang, B.; Chen, X. The Roles of TIF1γ in Cancer. Front. Oncol. 2019, 9, 979. [Google Scholar] [CrossRef] [PubMed]
- Kassem, L.; Deygas, M.; Fattet, L.; Lopez, J.; Goulvent, T.; Lavergne, E.; Chabaud, S.; Carrabin, N.; Chopin, N.; Bachelot, T.; et al. TIF1γ interferes with TGFβ1/SMAD4 signaling to promote poor outcome in operable breast cancer patients. BMC Cancer 2015, 15, 453. [Google Scholar] [CrossRef]
- Wang, E.; Kawaoka, S.; Roe, J.-S.; Shi, J.; Hohmann, A.F.; Xu, Y.; Bhagwat, A.S.; Suzuki, Y.; Kinney, J.B.; Vakoc, C.R. The transcriptional cofactor TRIM33 prevents apoptosis in B lymphoblastic leukemia by deactivating a single enhancer. eLife 2015, 4, e06377. [Google Scholar] [CrossRef] [PubMed]
- Massagué, J.; Xi, Q. TGF- b control of stem cell differentiation genes. FEBS Lett. 2012, 586, 1953–1958. [Google Scholar] [CrossRef]
- Rajderkar, S.; Panaretos, C.; Kaartinen, V. Trim33 regulates early maturation of mouse embryoid bodies in vitro. Biochem. Biophys. Rep. 2017, 12, 185–192. [Google Scholar] [CrossRef]
- Dai, H.-Y.; Ma, Y.; Da, Z.; Hou, X.-M. Knockdown of TRIM66 inhibits malignant behavior and epithelial-mesenchymal transition in non-small cell lung cancer. Pathol. Res. Pract. 2018, 214, 1130–1135. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Cui, J.; Wu, Y.; Sun, X.; Chen, N. Knockdown of TRIM66 inhibits cell proliferation, migration and invasion in colorectal cancer through JAK2/STAT3 pathway. Life Sci. 2019, 235, 116799. [Google Scholar] [CrossRef]
- Cao, H.; Gao, R.; Chen, L.; Feng, Y. TRIM66 promotes malignant progression of prostate carcinoma through the JAK/STAT pathway. FEBS Open Bio 2020, 10, 515–524. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Björling, E.; Agaton, C.; Szigyarto, C.A.-K.; Amini, B.; Andersen, E.; Andersson, A.-C.; Angelidou, P.; Asplund, A.; Asplund, C.; et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteom. 2005, 4, 1920–1932. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Choi, W.; Porten, S.; Kim, S.; Willis, D.; Plimack, E.R.; Hoffman-Censits, J.; Roth, B.; Cheng, T.; Tran, M.; Lee, I.-L.; et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 2014, 25, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-Y.; Kim, T.-J.; Kim, J.-Y.; Choi, C.H.; Do, I.-G.; Song, S.Y.; Sohn, I.; Jung, S.-H.; Bae, D.-S.; Lee, J.-W.; et al. Genetic profiling to predict recurrence of early cervical cancer. Gynecol. Oncol. 2013, 131, 650–654. [Google Scholar] [CrossRef] [PubMed]
- Bild, A.H.; Yao, G.; Chang, J.T.; Wang, Q.; Potti, A.; Chasse, D.; Joshi, M.-B.; Harpole, D.; Lancaster, J.M.; Berchuck, A.; et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2006, 439, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Landi, M.T.; Dracheva, T.; Rotunno, M.; Figueroa, J.D.; Liu, H.; Dasgupta, A.; Mann, F.E.; Fukuoka, J.; Hames, M.; Bergen, A.W.; et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS ONE 2008, 3, e1651. [Google Scholar] [CrossRef]
- Cirenajwis, H.; Ekedahl, H.; Lauss, M.; Harbst, K.; Carneiro, A.; Enoksson, J.; Rosengren, F.; Linda Werner-Hartman, L.; Törngren, T.; Kvist, A.; et al. Molecular stratification of metastatic melanoma using gene expression profiling: Prediction of survival outcome and benefit from molecular targeted therapy. Oncotarget 2015, 6, 12297–12309. [Google Scholar] [CrossRef]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.-M.; Gingras, M.-C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.C.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef]
- Pérez-Mancera, P.A.; Rust, A.G.; van der Weyden, L.; Kristiansen, G.; Li, A.; Sarver, A.L.; Silverstein, K.A.T.; Grützmann, R.; Aust, D.; Rümmele, P.; et al. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature 2012, 486, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Ross-Adams, H.; Lamb, A.D.; Dunning, M.J.; Halim, S.; Lindberg, J.; Massie, C.M.; Egevad, L.A.; Russell, R.; Ramos-Montoya, A.; Vowler, S.L.; et al. Integration of copy number and transcriptomics provides risk stratification in prostate cancer: A discovery and validation cohort study. EBioMedicine 2015, 2, 1133–1144. [Google Scholar] [CrossRef] [PubMed]
- Filion, C.; Motoi, T.; Olshen, A.B.; Laé, M.; Emnett, R.J.; Gutmann, D.H.; Perry, A.; Ladanyi, M.; Labelle, Y. The EWSR1/NR4A3 fusion protein of extraskeletal myxoid chondrosarcoma activates the PPARG nuclear receptor gene. J. Pathol. 2009, 217, 83–93. [Google Scholar] [CrossRef]
- Li, J.; Xi, Y.; Li, W.; McCarthy, R.L.; Stratton, S.A.; Zou, W.; Li, W.; Dent, S.Y.; Jain, A.K.; Barton, M.C. TRIM28 interacts with EZH2 and SWI/SNF to activate genes that promote mammosphere formation. Oncogene 2017, 36, 2991–3001. [Google Scholar] [CrossRef] [PubMed]
- Miranda, A.; Hamilton, P.T.; Zhang, A.W.; Becht, E.; Bruun, J.; Micke, P.; de Reynies, A.; Nelson, B.H. Cancer stemness, intratumoral heterogeneity, and immune response across cancers. Proc. Natl. Acad. Sci. USA 2019, 116, 9020–9029. [Google Scholar] [CrossRef]
- Pece, S.; Tosoni, D.; Confalonieri, S.; Mazzarol, G.; Vecchi, M.; Ronzoni, S.; Bernard, L.; Viale, G.; Pelicci, P.G.; Di Fiore, P.P. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell 2010, 140, 62–73. [Google Scholar] [CrossRef]
- Beier, D.; Hau, P.; Proescholdt, M.; Lohmeier, A.; Wischhusen, J.; Oefner, P.J.; Aigner, L.; Brawanski, A.; Bogdahn, U.; Beier, C.P. CD133+ and CD133- Glioblastoma-Derived Cancer Stem Cells Show Differential Growth Characteristics and Molecular Profiles. Cancer Res. 2007, 67, 4010–4015. [Google Scholar] [CrossRef]
- Pece, S.; Disalvatore, D.; Tosoni, D.; Vecchi, M.; Confalonieri, S.; Bertalot, G.; Viale, G.; Colleoni, M.; Veronesi, P.; Galimberti, V.; et al. Identification and clinical validation of a multigene assay that interrogates the biology of cancer stem cells and predicts metastasis in breast cancer: A retrospective consecutive study. EBioMedicine 2019, 42, 352–362. [Google Scholar] [CrossRef]
- Cheng, B.; Ren, X.; Kerppola, T.K. KAP1 represses differentiation-inducible genes in embryonic stem cells through cooperative binding with PRC1 and derepresses pluripotency-associated genes. Mol. Cell. Biol. 2014, 34, 2075–2091. [Google Scholar] [CrossRef]
- Miles, D.C.; de Vries, N.A.; Gisler, S.; Lieftink, C.; Akhtar, W.; Gogola, E.; Pawlitzky, I.; Hulsman, D.; Tanger, E.; Koppens, M.; et al. TRIM28 is an Epigenetic Barrier to Induced Pluripotent Stem Cell Reprogramming. Stem Cells 2017, 35, 147–157. [Google Scholar] [CrossRef]
- Koch, H.; Zhang, R.; Verdoodt, B.; Bailey, A.; Zhang, C.; Yates, J.R., 3rd; Menssen, A.; Hermeking, H. Large-scale identification of c-MYC-associated proteins using a combined TAP/MudPIT approach. Cell Cycle 2007, 6, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, G.J. Emerging roles of Myc in stem cell biology and novel tumor therapies. J. Exp. Clin. Cancer Res. 2018, 37, 173. [Google Scholar] [CrossRef] [PubMed]
- Czerwinska, P.; Mazurek, S.; Wiznerowicz, M. The complexity of TRIM28 contribution to cancer. J. Biomed. Sci. 2017, 24, 63–77. [Google Scholar] [CrossRef]
- Buckley, S.M.; Aranda-Orgilles, B.; Strikoudis, A.; Apostolou, E.; Loizou, E.; Moran-Crusio, K.; Farnsworth, C.L.; Koller, A.A.; Dasgupta, R.; Silva, J.C.; et al. Regulation of Pluripotency and Cellular Reprogramming by the Ubiquitin-Proteasome System. Cell Stem Cell 2012, 11, 783–798. [Google Scholar] [CrossRef] [PubMed]
- Pineda, C.T.; Potts, P.R. Oncogenic MAGEA-TRIM28 ubiquitin ligase downregulates autophagy by ubiquitinating and degrading AMPK in cancer. Autophagy 2015, 11, 844–846. [Google Scholar] [CrossRef] [PubMed]
- Pineda, C.T.; Ramanathan, S.; Fon Tacer, K.; Weon, J.L.; Potts, M.B.; Ou, Y.H.; White, M.A.; Potts, P.R. Degradation of AMPK by a cancer-specific ubiquitin ligase. Cell 2015, 160, 715–728. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Z.; Guo, X.; Li, F.; Wei, Q.; Chen, X.; Gong, D.; Xu, Y.; Chen, W.; Liu, Y. TRIM66 reads unmodified H3R2K4 and H3K56ac to respond to DNA damage in embryonic stem cells. Nat. Commun. 2019, 10, 4273. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaira, K.; Imai, H.; Kagamu, H. Perspective of Immune Checkpoint Inhibitors in Thymic Carcinoma. Cancers 2021, 13, 1065. https://doi.org/10.3390/cancers13051065
Kaira K, Imai H, Kagamu H. Perspective of Immune Checkpoint Inhibitors in Thymic Carcinoma. Cancers. 2021; 13(5):1065. https://doi.org/10.3390/cancers13051065
Chicago/Turabian StyleKaira, Kyoichi, Hisao Imai, and Hiroshi Kagamu. 2021. "Perspective of Immune Checkpoint Inhibitors in Thymic Carcinoma" Cancers 13, no. 5: 1065. https://doi.org/10.3390/cancers13051065
APA StyleKaira, K., Imai, H., & Kagamu, H. (2021). Perspective of Immune Checkpoint Inhibitors in Thymic Carcinoma. Cancers, 13(5), 1065. https://doi.org/10.3390/cancers13051065





