PDX-Derived Ewing’s Sarcoma Cells Retain High Viability and Disease Phenotype in Alginate Encapsulated Spheroid Cultures
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Samples Processing
2.3. Cell Culture and Spheroid Encapsulation
2.4. Spheroid Retrieval from Alginate Capsules and Diameter Analysis
2.5. Metabolic Viability Assessment
2.6. Cell Viability Analysis
2.7. Cell Proliferation Analysis
2.8. Exposure to Chemotherapeutic Drugs
2.9. RNA Extraction, RT-qPCR Analysis, and Detection of EWSR1-FLI1 Expression
2.10. Mouse Cells Contamination Analysis
2.11. Hypoxia Detection
2.12. Immunofluorescence Analysis
2.13. Flow Cytometry Analysis
3. Results
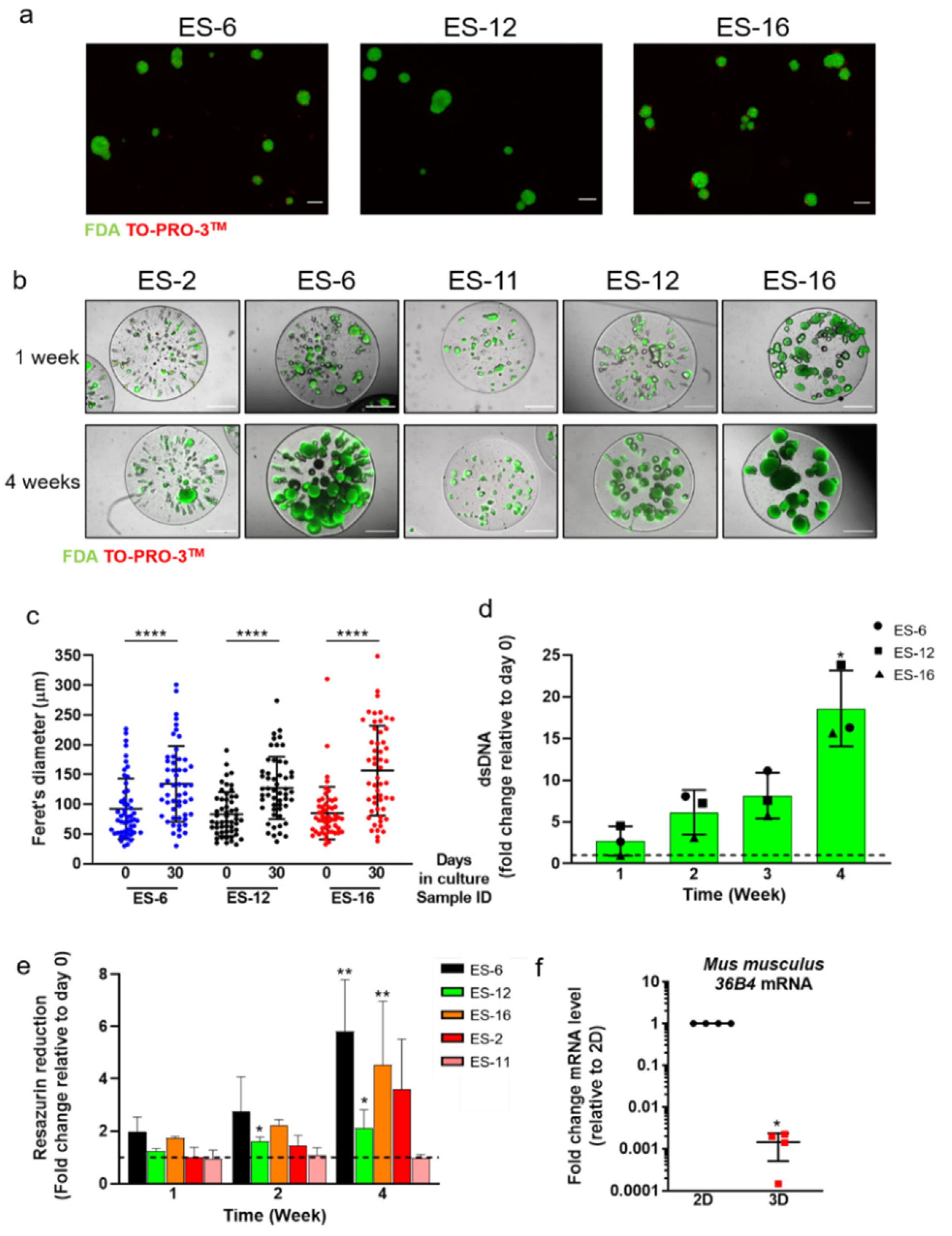
3.1. PDX-Derived ES Cell Spheroid Cultures Sustain Cell Proliferation and Reduce Mouse Cell Contamination
3.2. ES Cells in Encapsulated Spheroid Cultures Retain EWSR1-FLI1 Translocation
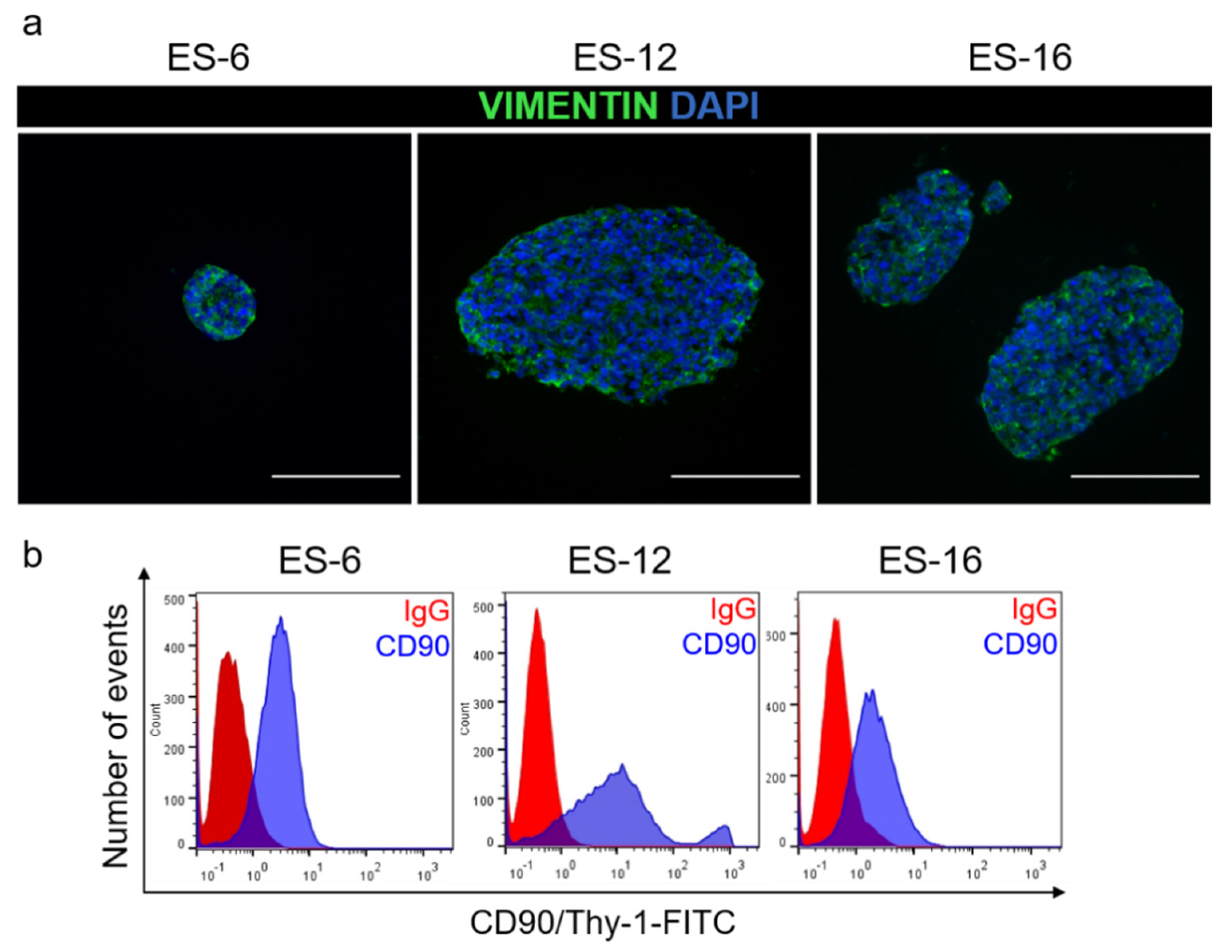
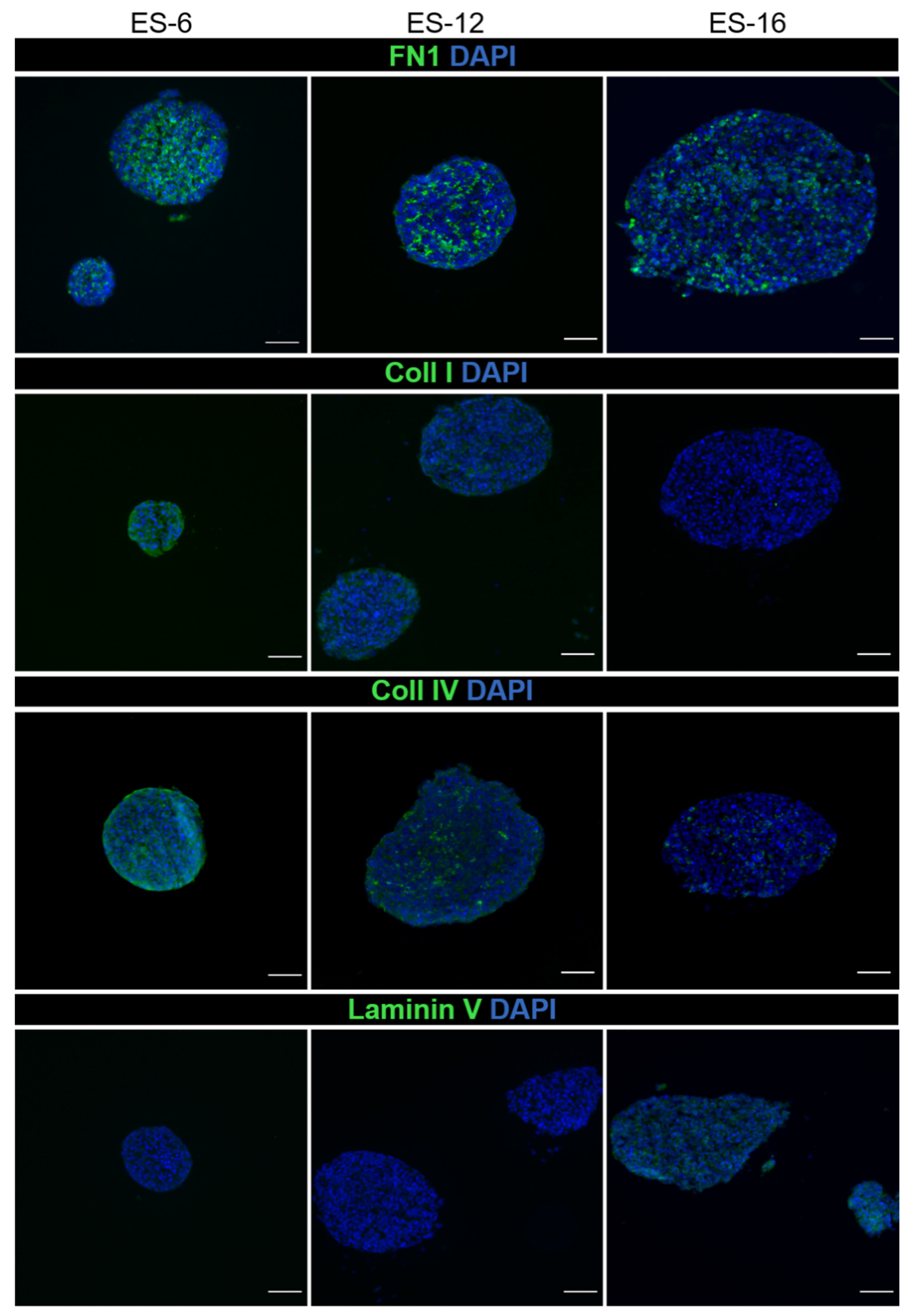
3.3. ES Cells in Encapsulated Spheroid Cultures Express Typical Mesenchymal Cell Markers and Secrete ECM Components
3.4. Proof of Concept of the Applicability of Encapsulated ES Spheroids Cultures in Drug Assays
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Mater, D.; Wagner, L. Management of recurrent Ewing sarcoma: Challenges and approaches. OncoTargets Ther. 2019, 12, 2279–2288. [Google Scholar] [CrossRef]
- Grünewald, T.G.P.; Cidre-Aranaz, F.; Surdez, D.; Tomazou, E.M.; De Álava, E.; Kovar, H.; Sorensen, P.H.; Delattre, O.; Dirksen, U. Ewing sarcoma. Nat. Rev. Dis. Prim. 2018, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Barr, F.G.; Chatten, J.; D’Cruz, C.M.; Wilson, A.E.; Nauta, L.E.; Nycum, L.M.; Biegel, J.A.; Womer, R.B. Molecular Assays for Chromosomal Translocations in the Diagnosis of Pediatric Soft Tissue Sarcomas. JAMA 1995, 273, 553–557. [Google Scholar] [CrossRef]
- Dasgupta, R.; Fuchs, J.; Rodeberg, D. Rhabdomyosarcoma. Semin. Pediatr. Surg. 2016, 25, 276–283. [Google Scholar] [CrossRef]
- Stegmaier, S.; Leuschner, I.; Aakcha-Rudel, E.; Münch, P.; Kazanowska, B.; Békàssy, A.; Treuner, J.; Koscielniak, E. Identification of Various Exon Combinations of theews/fli1Translocation: An Optimized RT-PCR Method for Paraffin Embedded Tissue. Klinische Pädiatrie 2004, 216, 315–322. [Google Scholar] [CrossRef]
- Kowalewski, A.A.; Randall, R.L.; Lessnick, S.L. Cell Cycle Deregulation in Ewing’s Sarcoma Pathogenesis. Sarcoma 2010, 2011, 1–10. [Google Scholar] [CrossRef]
- Lessnick, S.L.; Ladanyi, M. Molecular Pathogenesis of Ewing Sarcoma: New Therapeutic and Transcriptional Targets. Annu. Rev. Pathol. Mech. Dis. 2012, 7, 145–159. [Google Scholar] [CrossRef]
- Yu, H.; Ge, Y.; Guo, L.; Huang, L. Potential approaches to the treatment of Ewing’s sarcoma. Oncotarget 2016, 8, 5523–5539. [Google Scholar] [CrossRef] [PubMed]
- Rizk, V.T.; Walko, C.M.; Brohl, A.S. Precision medicine approaches for the management of Ewing sarcoma: Current per-spectives. Pharmgenomics Pers. Med. 2019, 12, 9–14. [Google Scholar] [CrossRef]
- Teicher, B.A.; Bagley, R.G.; Rouleau, C.; Kruger, A.; Ren, Y.; Kurtzberg, L. Characteristics of human Ewing/PNET sarcoma models. Ann. Saudi Med. 2011, 31, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Bukchin, A.; Pascual-Pasto, G.; Cuadrado-Vilanova, M.; Castillo-Ecija, H.; Monterrubio, C.; Olaciregui, N.G.; Vilà-Ubach, M.; Ordeix, L.; Mora, J.; Carcaboso, A.M.; et al. Glucosylated nanomicelles target glucose-avid pediatric patient-derived sarcomas. J. Control. Release 2018, 276, 59–71. [Google Scholar] [CrossRef]
- Puerto-Camacho, P.; Amaral, A.T.; Lamhamedi-Cherradi, S.-E.; Menegaz, B.A.; Castillo-Ecija, H.; Ordóñez, J.L.; Dominguez-Hormaetxe, S.; Jordan-Perez, C.; Diaz-Martin, J.; Romero-Pérez, L.; et al. Preclinical Efficacy of Endoglin-Targeting Antibody–Drug Conjugates for the Treatment of Ewing Sarcoma. Clin. Cancer Res. 2019, 25, 2228–2240. [Google Scholar] [CrossRef]
- Mitra, A.; Mishra, L.; Li, S. Technologies for deriving primary tumor cells for use in personalized cancer therapy. Trends Biotechnol. 2013, 31, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Salawu, A.; Fernando, M.; Hughes, D.; Reed, M.W.R.; Woll, P.; Greaves, C.; Day, C.; Alhajimohammed, M.; Sisley, K. Establishment and molecular characterisation of seven novel soft-tissue sarcoma cell lines. Br. J. Cancer 2016, 115, 1058–1068. [Google Scholar] [CrossRef]
- Ladanyi, M. EWS-FLI1 and Ewing’s sarcoma: Recent molecular data and new insights. Cancer Biol. Ther. 2002, 1, 329–335. [Google Scholar] [CrossRef]
- Stock, K.; Estrada, M.F.; Vidic, S.; Gjerde, K.; Rudisch, A.; Santo, V.E.; Barbier, M.; Blom, S.; Arundkar, S.C.; Selvam, I.; et al. Capturing tumor complexity in vitro: Comparative analysis of 2D and 3D tumor models for drug discovery. Sci. Rep. 2016, 6, 28951. [Google Scholar] [CrossRef]
- Vidic, S.; Estrada, M.F.; Gjerde, K.; Santo, V.E.; Osswald, A.; Barbier, M.; Chong, Y.T.; Sommergruber, W.; De Hoogt, R.; Brito, C.; et al. PREDECT Protocols for Complex 2D/3D Cultures. Adv. Struct. Saf. Stud. 2018, 1888, 1–20. [Google Scholar] [CrossRef]
- Koga, Y.; Ochiai, A. Systematic Review of Patient-Derived Xenograft Models for Preclinical Studies of Anti-Cancer Drugs in Solid Tumors. Cells 2019, 8, 418. [Google Scholar] [CrossRef] [PubMed]
- Workman, P.; An Ad Hoc Committee of the National Cancer Research Institute; Aboagye, E.O.; Balkwill, F.; Balmain, A.; Bruder, G.; Chaplin, D.J.; Double, J.A.; Everitt, J.; Farningham, D.A.H.; et al. Guidelines for the welfare and use of animals in cancer research. Br. J. Cancer 2010, 102, 1555–1577. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Seol, H.S.; Chang, S. The Generation and Application of Patient-Derived Xenograft Model for Cancer Research. Cancer Res. Treat. 2018, 50, 1–10. [Google Scholar] [CrossRef]
- Gaebler, M.; Silvestri, A.; Haybaeck, J.; Reichardt, P.; Lowery, C.D.; Stancato, L.F.; Zybarth, G.; Regenbrecht, C.R.A. Three-Dimensional Patient-Derived In Vitro Sarcoma Models: Promising Tools for Improving Clinical Tumor Management. Front. Oncol. 2017, 7, 203. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Shen, J.; Hornicek, F.; Duan, Z. Three-dimensional (3D) culture in sarcoma research and the clinical significance. Biofabrication 2017, 9, 032003. [Google Scholar] [CrossRef] [PubMed]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernández-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef]
- Tuveson, D.; Clevers, H. Cancer modeling meets human organoid technology. Science 2019, 364, 952–955. [Google Scholar] [CrossRef] [PubMed]
- Bruland, Ø.S.; Fodstad, Ø.; Pihl, A. The use of multicellular spheroids in establishing human sarcoma cell lines In vitro. Int. J. Cancer 1985, 35, 793–798. [Google Scholar] [CrossRef]
- Lawlor, E.R.; Scheel, C.; Irving, J.; Sorensen, P.H.B. Anchorage-independent multi-cellular spheroids as an in vitro model of growth signaling in Ewing tumors. Oncogene 2002, 21, 307–318. [Google Scholar] [CrossRef]
- Leuchte, K.; Altvater, B.; Hoffschlag, S.; Potratz, J.; Meltzer, J.; Clemens, D.; Luecke, A.; Hardes, J.; Dirksen, U.; Juergens, H.; et al. Anchorage-independent growth of Ewing sarcoma cells under serum-free conditions is not associated with stem-cell like phenotype and function. Oncol. Rep. 2014, 32, 845–852. [Google Scholar] [CrossRef]
- Estrada, M.F.; Rebelo, S.P.; Davies, E.J.; Pinto, M.T.; Pereira, H.; Santo, V.E.; Smalley, M.J.; Barry, S.T.; Gualda, E.J.; Alves, P.M.; et al. Modelling the tumour microenvironment in long-term microencapsulated 3D co-cultures recapitulates phenotypic features of disease progression. Biomaterials 2016, 78, 50–61. [Google Scholar] [CrossRef]
- Rebelo, S.P.; Pinto, C.; Martins, T.R.; Harrer, N.; Estrada, M.F.; Loza-Alvarez, P.; Cabeçadas, J.; Alves, P.M.; Gualda, E.J.; Sommergruber, W.; et al. 3D-3-culture: A tool to unveil macrophage plasticity in the tumour microenvironment. Biomaterials 2018, 163, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Dhamecha, D.; Movsas, R.; Sano, U.; Menon, J.U. Applications of alginate microspheres in therapeutics delivery and cell culture: Past, present and future. Int. J. Pharm. 2019, 569, 118627. [Google Scholar] [CrossRef]
- Domenici, G.; Aurrekoetxea-Rodríguez, I.; Simões, B.M.; Rábano, M.; Lee, S.Y.; Millán, J.S.; Comaills, V.; Oliemuller, E.; López-Ruiz, J.A.; Zabalza, I.; et al. A Sox2–Sox9 signalling axis maintains human breast luminal progenitor and breast cancer stem cells. Oncogene 2019, 38, 3151–3169. [Google Scholar] [CrossRef] [PubMed]
- Alcoser, S.Y.; Kimmel, D.J.; Borgel, S.D.; Carter, J.P.; Dougherty, K.M.; Hollingshead, M.G. Real-time PCR-based assay to quantify the relative amount of human and mouse tissue present in tumor xenografts. BMC Biotechnol. 2011, 11, 124. [Google Scholar] [CrossRef]
- Lin, M.-T.; Tseng, L.-H.; Kamiyama, H.; Kamiyama, M.; Lim, P.; Hidalgo, M.; Wheelan, S.; Eshleman, J. Quantifying the relative amount of mouse and human DNA in cancer xenografts using species-specific variation in gene length. Biotechniques 2010, 48, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Schneeberger, V.E.; Allaj, V.; Gardner, E.E.; Poirier, J.T.; Rudin, C.M. Quantitation of Murine Stroma and Selective Purification of the Human Tumor Component of Patient-Derived Xenografts for Genomic Analysis. PLoS ONE 2016, 11, e0160587. [Google Scholar] [CrossRef]
- Wykoff, C.C.; Beasley, N.J.; Watson, P.H.; Turner, K.J.; Pastorek, J.; Sibtain, A.; Wilson, G.D.; Turley, H.; Talks, K.L.; Maxwell, P.H.; et al. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res. 2000, 60, 7075–7083. [Google Scholar]
- García-Aragoncillo, E.; Carrillo, J.; Lalli, E.; Agra, N.; López, G.G.; Pestana, A.; Alonso, J. DAX1, a direct target of EWS/FLI1 oncoprotein, is a principal regulator of cell-cycle progression in Ewing’s tumor cells. Oncogene 2008, 27, 6034–6043. [Google Scholar] [CrossRef] [PubMed]
- Barpande, S.; Tupkari, J.; Deshingkar, S. Ewing’s sarcoma of zygoma. J. Oral Maxillofac. Pathol. 2009, 13, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Wiles, E.T.; Bell, R.; Thomas, D.; Beckerle, M.; Lessnick, S.L. ZEB2 Represses the Epithelial Phenotype and Facilitates Metastasis in Ewing Sarcoma. Genes Cancer 2013, 4, 486–500. [Google Scholar] [CrossRef] [PubMed]
- Marturano-Kruik, A.; Villasante, A.; Yaeger, K.; Ambati, S.R.; Chramiec, A.; Raimondi, M.; Vunjak-Novakovic, G. Biomechanical regulation of drug sensitivity in an engineered model of human tumor. Biomaterials 2018, 150, 150–161. [Google Scholar] [CrossRef]
- Hawkins, A.G.; Basrur, V.; LePrevost, F.D.V.; Pedersen, E.; Sperring, C.; Nesvizhskii, A.I.; Lawlor, E.R. The Ewing Sarcoma Secretome and Its Response to Activation of Wnt/beta-catenin Signaling. Mol. Cell. Proteom. 2018, 17, 901–912. [Google Scholar] [CrossRef] [PubMed]
- Scarpa, S.; Modesti, A.; Triche, T.J. Extracellular matrix synthesis by undifferentiated childhood tumor cell lines. Am. J. Pathol. 1987, 129, 74–85. [Google Scholar] [PubMed]
- Kuo, C.; Kent, P.M.; Logan, A.D.; Tamulonis, K.B.; Dalton, K.L.; Batus, M.; Fernandez, K.; McFall, R.E. Docetaxel, bevacizumab, and gemcitabine for very high risk sarcomas in adolescents and young adults: A single-center experience. Pediatr. Blood Cancer 2017, 64, e26265. [Google Scholar] [CrossRef]
- Manzanares, A.; Restrepo-Perdomo, C.A.; Botteri, G.; Castillo-Ecija, H.; Pascual-Pasto, G.; Cano, F.; Garcia-Alvarez, L.; Monterrubio, C.; Ruiz, B.; Vazquez-Carrera, M.; et al. Tissue Compatibility of SN-38-Loaded Anticancer Nanofiber Matrices. Adv. Health Mater. 2018, 7, 1800255. [Google Scholar] [CrossRef] [PubMed]
- Herzog, J.; Von Klot-Heydenfeldt, F.; Jabar, S.; Ranft, A.; Rossig, C.; Dirksen, U.; Brande, J.V.D.; D’Incalci, M.; Von Luettichau, I.; Grohar, P.J.; et al. Trabectedin Followed by Irinotecan Can Stabilize Disease in Advanced Translocation-Positive Sarcomas with Acceptable Toxicity. Sarcoma 2016, 2016, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Cosetti, M.; Wexler, L.H.; Calleja, E.; Trippett, T.; Laquaglia, M.; Huvos, A.G.; Gerald, W.; Healey, J.H.; Meyers, P.A.; Gorlick, R. Irinotecan for Pediatric Solid Tumors: The Memorial Sloan-Kettering Experience. J. Pediatr. Hematol. 2002, 24, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Casey, D.A.; Wexler, L.H.; Merchant, M.S.; Chou, A.J.; Merola, P.R.; Price, A.P.; Meyers, P.A. Irinotecan and temozolomide for Ewing sarcoma: The Memorial Sloan-Kettering experience. Pediatr. Blood Cancer 2009, 53, 1029–1034. [Google Scholar] [CrossRef]
- Yoon, J.H.; Kwon, M.M.; Park, H.J.; Park, S.Y.; Lim, K.Y.; Joo, J.; Park, B.-K. A study of docetaxel and irinotecan in children and young adults with recurrent or refractory Ewing sarcoma family of tumors. BMC Cancer 2014, 14, 1–9. [Google Scholar] [CrossRef]
- Castillo-Ecija, H.; Monterrubio, C.; Pascual-Pasto, G.; Gomez-Gonzalez, S.; Garcia-Dominguez, D.J.; Hontecillas-Prieto, L.; Resa-Pares, C.; Burgueño, V.; Paco, S.; Olaciregui, N.G.; et al. Treatment-driven selection of chemoresistant Ewing sarcoma tumors with limited drug distribution. J. Control. Release 2020, 324, 440–449. [Google Scholar] [CrossRef]
- Ueno, M.; Nonaka, S.; Yamazaki, R.; Deguchi, N.; Murai, M. SN-38 induces cell cycle arrest and apoptosis in human testicular cancer. Eur. Urol. 2002, 42, 390–397. [Google Scholar] [CrossRef]
- Pauli, C.; Hopkins, B.D.; Prandi, D.; Shaw, R.; Fedrizzi, T.; Sboner, A.; Sailer, V.; Augello, M.; Puca, L.; Rosati, R.; et al. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discov. 2017, 7, 462–477. [Google Scholar] [CrossRef]
- Colella, G.; Fazioli, F.; Gallo, M.; De Chiara, A.; Apice, G.; Ruosi, C.; Cimmino, A.; De Nigris, F. Sarcoma Spheroids and Organoids—Promising Tools in the Era of Personalized Medicine. Int. J. Mol. Sci. 2018, 19, 615. [Google Scholar] [CrossRef]
- Infanger, D.W.; Lynch, M.E.; Fischbach, C. Engineered Culture Models for Studies of Tumor-Microenvironment Interactions. Annu. Rev. Biomed. Eng. 2013, 15, 29–53. [Google Scholar] [CrossRef]
- Di Modugno, F.; Colosi, C.; Trono, P.; Antonacci, G.; Ruocco, G.; Nisticò, P. 3D models in the new era of immune oncology: Focus on T cells, CAF and ECM. J. Exp. Clin. Cancer Res. 2019, 38, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Fong, E.L.S.; Lamhamedi-Cherradi, S.-E.; Burdett, E.; Ramamoorthy, V.; Lazar, A.J.; Kasper, F.K.; Farach-Carson, M.C.; Vishwamitra, D.; Demicco, E.G.; Menegaz, B.A.; et al. Modeling Ewing sarcoma tumors in vitro with 3D scaffolds. Proc. Natl. Acad. Sci. USA 2013, 110, 6500–6505. [Google Scholar] [CrossRef]
- Santoro, M.; Lamhamedi-Cherradi, S.-E.; Menegaz, B.A.; Ludwig, J.A.; Mikos, A.G. Flow perfusion effects on three-dimensional culture and drug sensitivity of Ewing sarcoma. Proc. Natl. Acad. Sci. USA 2015, 112, 10304–10309. [Google Scholar] [CrossRef] [PubMed]
- Nanni, P.; Landuzzi, L.; Manara, M.C.; Righi, A.; Nicoletti, G.; Cristalli, C.; Pasello, M.; Parra, A.; Carrabotta, M.; Ferracin, M.; et al. Bone sarcoma patient-derived xenografts are faithful and stable preclinical models for molecular and therapeutic investigations. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Ordóñez, J.L.; Amaral, A.T.; Carcaboso, A.M.; Herrero-Martín, D.; García-Macías, M.D.C.; Sevillano, V.; Alonso, D.; Pascual-Pasto, G.; San-Segundo, L.; Vila-Ubach, M.; et al. The PARP inhibitor olaparib enhances the sensitivity of Ewing sarcoma to trabectedin. Oncotarget 2015, 6, 18875–18890. [Google Scholar] [CrossRef] [PubMed]
- Rokstad, A.M.; Donati, I.; Borgogna, M.; Oberholzer, J.; Strand, B.L.; Espevik, T.; Skjåk-Bræk, G. Cell-compatible covalently reinforced beads obtained from a chemoenzymatically engineered alginate. Biomaterials 2006, 27, 4726–4737. [Google Scholar] [CrossRef]
- García-Domínguez, D.J.; Hontecillas-Prieto, L.; Rodríguez-Núñez, P.; Pascual-Pasto, G.; Vila-Ubach, M.; García-Mejías, R.; Robles, M.J.; Tirado, O.M.; Mora, J.; Carcaboso, A.M.; et al. The combination of epigenetic drugs SAHA and HCI-2509 synergistically inhibits EWS-FLI1 and tumor growth in Ewing sarcoma. Oncotarget 2018, 9, 31397–31410. [Google Scholar] [CrossRef]
- Mendiola, M.; Carrillo, J.; García, E.; Lalli, E.; Hernández, T.; De Alava, E.; Tirode, F.; Delattre, O.; García-Miguel, P.; Lopez Barea, F.; et al. The orphan nuclear receptor DAX1 is up-regulated by the EWS/FLI1 oncoprotein and is highly expressed in Ewing tumors. Int. J. Cancer 2005, 118, 1381–1389. [Google Scholar] [CrossRef]
- Erkizan, H.V.; Uversky, V.N.; Toretsky, J.A. Oncogenic Partnerships: EWS-FLI1 Protein Interactions Initiate Key Pathways of Ewing’s Sarcoma. Clin. Cancer Res. 2010, 16, 4077–4083. [Google Scholar] [CrossRef] [PubMed]
- Akamine, R.; Yamamoto, T.; Watanabe, M.; Yamazaki, N.; Kataoka, M.; Ishikawa, M.; Ooie, T.; Baba, Y.; Shinohara, Y. Usefulness of the 5′ region of the cDNA encoding acidic ribosomal phosphoprotein P0 conserved among rats, mice, and humans as a standard probe for gene expression analysis in different tissues and animal species. J. Biochem. Biophys. Methods 2007, 70, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Brohem, C.A.; De Carvalho, C.M.; Radoski, C.L.; Santi, F.C.; Baptista, M.C.; Swinka, B.B.; Urban, C.D.A.; De Araujo, L.R.R.; Graf, R.M.; Feferman, I.H.S.; et al. Comparison between fibroblasts and mesenchymal stem cells derived from dermal and adipose tissue. Int. J. Cosmet. Sci. 2013, 35, 448–457. [Google Scholar] [CrossRef] [PubMed]
- Vlodavsky, I.; Lui, G.M.; Gospodarowicz, D. Morphological appearance, growth behavior and migratory activity of human tumor cells maintained on extracellular matrix versus plastic. Cell 1980, 19, 607–616. [Google Scholar] [CrossRef]
- Riffle, S.; Hegde, R.S. Modeling tumor cell adaptations to hypoxia in multicellular tumor spheroids. J. Exp. Clin. Cancer Res. 2017, 36, 1–10. [Google Scholar] [CrossRef]
- Giovagnoli, S.; Luca, G.; Blasi, P.; Mancuso, F.; Schoubben, A.; Arato, I.; Calvitti, M.; Falabella, G.; Basta, G.; Bodo, M.; et al. Alginates in Pharmaceutics and Biomedicine: Is the Future so Bright? Curr. Pharm. Des. 2015, 21, 4917–4935. [Google Scholar] [CrossRef]
- Aryee, D.N.; Niedan, S.; Kauer, M.; Schwentner, R.; Bennani-Baiti, I.M.; Ban, J.; Muehlbacher, K.; Kreppel, M.; Walker, R.L.; Meltzer, P.; et al. Hypoxia Modulates EWS-FLI1 Transcriptional Signature and Enhances the Malignant Properties of Ewing’s Sarcoma Cells In vitro. Cancer Res. 2010, 70, 4015–4023. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, W.; Li, S.; Tu, C. Prognosis value of Hypoxia-inducible factor-1α expression in patients with bone and soft tissue sarcoma: A meta-analysis. SpringerPlus 2016, 5, 1–10. [Google Scholar] [CrossRef]
- Eehnman, M.; Elarsson, O. Microenvironmental Targets in Sarcoma. Front. Oncol. 2015, 5, 248. [Google Scholar] [CrossRef]
- Bai, C.; Yang, M.; Fan, Z.; Li, S.; Gao, T.; Fang, Z. Associations of chemo- and radio-resistant phenotypes with the gap junction, adhesion and extracellular matrix in a three-dimensional culture model of soft sarcoma. J. Exp. Clin. Cancer Res. 2015, 34, 1–10. [Google Scholar] [CrossRef]
- Junttila, M.R.; De Sauvage, F.J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nat. Cell Biol. 2013, 501, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, H.; Murakami, T.; Tsuchida, K.; Sugino, H.; Miyake, H.; Tashiro, S. Tumor-Stroma Interaction of Human Pancreatic Cancer: Acquired Resistance to Anticancer Drugs and Proliferation Regulation Is Dependent on Extracellular Matrix Proteins. Pancreas 2004, 28, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Berghuis, D.; Santos, S.J.; Baelde, H.J.; Taminiau, A.H.; Egeler, R.M.; Schilham, M.W.; Hogendoorn, P.C.; Lankester, A.C. Pro-inflammatory chemokine-chemokine receptor interactions within the Ewing sarcoma microenvironment determine CD8+ T-lymphocyte infiltration and affect tumour progression. J. Pathol. 2010, 223, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Fukushi, J.-I.; Yamamoto, S.; Matsumoto, Y.; Setsu, N.; Oda, Y.; Yamada, H.; Okada, S.; Watari, K.; Ono, M.; et al. Macrophage Infiltration Predicts a Poor Prognosis for Human Ewing Sarcoma. Am. J. Pathol. 2011, 179, 1157–1170. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Domenici, G.; Eduardo, R.; Castillo-Ecija, H.; Orive, G.; Montero Carcaboso, Á.; Brito, C. PDX-Derived Ewing’s Sarcoma Cells Retain High Viability and Disease Phenotype in Alginate Encapsulated Spheroid Cultures. Cancers 2021, 13, 879. https://doi.org/10.3390/cancers13040879
Domenici G, Eduardo R, Castillo-Ecija H, Orive G, Montero Carcaboso Á, Brito C. PDX-Derived Ewing’s Sarcoma Cells Retain High Viability and Disease Phenotype in Alginate Encapsulated Spheroid Cultures. Cancers. 2021; 13(4):879. https://doi.org/10.3390/cancers13040879
Chicago/Turabian StyleDomenici, Giacomo, Rodrigo Eduardo, Helena Castillo-Ecija, Gorka Orive, Ángel Montero Carcaboso, and Catarina Brito. 2021. "PDX-Derived Ewing’s Sarcoma Cells Retain High Viability and Disease Phenotype in Alginate Encapsulated Spheroid Cultures" Cancers 13, no. 4: 879. https://doi.org/10.3390/cancers13040879
APA StyleDomenici, G., Eduardo, R., Castillo-Ecija, H., Orive, G., Montero Carcaboso, Á., & Brito, C. (2021). PDX-Derived Ewing’s Sarcoma Cells Retain High Viability and Disease Phenotype in Alginate Encapsulated Spheroid Cultures. Cancers, 13(4), 879. https://doi.org/10.3390/cancers13040879








