Prognostic Significance of Gene Expression and DNA Methylation Markers in Circulating Tumor Cells and Paired Plasma Derived Exosomes in Metastatic Castration Resistant Prostate Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
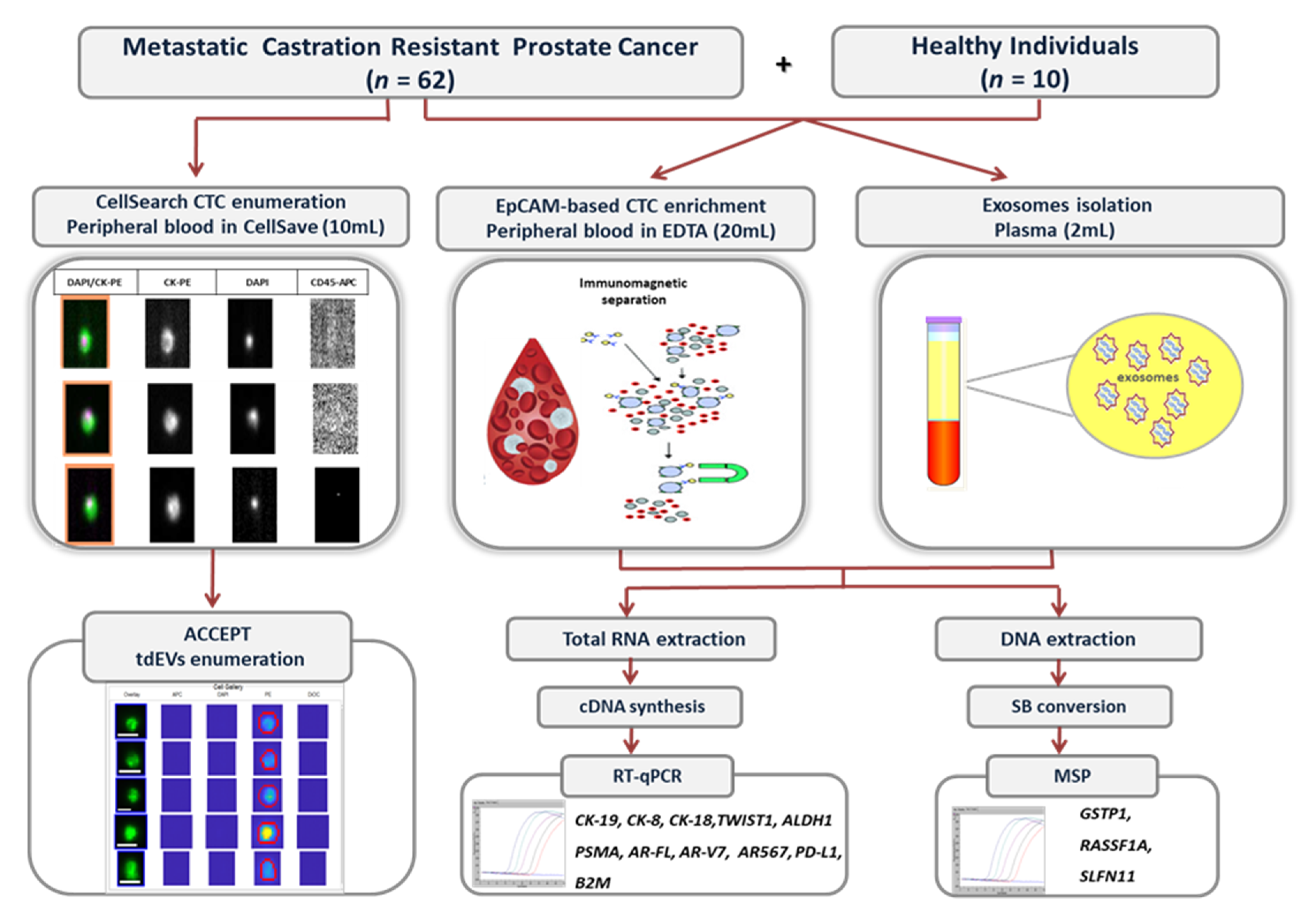
2. Materials and Methods
2.1. Peripheral Blood Samples Collection
2.2. Isolation of EpCAM-Positive CTCs and Exosomes
2.3. RNA Extraction and cDNA Synthesis
2.4. RT-qPCR
2.5. DNA Isolation
2.6. Sodium Bisulfite (SB) Treatment
2.7. Real-Time Methylation Specific PCR (Real-Time MSP)
2.8. CTCs and Tumor-Derived EVs (tdEVs) Enumeration
2.9. Statistical Analysis
3. Results
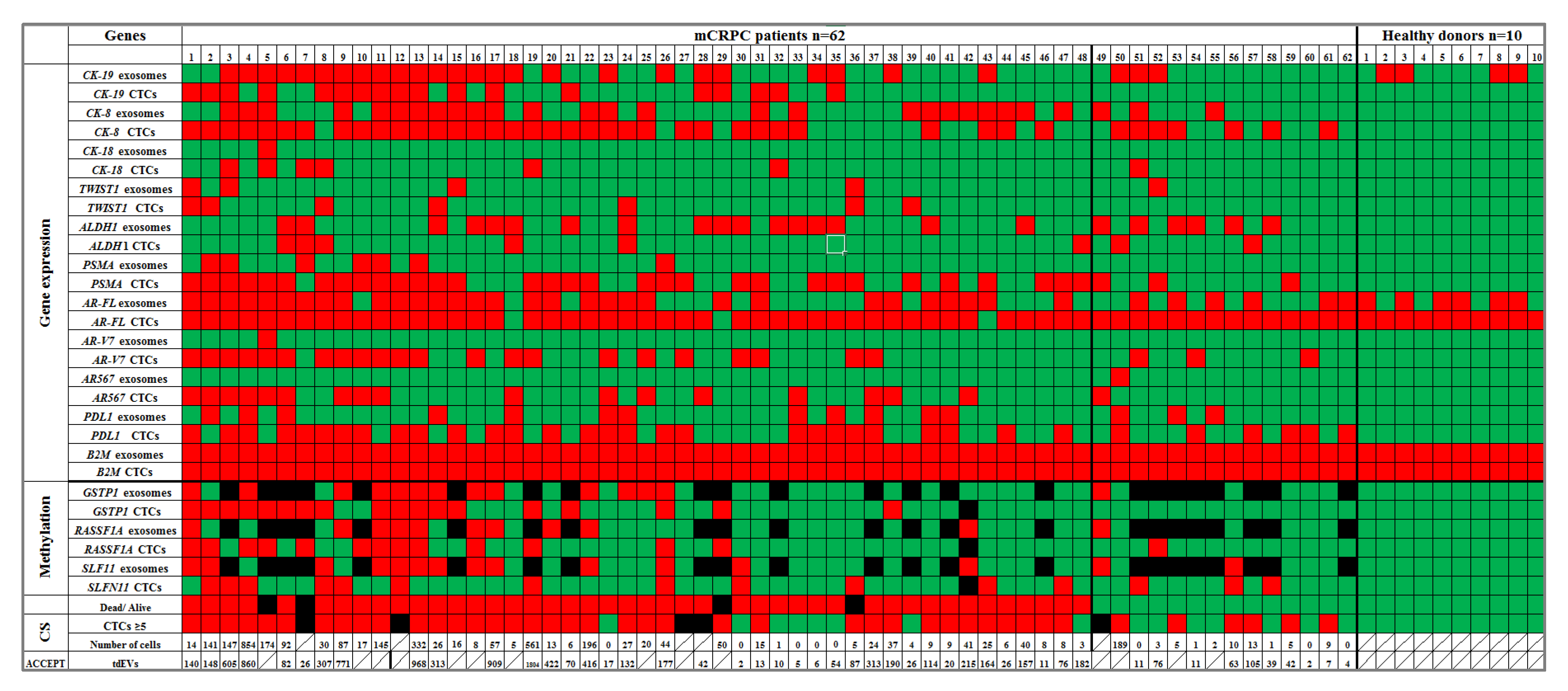
3.1. Gene Expression Analysis
3.2. DNA Methylation
3.3. Direct Comparison of Gene Expression Markers between Plasma-Derived Exosomes and Paired EpCAM-Positive CTCs
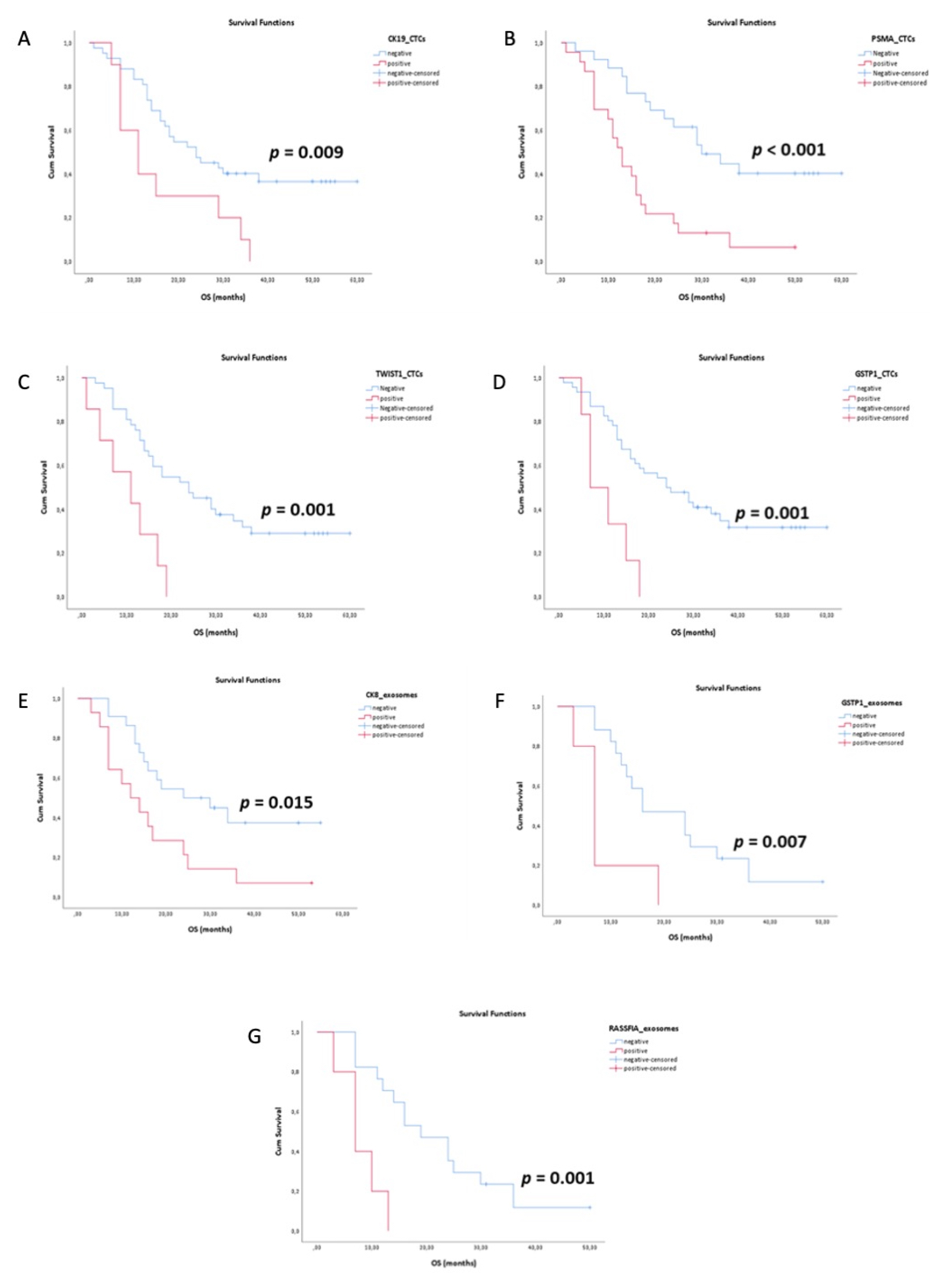
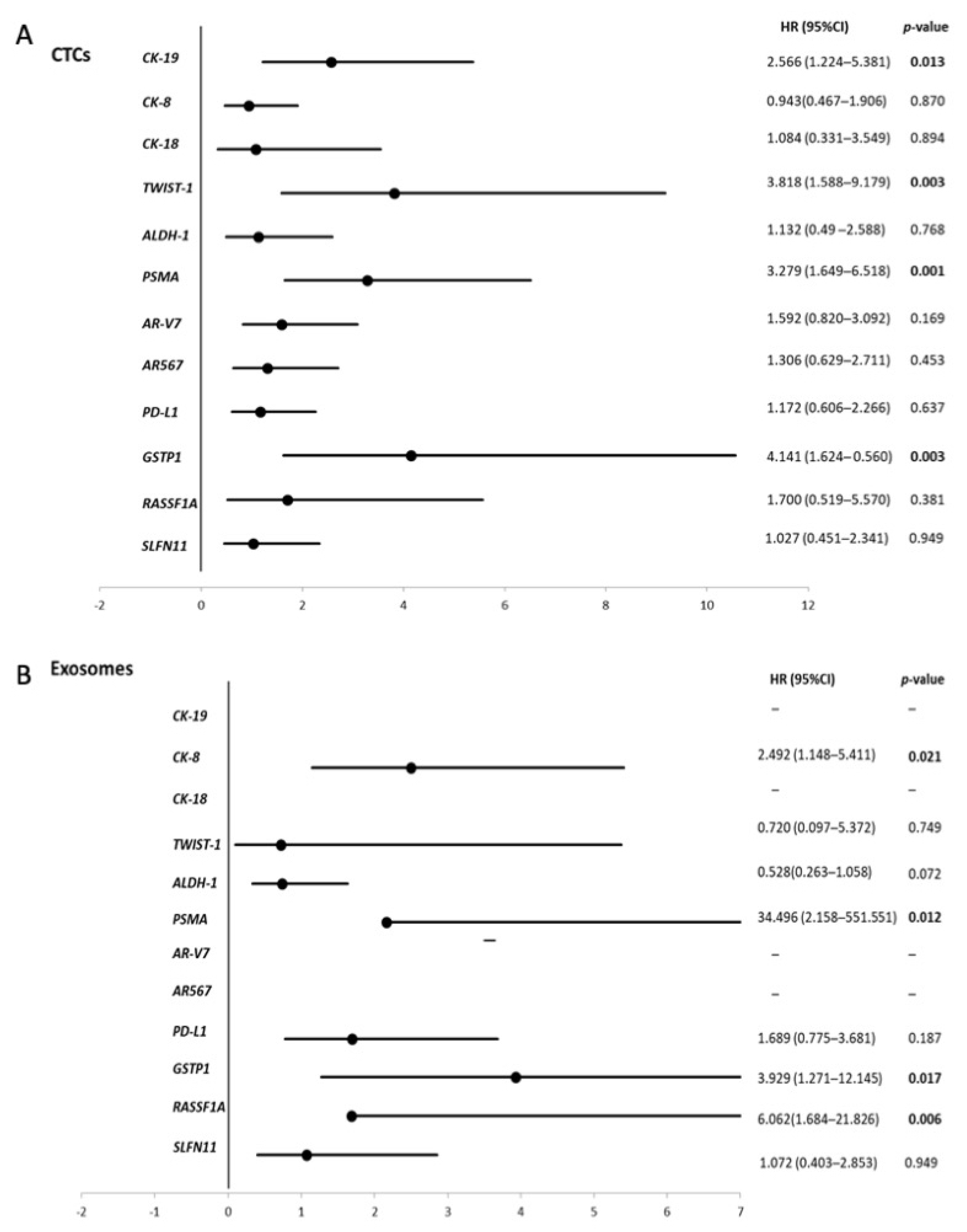
3.4. Survival Analysis
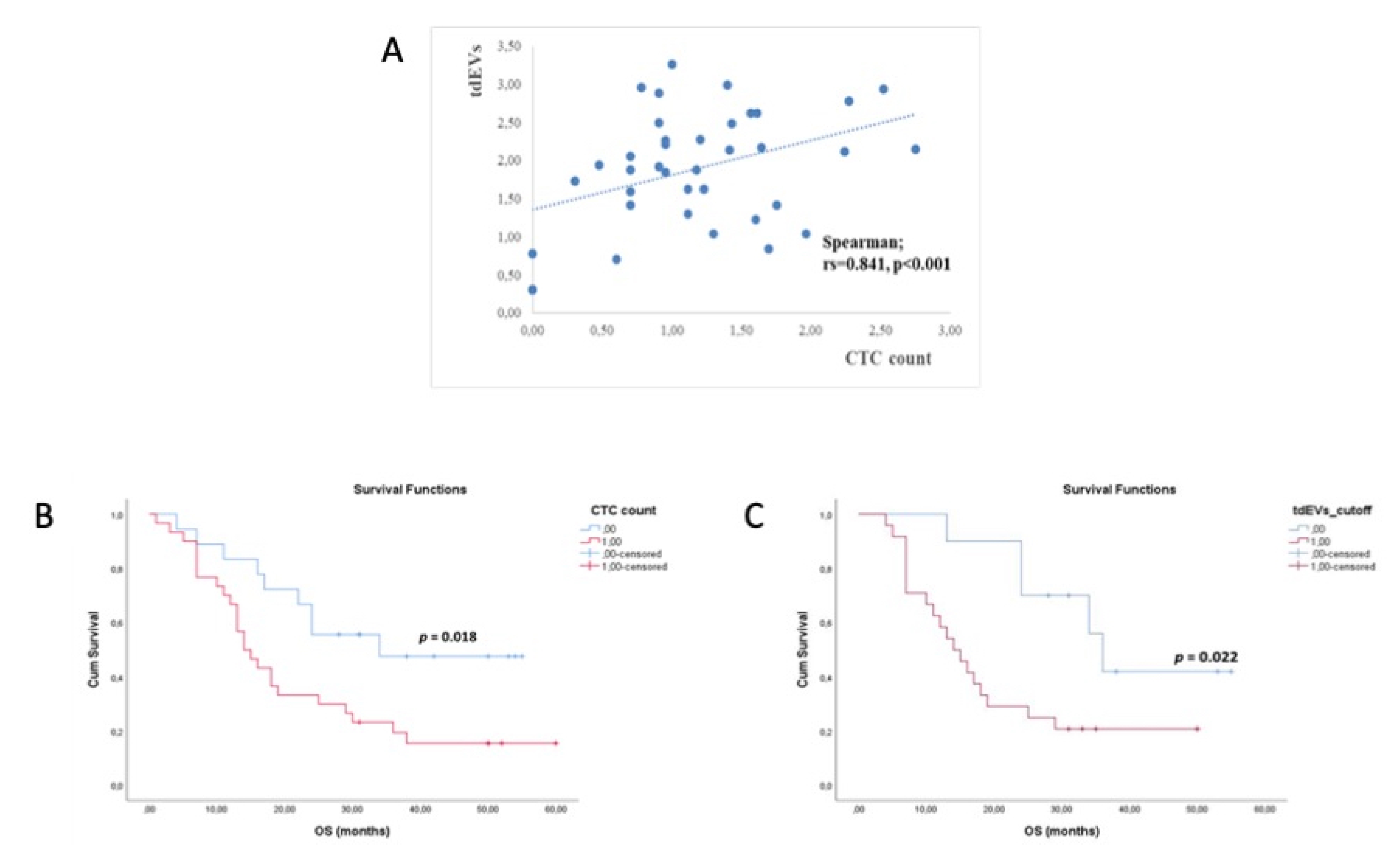
3.5. CTCs and tdEVs Enumeration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sathianathen, N.J.; Konety, B.R.; Crook, J.; Saad, F.; Lawrentschuk, N. Landmarks in prostate cancer. Nat. Rev. Urol. 2018, 15, 627–642. [Google Scholar] [CrossRef]
- Pantel, K.; Hille, C.; Scher, H.I. Circulating Tumor Cells in Prostate Cancer: From Discovery to Clinical Utility. Clin. Chem. 2019, 65, 87–99. [Google Scholar] [CrossRef] [Green Version]
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2019, 20, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Lianidou, E.; Hoon, D. Circulating Tumor Cells and circulating Tumor DNA. In Tietz Textbook of Clinical Chemistry and Molecular Diagnostics, 6th ed.; Rifai, N., Horvath, A.-R., Wittwer, C., Eds.; Elsevier Ltd.: Amsterdam, The Netherland, 2017; pp. 1111–1144. [Google Scholar]
- Lianidou, E.; Pantel, K. Liquid biopsies. Genes Chromosom Cancer 2019, 58, 219–232. [Google Scholar] [CrossRef] [PubMed]
- van der Toom, E.E.; Axelrod, H.D.; de la Rosette, J.J.; de Reijke, T.M.; Pienta, K.J.; Valkenburg, K.C. Prostate-specific markers to identify rare prostate cancer cells in liquid biopsies. Nat. Rev. Urol. 2019, 16, 7–22. [Google Scholar] [CrossRef]
- Markou, A.; Lazaridou, M.; Paraskevopoulos, P.; Chen, S.; Świerczewska, M.; Budna, J.; Kuske, A.; Tobias, M.G.; Simon, A.J.; Thomas, K.; et al. Multiplex Gene Expression Profiling of In Vivo Isolated Circulating Tumor Cells in High-Risk Prostate Cancer Patients. Clin. Chem. 2018, 64, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Dagogo-Jack, I.; Shaw, A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2017, 15, 81–94. [Google Scholar] [CrossRef]
- Brabletz, T.; Kalluri, R.; Nieto, M.A.; Weinberg, R.A. EMT in cancer. Nat. Rev. Cancer 2018, 18, 128–134. [Google Scholar] [CrossRef] [PubMed]
- de Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.; Pienta, K.J.; Raghavan, D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef] [Green Version]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Nanou, A.; Coumans, F.A.W.; van Dalum, G.; Zeune, L.L.; Dolling, D.; Onstenk, W.; Crespo, M.; Fontes, M.S.; Rescigno, P.; Fowler, G.; et al. Circulating tumor cells, tumor-derived extracellular vesicles and plasma cytokeratins in castration-resistant prostate cancer patients. Oncotarget 2018, 9, 19283–19293. [Google Scholar] [CrossRef] [Green Version]
- Keup, C.; Mach, P.; Aktas, B.; Tewes, M.; Kolberg, H.-C.; Hauch, S.; Sprenger-Haussels, M.; Kimmig, R.; Kasimir-Bauer, S. RNA Profiles of Circulating Tumor Cells and Extracellular Vesicles for Therapy Stratification of Metastatic Breast Cancer Patients. Clin. Chem. 2018, 64, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Antonarakis, E.S.; Lu, C.; Wang, H.; Luber, B.; Nakazawa, M.; Roeser, J.C.; Chen, Y.; Mohammad, T.A.; Chen, Y.; Fedor, H.L.; et al. AR-V7 and Resistance to Enzalutamide and Abiraterone in Prostate Cancer. N. Engl. J. Med. 2014, 371, 1028–1038. [Google Scholar] [CrossRef] [Green Version]
- Antonarakis, E.S.; Lu, C.; Luber, B.; Wang, H.; Chen, Y.; Nakazawa, M.; Nadal, R.; Paller, C.J.; Denmeade, S.R.; Carducci, M.A.; et al. Androgen Receptor Splice Variant 7 and Efficacy of Taxane Chemotherapy in Patients With Metastatic Castration-Resistant Prostate Cancer. JAMA Oncol. 2015, 1, 582. [Google Scholar] [CrossRef] [Green Version]
- Lianidou, E.S. Gene expression profiling and DNA methylation analyses of CTCs. Mol. Oncol. 2016, 10, 431–442. [Google Scholar] [CrossRef]
- Zavridou, M.; Mastoraki, S.; Strati, A.; Tzanikou, E.; Chimonidou, M.; Lianidou, E. Evaluation of Preanalytical Conditions and Implementation of Quality Control Steps for Reliable Gene Expression and DNA Methylation Analyses in Liquid Biopsies. Clin. Chem. 2018, 64, 1522–1533. [Google Scholar] [CrossRef]
- Zavridou, M.; Mastoraki, S.; Strati, A.; Koutsodontis, G.; Klinakis, A.; Psyrri, A.; Lianidou, E. Direct comparison of size-dependent versus EpCAM-dependent CTC enrichment at the gene expression and DNA methylation level in head and neck squamous cell carcinoma. Sci. Rep. 2020, 10, 6551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strati, A.; Zavridou, M.; Bournakis, E.; Mastoraki, S.; Lianidou, E. Expression pattern of androgen receptors,: Ar-v7 and ar-567es, in circulating tumor cells and paired plasma-derived extracellular vesicles in metastatic castration resistant prostate cancer. Analyst 2019, 144, 6671–6680. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chimonidou, M.; Strati, A.; Tzitzira, A.; Sotiropoulou, G.; Malamos, N.; Georgoulias, V.; Lianidou, E. DNA methylation of tumor suppressor and metastasis suppressor genes in circulating tumor cells. Clin. Chem. 2011, 57, 1169–1177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giannopoulou, L.; Chebouti, I.; Pavlakis, K.; Kasimir-Bauer, S.; Lianidou, E.S. RASSF1A promoter methylation in high-grade serous ovarian cancer: A direct comparison study in primary tumors, adjacent morphologically tumor cell-free tissues and paired circulating tumor DNA. Oncotarget 2017, 8, 21429–21443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanou, A.; Miller, M.C.; Zeune, L.L.; de Wit, S.; Punt, C.J.A.; Groen, H.J.M.; Hayes, D.F.; de Bono, J.S.; Terstappen, L.W.M.M. Tumour-derived extracellular vesicles in blood of metastatic cancer patients associate with overall survival. Br. J. Cancer 2020, 122, 801–811. [Google Scholar] [CrossRef] [Green Version]
- Wu, A.; Attard, G. Plasma DNA Analysis in Prostate Cancer: Opportunities for Improving Clinical Management. Clin. Chem. 2019, 65, 100–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kilgour, E.; Rothwell, D.G.; Brady, G.; Dive, C. Liquid Biopsy-Based Biomarkers of Treatment Response and Resistance. Cancer Cell 2020, 37, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Gorges, T.M.; Kuske, A.; Röck, K.; Mauermann, O.; Müller, V.; Peine, S.; Verpoort, K.; Novosadova, V.; Kubista, M.; Riethdorf, S.; et al. Accession of Tumor Heterogeneity by Multiplex Transcriptome Profiling of Single Circulating Tumor Cells. Clin. Chem. 2016, 62, 1504–1515. [Google Scholar] [CrossRef]
- Gorges, T.M.; Riethdorf, S.; von Ahsen, O.; Nastaly, P.; Röck, K.; Boede, M.; Peine, S.; Kuske, A.; Schmid, E.; Kneip, C.; et al. Heterogeneous PSMA expression on circulating tumor cells—A potential basis for stratification and monitoring of PSMA-directed therapies in prostate cancer. Oncotarget 2016, 7, 34930–34941. [Google Scholar] [CrossRef] [Green Version]
- Kessel, K.; Seifert, R.; Weckesser, M.; Roll, W.; Humberg, V.; Schlack, K.; Bögemann, M.; Bernemann, C.; Rahbar, K. Molecular analysis of circulating tumor cells of metastatic castration-resistant Prostate Cancer Patients receiving 177Lu-PSMA-617 Radioligand Therapy. Theranostics 2020, 10, 7645–7655. [Google Scholar] [CrossRef]
- Paller, C.J.; Piana, D.; Eshleman, J.R.; Riel, S.; Denmeade, S.R.; Isaacsson Velho, P.; Rowe, S.P.; Pomper, M.G.; Antonarakis, E.S.; Luo, J.; et al. A pilot study of prostate-specific membrane antigen (PSMA) dynamics in men undergoing treatment for advanced prostate cancer. Prostate 2019, 79, 1597–1603. [Google Scholar] [CrossRef]
- Scher, H.I.; Lu, D.; Schreiber, N.A.; Louw, J.; Graf, R.P.; Vargas, H.A.; Johnson, A.; Jendrisak, A.; Bambury, R.; Danila, D.; et al. Association of AR-V7 on Circulating Tumor Cells as a Treatment-Specific Biomarker With Outcomes and Survival in Castration-Resistant Prostate Cancer. JAMA Oncol. 2016, 2, 1441. [Google Scholar] [CrossRef] [PubMed]
- Scher, H.I.; Graf, R.P.; Schreiber, N.A.; Jayaram, A.; Winquist, E.; McLaughlin, B.; Lu, D.; Fleisher, M.; Orr, S.; Lowes, L.; et al. Assessment of the Validity of Nuclear-Localized Androgen Receptor Splice Variant 7 in Circulating Tumor Cells as a Predictive Biomarker for Castration-Resistant Prostate Cancer. JAMA Oncol. 2018, 4, 1179–1186. [Google Scholar] [CrossRef] [PubMed]
- Lokhandwala, P.M.; Riel, S.L.; Haley, L.; Lu, C.; Chen, Y.; Silberstein, J.; Zhu, Y.; Zheng, G.; Lin, M.T.; Gocke, C.D.; et al. Analytical Validation of Androgen Receptor Splice Variant 7 Detection in a Clinical Laboratory Improvement Amendments (CLIA) Laboratory Setting. J. Mol. Diagn. 2017, 19, 115–125. [Google Scholar] [CrossRef] [Green Version]
- El-Heliebi, A.; Hille, C.; Laxman, N.; Svedlund, J.; Haudum, C.; Ercan, E.; Kroneis, T.; Chen, S.; Smolle, M.; Rossmann, C.; et al. In Situ Detection and Quantification of AR-V7, AR-FL, PSA, andKRASPoint Mutations in Circulating Tumor Cells. Clin. Chem. 2018, 64, 536–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharp, A.; Welti, J.C.; Lambros, M.B.K.; Dolling, D.; Rodrigues, D.N.; Pope, L.; Aversa, C.; Figueiredo, I.; Fraser, J.; Ahmad, Z.; et al. Clinical Utility of Circulating Tumour Cell Androgen Receptor Splice Variant-7 Status in Metastatic Castration-resistant Prostate Cancer. Eur. Urol. 2019, 76, 676–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nimir, M.; Ma, Y.; Jeffreys, S.A.; Opperman, T.; Young, F.; Khan, T.; Ding, P.; Chua, W.; Balakrishnar, B.; Cooper, A.; et al. Detection of AR-V7 in Liquid Biopsies of Castrate Resistant Prostate Cancer Patients: A Comparison of AR-V7 Analysis in Circulating Tumor Cells, Circulating Tumor RNA and Exosomes. Cells 2019, 8, 688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strati, A.; Koutsodontis, G.; Papaxoinis, G.; Angelidis, I.; Zavridou, M.; Economopoulou, P.; Kotsantis, I.; Avgeris, M.; Mazel, M.; Perisanidis, C.; et al. Prognostic significance of PD-L1 expression on circulating tumor cells in patients with head and neck squamous cell carcinoma. Ann. Oncol. 2017, 28, 1923–1933. [Google Scholar] [CrossRef]
- Antonarakis, E.S.; Piulats, J.M.; Gross-Goupil, M.; Goh, J.; Ojamaa, K.; Hoimes, C.J.; Vaishampayan, U.; Berger, R.; Sezer, A.; Alanko, T.; et al. Pembrolizumab for treatment-refractory metastatic castration-resistant prostate cancer: Multicohort, open-label phase II KEYNOTE-199 study. J. Clin. Oncol. 2020, 38, 395–405. [Google Scholar] [CrossRef]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef]
- Zoppoli, G.; Regairaz, M.; Leo, E.; Reinhold, W.C.; Varma, S.; Ballestrero, A.; Doroshow, J.H.; Pommier, Y. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc. Natl. Acad. Sci. USA 2012, 109, 15030–15035. [Google Scholar] [CrossRef] [Green Version]
- Mattox, A.K.; Bettegowda, C.; Zhou, S.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Applications of liquid biopsies for cancer. Sci. Transl. Med. 2019, 11, eaay1984. [Google Scholar] [CrossRef] [Green Version]
- Conteduca, V.; Ku, S.Y.; Puca, L.; Slade, M.; Fernandez, L.; Hess, J.; Bareja, R.; Vlachostergios, P.J.; Sigouros, M.; Mosquera, J.M.; et al. SLFN11 Expression in Advanced Prostate Cancer and Response to Platinum-based Chemotherapy. Mol. Cancer Ther. 2020, 19, 1157–1164. [Google Scholar] [CrossRef] [Green Version]
- Nogales, V.; Reinhold, W.C.; Varma, S.; Martinez-Cardus, A.; Moutinho, C.; Moran, S.; Heyn, H.; Sebio, A.; Barnadas, A.; Pommier, Y.; et al. Epigenetic inactivation of the putative DNA/RNA helicase SLFN11 in human cancer confers resistance to platinum drugs. Oncotarget 2016, 7, 3084–3097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markowski, M.C.; Silberstein, J.L.; Eshleman, J.R.; Eisenberger, M.A.; Luo, J.; Antonarakis, E.S. Clinical Utility of CLIA-Grade AR-V7 Testing in Patients With Metastatic Castration-Resistant Prostate Cancer. JCO Precis. Oncol. 2017, 1, PO.17.00127. [Google Scholar]
| Covariates | Covariates Value | Multivariate Cox Regression Analysis | ||
|---|---|---|---|---|
| HR a | 95%CI b | p-Value | ||
| CTC count | ≥5 vs. <5 | 2.202 | 1.013–4.786 | 0.046 |
| Gleason score | >8 vs. ≤8 | 1.230 | 0.616–2.456 | 0.558 |
| CK19 | Yes vs. No | 2.609 | 1.176–5.790 | 0.018 |
| CTC count | ≥5 vs. <5 | 1.957 | 0.885–4.324 | 0.097 |
| Gleason score | >8 vs. ≤8 | 1.025 | 0.500–2.101 | 0.946 |
| PSMA | Yes vs. No | 3.242 | 1.536–6.846 | 0.002 |
| CTC count | ≥5 vs. <5 | 2.244 | 1.024–4.921 | 0.044 |
| Gleason score | >8 vs. ≤8 | 1.610 | 0.774–3.350 | 0.202 |
| TWIST1 | Yes vs. No | 4.421 | 1.744–11.209 | 0.002 |
| CTC count | ≥5 vs. <5 | 2.024 | 0.906–4.518 | 0.085 |
| Gleason score | >8 vs. ≤8 | 1.275 | 0.641–2.539 | 0.489 |
| GSTP1 | Yes vs. No | 3.111 | 1.163–8.321 | 0.024 |
| CTC count | ≥5 vs. <5 | 1.937 | 0.834–4.496 | 0.124 |
| Gleason score | >8 vs. ≤8 | 1.234 | 0.572–2.663 | 0.592 |
| CK19 | Yes vs. No | 1.418 | 0.553–3.634 | 0.467 |
| PSMA | Yes vs. No | 2.669 | 1.152–6.186 | 0.022 |
| TWIST1 | Yes vs. No | 3.722 | 1.412–9.812 | 0.008 |
| GSTP1 | Yes vs. No | 1.277 | 0.402–4.050 | 0.679 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zavridou, M.; Strati, A.; Bournakis, E.; Smilkou, S.; Tserpeli, V.; Lianidou, E. Prognostic Significance of Gene Expression and DNA Methylation Markers in Circulating Tumor Cells and Paired Plasma Derived Exosomes in Metastatic Castration Resistant Prostate Cancer. Cancers 2021, 13, 780. https://doi.org/10.3390/cancers13040780
Zavridou M, Strati A, Bournakis E, Smilkou S, Tserpeli V, Lianidou E. Prognostic Significance of Gene Expression and DNA Methylation Markers in Circulating Tumor Cells and Paired Plasma Derived Exosomes in Metastatic Castration Resistant Prostate Cancer. Cancers. 2021; 13(4):780. https://doi.org/10.3390/cancers13040780
Chicago/Turabian StyleZavridou, Martha, Areti Strati, Evangelos Bournakis, Stavroula Smilkou, Victoria Tserpeli, and Evi Lianidou. 2021. "Prognostic Significance of Gene Expression and DNA Methylation Markers in Circulating Tumor Cells and Paired Plasma Derived Exosomes in Metastatic Castration Resistant Prostate Cancer" Cancers 13, no. 4: 780. https://doi.org/10.3390/cancers13040780
APA StyleZavridou, M., Strati, A., Bournakis, E., Smilkou, S., Tserpeli, V., & Lianidou, E. (2021). Prognostic Significance of Gene Expression and DNA Methylation Markers in Circulating Tumor Cells and Paired Plasma Derived Exosomes in Metastatic Castration Resistant Prostate Cancer. Cancers, 13(4), 780. https://doi.org/10.3390/cancers13040780








