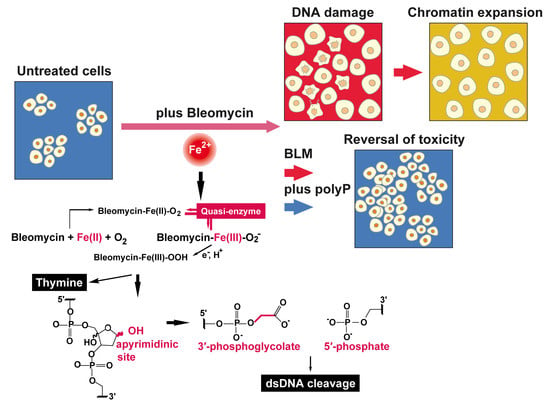
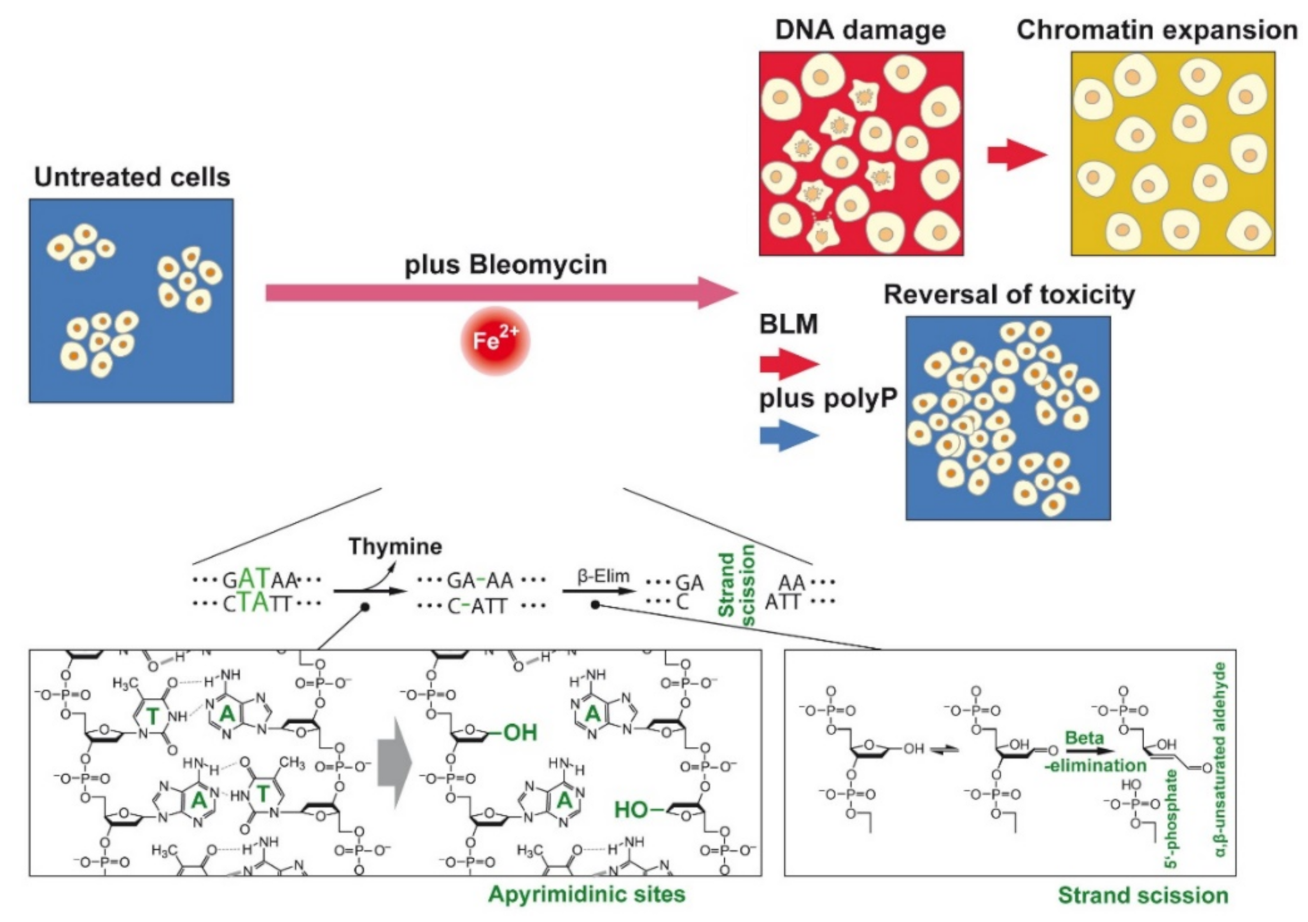
Polyphosphate Reverses the Toxicity of the Quasi-Enzyme Bleomycin on Alveolar Endothelial Lung Cells In Vitro †
Abstract
:Simple Summary
Abstract
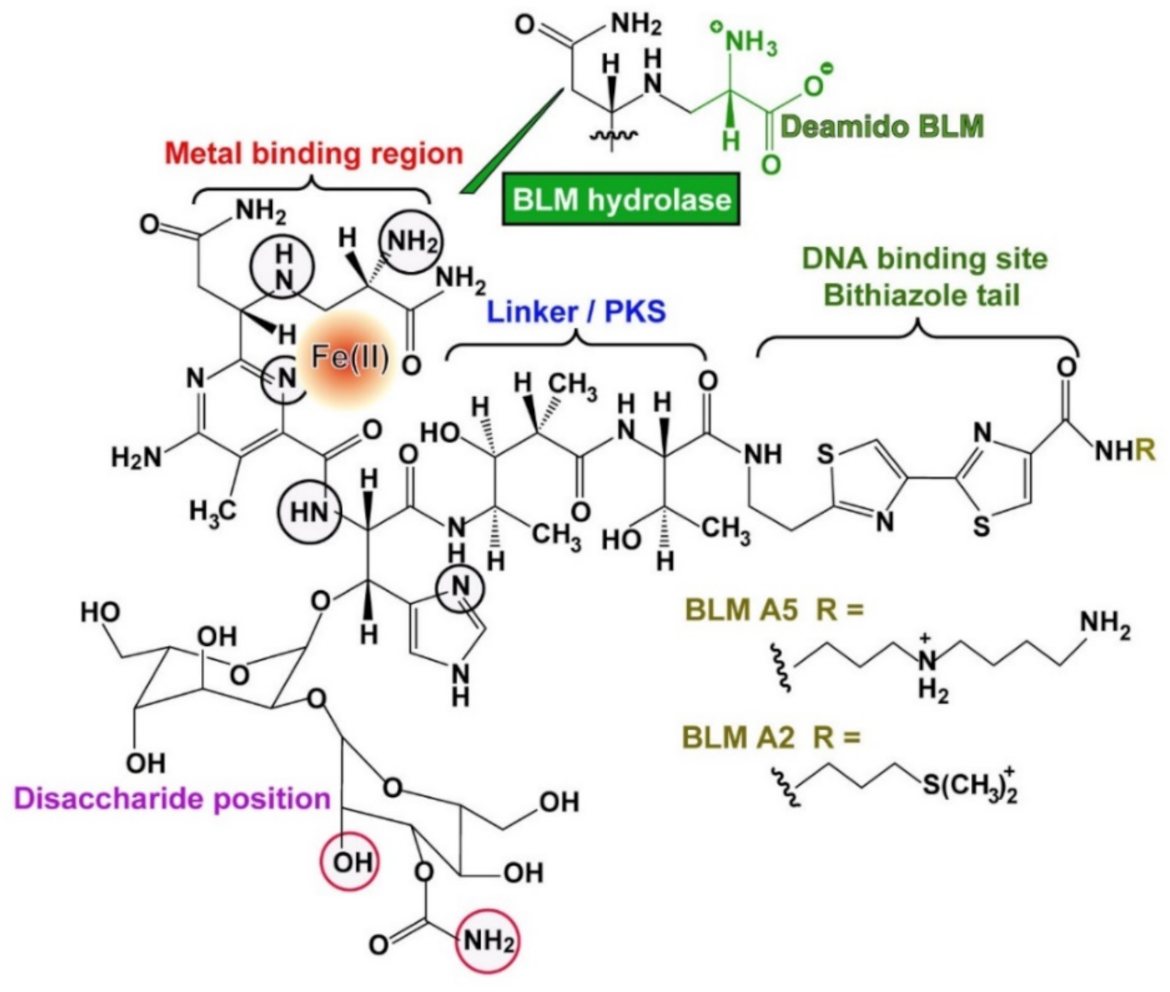
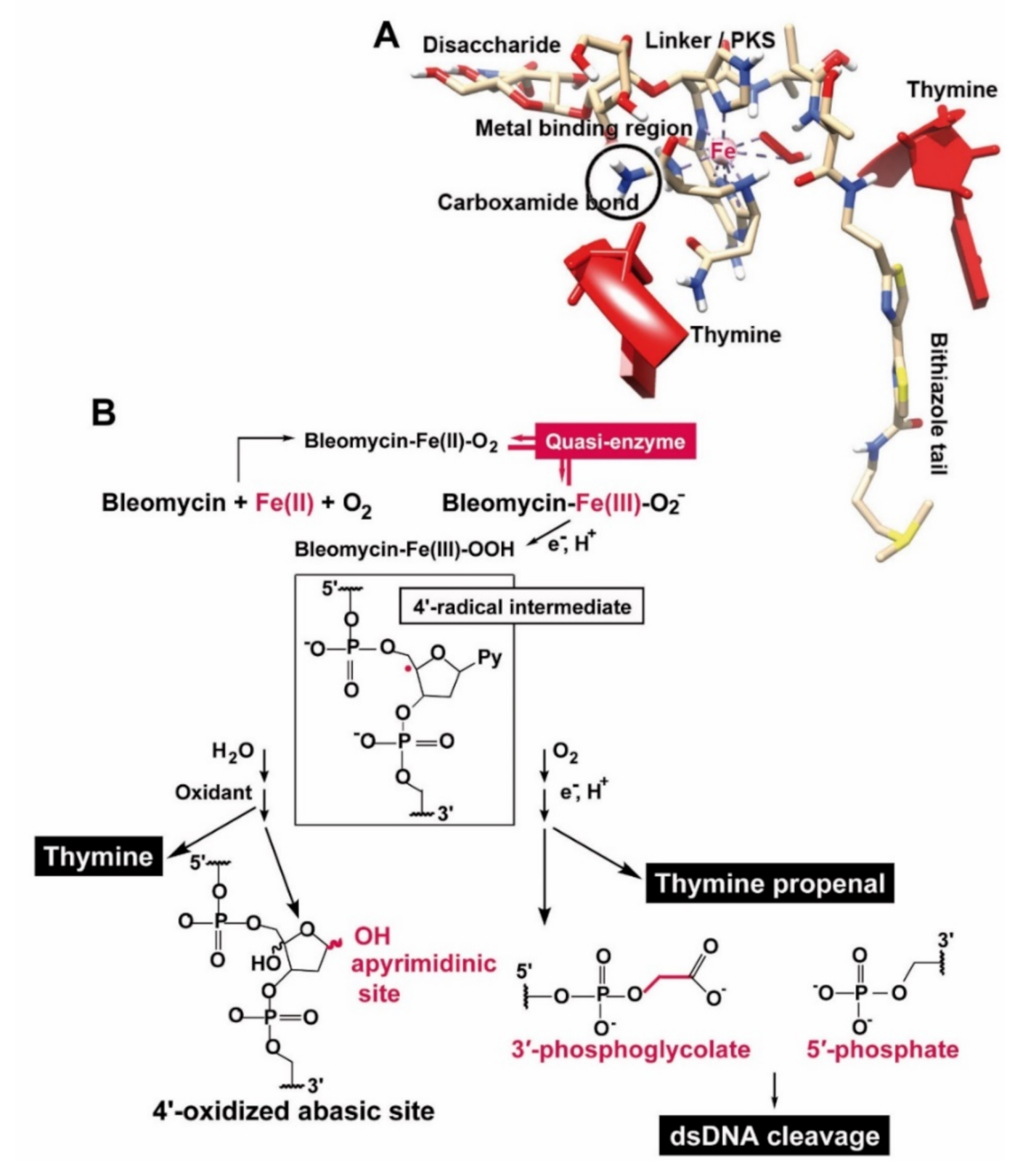
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. A549 Cells
2.3. BEAS-2B Cells
2.4. Staining of the Cells: Immunocytochemistry
2.5. Cytotoxicity Assays: MTT
2.6. DNA Stand Break Assay
2.7. Quantitative Real-Time Polymerase Chain Reaction: BLM Hydrolase
2.8. Statistical Analysis
3. Results
3.1. Effect of BLM on Cell Growth
3.2. Effect of BLM and polyP Alone and in Combination on BEAS-2B Cell Viability
3.3. DNA Strand Breaks Caused by BLM
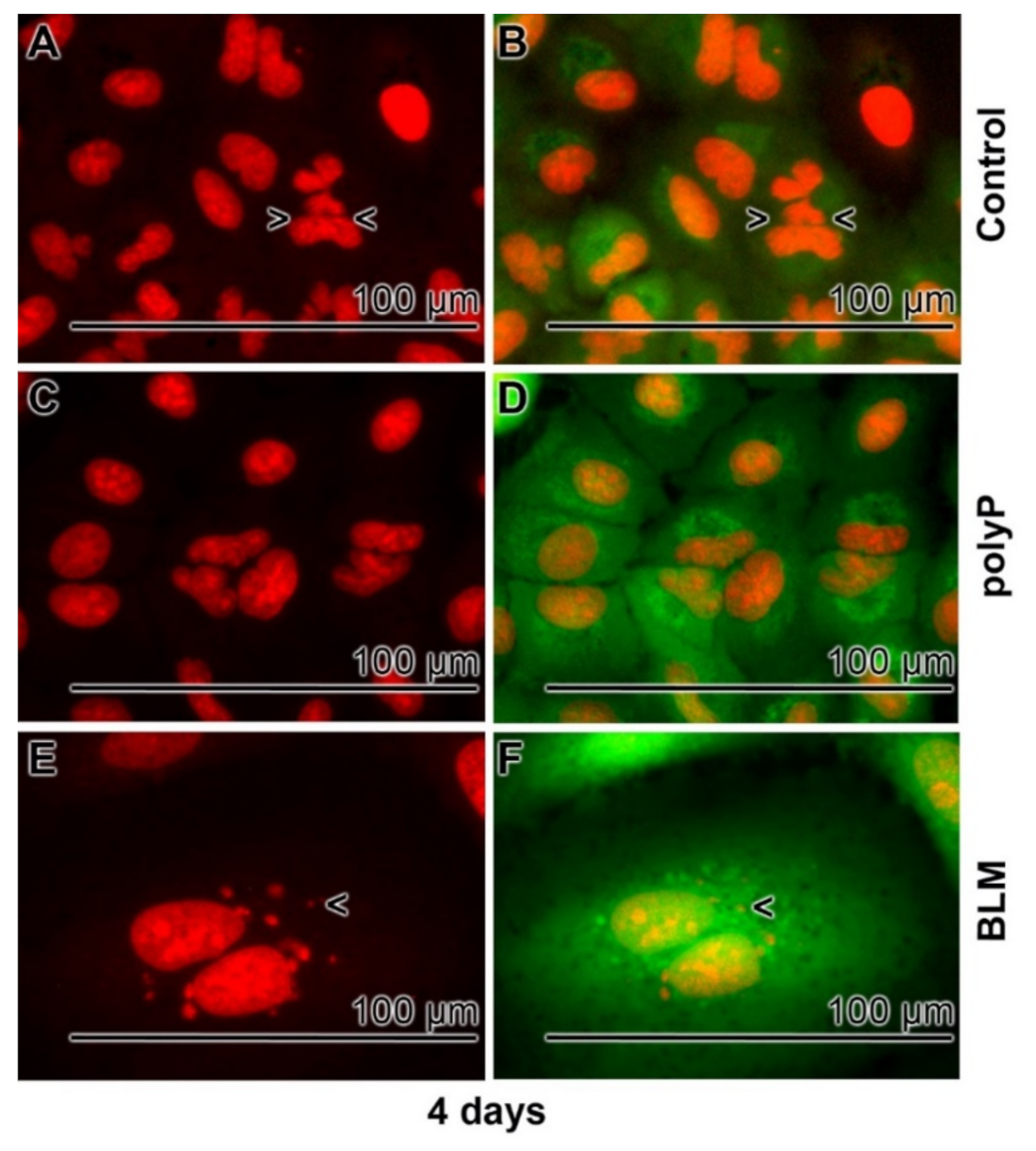
3.4. Severe effects of BLM on Cell Morphology
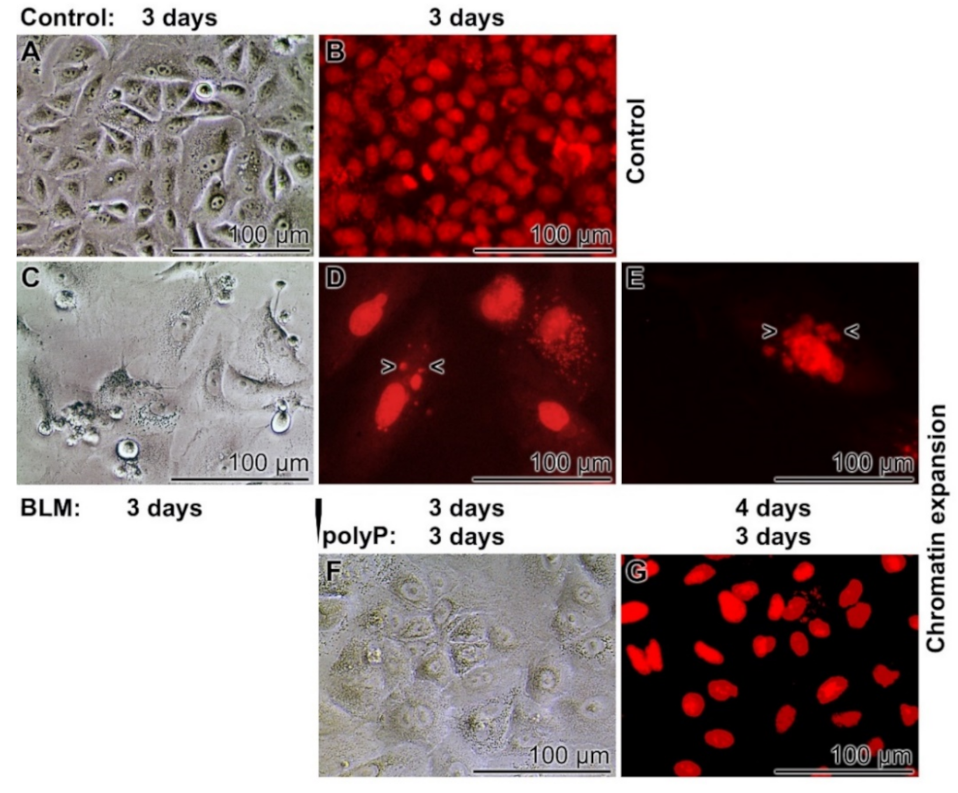
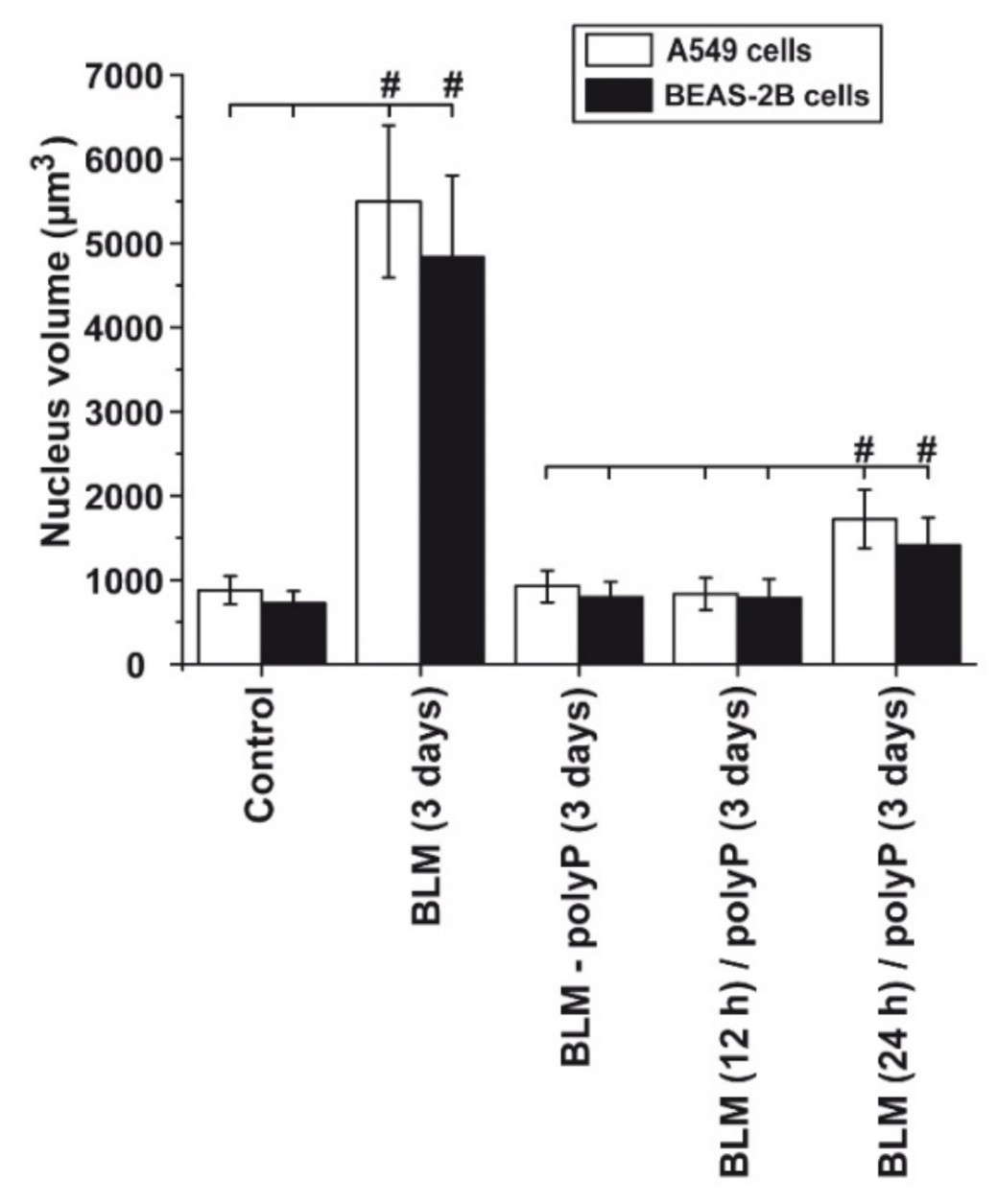
3.5. Prevention of Nuclear Enlargement in BLM Treated Cells During Co-Incubation with polyP
3.6. Effect of Sequential Application of BLM and polyP
3.7. Time Dependent Deleterious Influence of BLM on of the polyP Rescue Activity: A549 Cells and BEAS-2B Cells
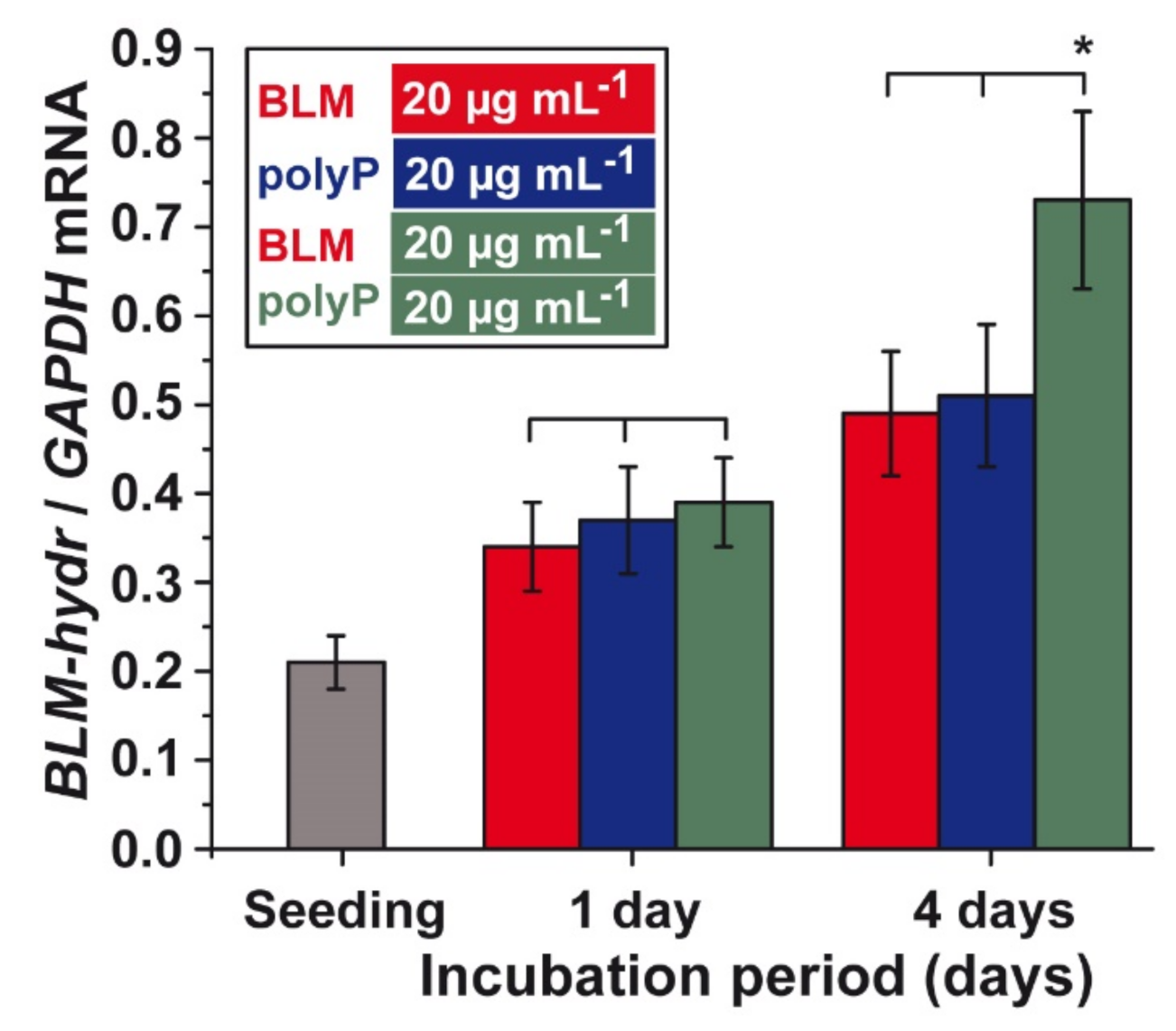
3.8. Effect of BLM and polyP Alone or in Combination on BLM Hydrolase Gene Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Umezawa, H. Bleomycin. In Antibiotics, Volume 3, Mechanism of Action of Antimicrobial and Antitumor Agents; Corcoran, J.W., Hahn, F.E., Snell, J.F., Arora, K.L., Eds.; Springer: Berlin/Heidelberg, Germany, 1975; pp. 21–33. [Google Scholar]
- Umezawa, H.; Maeda, K.; Takeuchi, T.; Okami, Y. New antibiotics, bleomycin A and B. J. Antibiot. 1966, 19, 200–209. [Google Scholar]
- Suzuki, H.; Nagai, K.; Yamaki, H.; Tanaka, N.; Umezawa, H. On the mechanism of action of bleomycin, scission of DNA strands in vitro and in vivo. J. Antibiot. 1969, 22, 446–448. [Google Scholar]
- Ichikawa, T.; Umezawa, H.; Ohashi, S.; Takeuchi, T.; Ishizuka, M.; Hori, S. Animal experiments confirming the specific effect of bleomycin against squamous cell carcinoma. In Proceedings of the 6th International Congress of Chemotherapy, Tokyo, Japan, 10–15 August 1969, 16th ed.; Scientific Committee: Tokyo, Japan, 1970; Volume 2, pp. 315–316. [Google Scholar]
- Umezawa, H.; Takeuchi, T.; Hori, S.; Sawa, T.; Ishizuka, M. Studies on the mechanism of antitumor effect of bleomycin of squamous cell carcinoma. J. Antibiot. 1972, 25, 409–420. [Google Scholar]
- ksCrooke, S.T.; Bradner, W.T. Bleomycin, a review. J. Med. 1976, 7, 333–428. [Google Scholar]
- Du, L.; Sánchez, C.; Chen, M.; Edwards, D.J.; Shen, B. The biosynthetic gene cluster for the antitumor drug bleomycin from Streptomyces verticillus ATCC15003 supporting functional interactions between nonribosomal peptide synthetases and a polyketide synthase. Chem. Biol. 2000, 7, 623–642. [Google Scholar]
- Rajani, C.; Kincaid, J.R.; Petering, D.H. A systematic approach toward the analysis of drug-DNA interactions using Raman spectroscopy, the binding of metal-free bleomycins A2 and B2 to calf thymus DNA. Biopolymers 1999, 52, 110–128. [Google Scholar]
- Goodwin, K.D.; Lewis, M.A.; Long, E.C.; Georgiadis, M.M. Crystal structure of DNA-bound Co(III) bleomycin B2: Insights on intercalation and minor groove binding. Proc. Natl. Acad. Sci. USA 2008, 105, 5052–5056. [Google Scholar]
- Lehmann, T.; Topchiy, E. Contributions of NMR to the understanding of the coordination chemistry and DNA interactions of metallo-bleomycins. Molecules 2013, 18, 9253–9277. [Google Scholar]
- Müller, W.; Yamazaki, Z.; Breter, H.J.; Zahn, R.K. Action of Bleomycin on DNA and RNA. Eur. J. Biochem. 1972, 31, 518–525. [Google Scholar]
- Müller, W.E.G.; Zahn, R.K. Bleomycin: Mode of action on DNA. Gann Monogr Cancer Res. 1976, 19, 51–62. [Google Scholar]
- Burger, R.M.; Peisach, J.; Horwitz, S.B. Activated bleomycin. A transient complex of drug, iron, and oxygen that degrades DNA. J. Biol. Chem. 1981, 256, 11636–11644. [Google Scholar]
- McLean, M.J.; Dar, A.; Waring, M.J. Differences between sites of binding to DNA and strand cleavage for complexes of bleomycin with iron or cobalt. J. Mol. Recognit. 1989, 1, 184–192. [Google Scholar]
- Chen, J.; Stubbe, J. Bleomycins: Towards better therapeutics. Nat. Rev. Cancer 2005, 5, 102–112. [Google Scholar]
- Müller, W.E.G.; Zahn, R.K. Bleomycin, an antibiotic that removes thymine from double-stranded DNA. In Progress in Nucleic Acid Research and Molecular Biology; Cohn, W.E., Ed.; Academic Press: Cambridge, MA, USA, 1977; Volume 20, pp. 21–57. [Google Scholar]
- Zarytova, V.P.; Sergeyev, D.S.; Godovikova, T.S. ‘Quasi-enzyme’ cleavage of a DNA target by a bleomycin A5 oligonucleotide derivative. Nucleic Acids Symp. Ser. 1991, 1991, 249. [Google Scholar]
- Roy, B.; Hecht, S.M. Hairpin DNA sequences bound strongly by bleomycin exhibit enhanced double-strand cleavage. J. Am. Chem. Soc. 2014, 136, 4382–4393. [Google Scholar]
- Ebrahimi, K.H.; Hagedoorn, P.L.; Hagen, W.R. Unity in the biochemistry of the iron-storage proteins ferritin and bacterioferritin. Chem. Rev. 2015, 115, 295–326. [Google Scholar]
- Gilbert, W.; Maxam, A. The nucleotide sequence of the lac operator. Proc. Natl. Acad. Sci. USA 1973, 70, 3581–3584. [Google Scholar]
- Yang, Z.; Price, N.E.; Johnson, K.M.; Wang, Y.; Gates, K.S. Interstrand cross-links arising from strand breaks at true abasic sites in duplex DNA. Nucleic Acids Res. 2017, 45, 6275–6283. [Google Scholar]
- Murray, V.; Martin, R.F. The sequence specificity of bleomycin-induced DNA damage in intact cells. J. Biol. Chem. 1985, 260, 10389–10391. [Google Scholar]
- Paul, M.; Murray, V. Use of an automated capillary DNA sequencer to investigate the interaction of cisplatin with telomeric DNA sequences. Biomed. Chromatogr. 2012, 26, 350–354. [Google Scholar]
- Galea, A.M.; Murray, V. The influence of chromatin structure on DNA damage induced by nitrogen mustard and cisplatin analogues. Chem. Biol. Drug Des. 2010, 75, 578–589. [Google Scholar]
- Murray, V.; Chen, J.K.; Galea, A.M. The anti-tumor drug bleomycin preferentially cleaves at the transcription start sites of actively transcribed genes in human cells. Cell. Mol. Life Sci. 2014, 71, 1505–1512. [Google Scholar]
- Cloos, J.; Reid, C.B.; van der Sterre, M.L.; Tobi, H.; Leemans, C.R.; Snow, G.B.; Braakhuis, B.J. A comparison of bleomycin-induced damage in lymphocytes and primary oral fibroblasts and keratinocytes in 30 subjects. Mutagenesis 1999, 14, 87–93. [Google Scholar]
- Latta, V.D.; Cecchettini, A.; Del Ry, S.; Morales, M.A. Bleomycin in the setting of lung fibrosis induction: From biological mechanisms to counteractions. Pharmacol. Res. 2015, 97, 122–130. [Google Scholar]
- Fyfe, A.J.; McKay, P. Toxicities associated with bleomycin. J. R. Coll. Physicians Edinb. 2010, 40, 213–215. [Google Scholar]
- Azambuja, E.; Fleck, J.F.; Batista, R.G.; Menna Barreto, S.S. Bleomycin lung toxicity: Who are the patients with increased risk? Pulm. Pharmacol. Ther. 2005, 18, 363–366. [Google Scholar]
- Gouda, M.M.; Prabhu, A.; Bhandary, Y.P. Curcumin alleviates IL-17A-mediated p53-PAI-1 expression in bleomycin-induced alveolar basal epithelial cells. J. Cell. Biochem. 2018, 119, 2222–2230. [Google Scholar]
- Albright, C.D.; Jones, R.T.; Hudson, E.A.; Fontana, J.A.; Trump, B.F.; Resau, J.H. Transformed human bronchial epithelial cells (BEAS-2B) alter the growth and morphology of normal human bronchial epithelial cells in vitro. Cell Biol. Toxicol. 1990, 6, 379–398. [Google Scholar]
- Beers, M.F.; Moodley, Y. When is an alveolar type 2 cell an alveolar type 2 cell? A conundrum for lung stem cell biology and regenerative medicine. Am. J. Respir. Cell Mol. Biol. 2017, 57, 18–27. [Google Scholar]
- Jang, S.; Ryu, S.M.; Lee, J.; Lee, H.; Hong, S.H.; Ha, K.S.; Park, W.S.; Han, E.T.; Yang, S.R. Bleomycin inhibits proliferation via Schlafen-mediated cell cycle arrest in mouse alveolar epithelial cells. Tuberc. Respir. Dis. 2019, 82, 133–142. [Google Scholar]
- Mata, M.; Ruíz, A.; Cerdá, M.; Martinez-Losa, M.; Cortijo, J.; Santangelo, F.; Serrano-Mollar, A.; Llombart-Bosch, A.; Morcillo, E.J. Oral N-acetylcysteine reduces bleomycin-induced lung damage and mucin Muc5ac expression in rats. Eur. Respir. J. 2003, 22, 900–905. [Google Scholar]
- Umezawa, H.; Hori, S.; Sawa, T.; Yoshioka, T.; Takeuchi, T. A bleomycin-inactivating enzyme in mouse liver. J. Antibiot. 1974, 27, 419–424. [Google Scholar]
- Ferrando, A.A.; Velasco, G.; Campo, E.; Lopez-Otin, C. Cloning and expression analysis of human bleomycin hydrolase, a cysteine proteinase involved in chemotherapy resistance. Cancer Res. 1996, 56, 1746–1750. [Google Scholar]
- Müller, W.E.G.; Zahn, R.K. Determination of the Bleomycin inactivating enzyme in biopsies. Gann 1976, 67, 425–430. [Google Scholar]
- Müller, W.E.G.; Schmidseder, R.; Rohde, H.J.; Zahn, R.K.; Scheunemann, H. Bleomycin-sensitivity test: Application for human squamous cell carcinoma. Cancer 1977, 40, 2787–2791. [Google Scholar]
- George, P.M.; Wells, A.U.; Jenkins, R.G. Pulmonary fibrosis and COVID-19: The potential role for antifibrotic therapy. Lancet Respir. Med. 2020, 8, 807–815. [Google Scholar]
- Neufurth, M.; Wang, X.H.; Tolba, E.; Lieberwirth, I.; Wang, S.; Schröder, H.C.; Müller, W.E.G. The inorganic polymer, polyphosphate, blocks binding of SARS-CoV-2 spike protein to ACE2 receptor at physiological concentrations. Biochem. Pharmacol. 2020, 182, 114215. [Google Scholar]
- Müller, W.E.G.; Neufurth, M.; Schepler, H.; Wang, S.; Tolba, E.; Schröder, H.C.; Wang, X.H. The biomaterial polyphosphate blocks stoichiometrically binding of the SARS-CoV-2 S-protein to the cellular ACE2 receptor. Biomater. Sci. 2020, 8, 6603–6610. [Google Scholar]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020, 94, e00127-20. [Google Scholar]
- Wang, L.; Wang, Y.; Yang, T.; Goo, Y.; Sun, T. Angiotensin-converting enzyme 2 attenuates bleomycin-induced lung fibrosis in mice. Cell. Physiol. Biochem. 2015, 36, 697–711. [Google Scholar]
- Kashima, S.; Fujiya, M.; Konishi, H.; Ueno, N.; Inaba, Y.; Moriichi, K.; Tanabe, H.; Ikuta, K.; Ohtake, T.; Kohgo, Y. Polyphosphate, an active molecule derived from probiotic Lactobacillus brevis, improves the fibrosis in murine colitis. Transl. Res. 2015, 166, 163–175. [Google Scholar]
- Fujiya, M.; Ueno, N.; Kashima, S.; Tanaka, K.; Sakatani, A.; Ando, K.; Moriichi, K.; Konishi, H.; Kamiyama, N.; Tasaki, Y.; et al. Long-chain polyphosphate is a potential agent for inducing mucosal healing of the colon in ulcerative colitis. Clin. Pharmacol. Ther. 2020, 107, 452–461. [Google Scholar]
- Wang, X.H.; Schröder, H.C.; Müller, W.E.G. Amorphous polyphosphate, a smart bioinspired nano-/bio-material for bone and cartilage regeneration: Towards a new paradigm in tissue engineering. J. Mater. Chem. B 2018, 6, 2385–2412. [Google Scholar]
- Aoshiba, K.; Tsuji, T.; Nagai, A. Bleomycin induces cellular senescence in alveolar epithelial cells. Eur. Respir. J. 2003, 22, 436–443. [Google Scholar]
- Cooper, J.R.; Abdullatif, M.B.; Burnett, E.C.; Kempsell, K.E.; Conforti, F.; Tolley, H.; Collins, J.E.; Davies, D.E. Long term culture of the A549 cancer cell line promotes multilamellar body formation and differentiation towards an alveolar type II pneumocyte phenotype. PLoS ONE 2016, 11, e0164438. [Google Scholar]
- Leyhausen, G.; Lorenz, B.; Zhu, H.; Geurtsen, W.; Bohnensack, R.; Müller, W.E.G.; Schröder, H.C. Inorganic polyphosphate in human osteoblast-like cells. J. Bone Miner. Res. 1998, 13, 803–812. [Google Scholar]
- Michelson, P.H.; Tigue, M.; Panos, R.J.; Sporn, P.H. Keratinocyte growth factor stimulates bronchial epithelial cell proliferation in vitro and in vivo. Am. J. Physiol. 1999, 277, L737–L742. [Google Scholar]
- Müller, W.E.G.; Belikov, S.I.; Tremel, W.; Perry, C.C.; Gieskes, W.W.C.; Boreiko, A.; Schröder, H.C. Siliceous spicules in marine demosponges (example Suberites domuncula). Micron 2006, 37, 107–120. [Google Scholar]
- Smith, P.J.; Wiltshire, M.; Errington, R.J. DRAQ5 labeling of nuclear DNA in live and fixed cells. Curr. Protoc. Cytom. 2004, 7. [Google Scholar] [CrossRef]
- Müller, W.E.G.; Wang, S.F.; Ackermann, M.; Gerich, T.; Wiens, M.; Neufurth, M.; Schröder, H.C.; Wang, X.H. Biologization of allogeneic bone grafts with polyphosphate: A route to a biomimetic periosteum. Adv. Funct. Mater. 2019, 29, 1905220. [Google Scholar]
- Müller, W.E.G.; Tolba, E.; Schröder, H.C.; Muñoz-Espí, R.; Diehl-Seifert, B.; Wang, X.H. Amorphous polyphosphate-hydroxyapatite: A morphogenetically active substrate for bone-related SaOS-2 cells in vitro. Acta Biomater. 2016, 31, 358–367. [Google Scholar]
- Müller, W.E.G.; Wang, S.; Tolba, E.; Neufurth, M.; Ackermann, M.; Muñoz-Espí, R.; Lieberwirth, I.; Glasser, G.; Schröder, H.C.; Wang, X.H. Transformation of amorphous polyphosphate nanoparticles into coacervate complexes: An approach for the encapsulation of mesenchymal stem cells. Small 2018, 14, 1801170. [Google Scholar]
- Batel, R.; Jaksic, Z.; Bihari, N.; Hamer, B.; Fafandel, M.; Chauvin, C.; Schröder, H.C.; Müller, W.E.G.; Zahn, R.K. A microplate assay for DNA damage determination (fast micromethod) in cell suspensions and solid tissues. Anal. Biochem. 1999, 270, 195–200. [Google Scholar]
- Cosa, G.; Focsaneanu, K.S.; McLean, J.R.; McNamee, J.P.; Scaiano, J.C. Photophysical properties of fluorescent DNA-dyes bound to single- and double-stranded DNA in aqueous buffered solution. Photochem. Photobiol. 2001, 73, 585–599. [Google Scholar]
- Jaksić, Z.; Batel, R. DNA integrity determination in marine invertebrates by Fast Micromethod. Aquat. Toxicol. 2003, 65, 361–376. [Google Scholar]
- Wiens, M.; Wang, X.H.; Schloßmacher, U.; Lieberwirth, I.; Glasser, G.; Ushijima, H.; Schröder, H.C.; Müller, W.E.G. Osteogenic potential of bio-silica on human osteoblast-like (SaOS-2) cells. Calcif. Tissue Int. 2010, 87, 513–524. [Google Scholar]
- Takeda, A.; Masuda, Y.; Yamamoto, T.; Hirabayashi, T.; Nakamura, Y.; Nakaya, K. Cloning and analysis of cDNA encoding rat bleomycin hydrolase, a DNA-binding cysteine protease. J. Biochem. 1996, 120, 353–359. [Google Scholar]
- Chen, J.; Chen, Y.; He, Q. Action of bleomycin is affected by bleomycin hydrolase but not by caveolin-1. Int. J. Oncol. 2012, 41, 2245–2252. [Google Scholar]
- Riise, R.; Odqvist, L.; Mattsson, J.; Monkley, S.; Abdillahi, S.M.; Tyrchan, C.; Muthas, D.; Yrlid, L.F. Bleomycin hydrolase regulates the release of chemokines important for inflammation and wound healing by keratinocytes. Sci. Rep. 2019, 9, 20407. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar]
- Müller, W.E.G.; Rohde, H.J.; Steffen, R.; Maidhof, A.; Zahn, R.K. Potentiation of the effectiveness of Bleomycin by A T-specific DNA ligands in vitro as well as in vivo. Cancer Lett. 1976, 1, 127–132. [Google Scholar]
- Nicolay, N.H.; Rühle, A.; Perez, R.L.; Trinh, T.; Sisombath, S.; Weber, K.J.; Ho, A.D.; Debus, J.; Saffrich, R.; Huber, P.E. Mesenchymal stem cells are sensitive to bleomycin treatment. Sci. Rep. 2016, 6, 26645. [Google Scholar]
- Wang, X.H.; Schröder, H.C.; Diehl-Seifert, B.; Kropf, K.; Schlossmacher, U.; Wiens, M.; Müller, W.E.G. Dual effect of inorganic polymeric phosphate/polyphosphate on osteoblasts and osteoclasts in vitro. J. Tissue Eng. Regen. Med. 2013, 7, 767–776. [Google Scholar]
- He, B.; Lu, N.; Zhou, Z. Cellular and nuclear degradation during apoptosis. Curr. Opin. Cell Biol. 2009, 21, 900–912. [Google Scholar]
- Zhang, X.H.; Zhang, N.; Lu, J.M.; Kong, Q.Z.; Zhao, Y.F. Tetrazolium violet induced apoptosis and cell cycle arrest in human lung cancer A549 cells. Biomol. Ther. 2012, 20, 177–182. [Google Scholar]
- Peter, Y.; Comellas, A.; Levantini, E.; Ingenito, E.P.; Shapiro, S.D. Epidermal growth factor receptor and claudin-2 participate in A549 permeability and remodeling: Implications for non-small cell lung cancer tumor colonization. Mol. Carcinog. 2009, 48, 488–497. [Google Scholar]
- Wang, D.D.; Liu, W.; Chang, J.J.; Cheng, X.; Zhang, X.Z.; Xu, H.; Feng, D.; Yu, L.J.; Wang, X.L. Bioengineering three-dimensional culture model of human lung cancer cells: An improved tool for screening EGFR targeted inhibitors. RSC Adv. 2016, 6, 24083–24090. [Google Scholar]
- Lindahl, T.; Andersson, A. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry 1972, 11, 3618–3623. [Google Scholar]
- Müller, W.E.G.; Hanske, W.; Maidhof, A.; Zahn, R.K. Influence of apurinic acid on programmed synthesis in different in vitro systems. Cancer Res. 1973, 33, 2330–2337. [Google Scholar]
- Sebti, S.M.; Jani, J.P.; Mistry, J.S.; Gorelik, E.; Lazo, J.S. Metabolic inactivation: A mechanism on human tumor resistance to bleomycin. Cancer Res. 1991, 51, 227–232. [Google Scholar]
- Murray, V.; Chen, J.K.; Chung, L.H. The interaction of the metallo-glycopeptide anti-tumour drug Bleomycin with DNA. Int. J. Mol. Sci. 2018, 19, 1372. [Google Scholar]
- Müller, W.E.G.; Schröder, H.C.; Wang, X.H. Inorganic polyphosphates as storage for and generator of metabolic energy in the extracellular matrix. Chem. Rev. 2019, 119, 12337–12374. [Google Scholar]
- Liu, F.; Zhou, P.; Wang, Q.; Zhang, M.; Li, D. The Schlafen family, complex roles in different cell types and virus replication. Cell Biol. Int. 2018, 42, 2–8. [Google Scholar]
- Mungunsukh, O.; Griffin, A.J.; Lee, Y.H.; Day, R.M. Bleomycin induces the extrinsic apoptotic pathway in pulmonary endothelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2010, 298, L696–L703. [Google Scholar]
- Toyokuni, S.; Yanatori, I.; Kong, Y.; Zheng, H.; Motooka, Y.; Jiang, L. Ferroptosis at the crossroads of infection, aging and cancer. Cancer Sci. 2020, 111, 2665–2671. [Google Scholar]
- Takashima, T.; Sakamoto, N.; Murai, J.; Taniyama, D.; Honma, R.; Ukai, S.; Maruyama, R.; Kuraoka, K.; Rajapakse, V.N.; Pommier, Y.; et al. Immunohistochemical analysis of SLFN11 expression uncovers potential non-responders to DNA-damaging agents overlooked by tissue RNA-seq. Virchows Arch. 2020. [Google Scholar] [CrossRef]
- Li, M.; Kao, E.; Gao, X.; Sandig, H.; Limmer, K.; Pavon-Eternod, M.; Jones, T.E.; Landry, S.; Pan, T.; Weitzman, M.D.; et al. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature 2012, 491, 125–128. [Google Scholar]
- Bustos, O.; Naik, S.; Ayers, G.; Casola, C.; Perez-Lamigueiro, M.A.; Chippindale, P.T.; Pritham, E.J.; de la Casa-Esperón, E. Evolution of the Schlafen genes, a gene family associated with embryonic lethality, meiotic drive, immune processes and orthopoxvirus virulence. Gene 2009, 447, 1–11. [Google Scholar]
- Li, M.L.; Yuan, G.; Greenberg, R.A. Chromatin yo-yo, expansion and condensation during DNA repair. Trends Cell Biol. 2014, 24, 616–618. [Google Scholar]
- Soria, G.; Polo, S.E.; Almouzni, G. Prime, repair, restore, the active role of chromatin in the DNA damage response. Mol. Cell 2012, 46, 722–734. [Google Scholar]
- Webster, M.; Witkin, K.L.; Cohen-Fix, O. Sizing up the nucleus, nuclear shape, size and nuclear-envelope assembly. J. Cell Sci. 2009, 122, 1477–1486. [Google Scholar]
- Takemoto, A.; Kawashima, S.A.; Li, J.J.; Jeffery, L.; Yamatsugu, K.; Elemento, O.; Nurse, P. Nuclear envelope expansion is crucial for proper chromosomal segregation during a closed mitosis. J. Cell Sci. 2016, 129, 1250–1259. [Google Scholar]
- Imle, A.; Polzer, B.; Alexander, S.; Klein, C.A.; Friedl, P. Genomic instability of micronucleated cells revealed by single-cell comparative genomic hybridization. Cytom. A 2009, 75, 562–568. [Google Scholar]
- Ferrando, A.A.; Pendás, A.M.; Llano, E.; Velasco, G.; Lidereau, R.; López-Otín, C. Gene characterization, promoter analysis, and chromosomal localization of human bleomycin hydrolase. J. Biol. Chem. 1997, 272, 33298–33304. [Google Scholar]
- Zabidi, M.A.; Arnold, C.D.; Schernhuber, K.; Pagani, M.; Rath, M.; Frank, O.; Stark, A. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature 2015, 518, 556–559. [Google Scholar]
- Lindahl, T.L.; Ramström, S.; Boknäs, N.; Faxälv, L. Caveats in studies of the physiological role of polyphosphates in coagulation. Biochem. Soc. Trans. 2016, 44, 35–39. [Google Scholar]
- Weitz, J.I.; Fredenburgh, J.C. Platelet polyphosphate, the long and the short of it. Blood 2017, 129, 1574–1575. [Google Scholar]
- Morrissey, J.H.; Choi, S.H.; Smith, S.A. Polyphosphate, an ancient molecule that links platelets, coagulation, and inflammation. Blood 2012, 119, 5972–5979. [Google Scholar]
- Wang, X.H.; Gericke, A.; Ackermann, M.; Wang, S.; Neufurth, M.; Schröder, H.C.; Pfeiffer, N.; Müller, W.E.G. Polyphosphate, the physiological metabolic fuel for corneal cells: A potential biomaterial for ocular surface repair. Biomaterials Sci. 2019, 7, 5506–5515. [Google Scholar]
- Rodriguez, L.R.; Emblom-Callahan, M.; Chhina, M.; Bui, S.; Aljeburry, B.; Tran, L.H.; Novak, R.; Lemma, M.; Nathan, S.D.; Grant, G.M. Global Gene Expression Analysis in an in vitro Fibroblast Model of Idiopathic Pulmonary Fibrosis Reveals Potential Role for CXCL14/CXCR4. Sci. Rep. 2018, 8, 3983. [Google Scholar]
- Weng, D.; Chen, J.X.; Li, H.H.; Liu, F.; Zhou, L.D.; Liu, H.P.; Zheng, R.J.; Jiang, Y.; Liu, Z.H.; Ge, B. 2-aminopurine suppresses the TGF-β1-induced epithelial-mesenchymal transition and attenuates bleomycin-induced pulmonary fibrosis. Cell Death Discov. 2018, 4, 17. [Google Scholar]
- Müller, W.E.G.; Neufurth, M.; Wang, S.; Tan, R.; Schröder, H.C.; Wang, X.H. Morphogenetic (mucin expression) as well as potential anti-corona viral activity of the marine secondary metabolite polyphosphate on A549 cells. Mar. Drugs 2020, 18, 639. [Google Scholar]
- Reeves, S.R.; Kang, I.; Chan, C.K.; Barrow, K.A.; Kolstad, T.K.; White, M.P.; Ziegler, S.F.; Wight, T.N.; Debley, J.S. Asthmatic bronchial epithelial cells promote the establishment of a Hyaluronan-enriched, leukocyte-adhesive extracellular matrix by lung fibroblasts. Respir. Res. 2018, 19, 146. [Google Scholar]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Müller, W.E.G.; Neufurth, M.; Wang, S.; Schröder, H.C.; Wang, X. Polyphosphate Reverses the Toxicity of the Quasi-Enzyme Bleomycin on Alveolar Endothelial Lung Cells In Vitro. Cancers 2021, 13, 750. https://doi.org/10.3390/cancers13040750
Müller WEG, Neufurth M, Wang S, Schröder HC, Wang X. Polyphosphate Reverses the Toxicity of the Quasi-Enzyme Bleomycin on Alveolar Endothelial Lung Cells In Vitro. Cancers. 2021; 13(4):750. https://doi.org/10.3390/cancers13040750
Chicago/Turabian StyleMüller, Werner E. G., Meik Neufurth, Shunfeng Wang, Heinz C. Schröder, and Xiaohong Wang. 2021. "Polyphosphate Reverses the Toxicity of the Quasi-Enzyme Bleomycin on Alveolar Endothelial Lung Cells In Vitro" Cancers 13, no. 4: 750. https://doi.org/10.3390/cancers13040750
APA StyleMüller, W. E. G., Neufurth, M., Wang, S., Schröder, H. C., & Wang, X. (2021). Polyphosphate Reverses the Toxicity of the Quasi-Enzyme Bleomycin on Alveolar Endothelial Lung Cells In Vitro. Cancers, 13(4), 750. https://doi.org/10.3390/cancers13040750