Regulation of p53 by E3s
Abstract
Simple Summary
Abstract
1. Introduction
2. The Tumor Suppressor Protein p53
3. E3s and The Ubiquitin Proteasome System
4. E3s That Target Wild-Type p53
4.1. E3s That Ubiquitinate p53 and Target. It for Degradation
4.1.1. RING-Domain E3s
4.1.2. HECT-Domain and U-Box-Domain E3s
4.1.3. E3s That Require Complex Formation
4.2. E3s That Mediate p53 Ubiquitination without Promoting Degradation
4.3. E3s That Mediate Neddylation and Sumoylation of p53
4.4. E3s That Act. on p53 without Modifying It
5. Tumor Virus-Associated E3s
6. E3s Targeting Mutated p53
7. E3s Targeting p53 Isoforms
8. E3s and Tumorigenesis
9. Targeting E3s for Tumor Therapy
10. Discussion
11. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lane, D.P.; Crawford, L.V. T antigen is bound to a host protein in SY40-transformed cells. Nature 1979, 278, 261–263. [Google Scholar] [CrossRef]
- Linzer, D.I.; Maltzman, W.; Levine, A.J. The SV40 a gene product is required for the production of a 54,000 MW cellular tumor antigen. Virology 1979, 98, 308–318. [Google Scholar] [CrossRef]
- Dippold, W.G.; Jay, G.; DeLeo, A.B.; Khoury, G.; Old, L.J. p53 transformation-related protein: Detection by monoclonal antibody in mouse and human cells. Proc. Natl. Acad. Sci. USA 1981, 78, 1695–1699. [Google Scholar] [CrossRef] [PubMed]
- Rotter, V. p53, a transformation-related cellular-encoded protein, can be used as a biochemical marker for the detection of primary mouse tumor cells. Proc. Natl. Acad. Sci. USA 1983, 80, 2613–2617. [Google Scholar] [CrossRef] [PubMed]
- Parada, L.F.; Land, H.; Weinberg, R.A.; Wolf, D.; Rotter, V. Cooperation between gene encoding p53 tumour antigen and ras in cellular trans-formation. Nature 1984, 312, 649–651. [Google Scholar] [CrossRef] [PubMed]
- Rovinski, B.; Benchimol, S. Immortalization of rat embryo fibroblasts by the cellular p53 oncogene. Oncogene 1988, 2, 445–452. [Google Scholar]
- Cathy, A.; Finlay, P.W.H.; Levine, A.J. The p53 proto-Oncogene can act as a suppressor of transformation. Cell 1989, 57, 1083–1093. [Google Scholar]
- Lane, D.P. p53, guardian of the genome. Nature 1992, 358, 15–16. [Google Scholar] [CrossRef]
- Efeyan, A.; Serrano, M. p53: Guardian of the Genome and Policeman of the Oncogenes. Cell Cycle 2007, 6, 1006–1010. [Google Scholar] [CrossRef]
- Palmero, I.; Pantoja, C.; Serrano, M. p19ARF links the tumour suppressor p53 to Ras. Nature 1998, 395, 125–126. [Google Scholar] [CrossRef]
- Miciak, J.; Bunz, F. Long story short: p53 mediates innate immunity. Biochim. Biophys. Acta 2016, 1865, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Agupitan, A.D.; Neeson, P.; Williams, S.; Howitt, J.; Haupt, S.; Haupt, Y. P53: A Guardian of Immunity Becomes Its Saboteur through Mutation. Int. J. Mol. Sci. 2020, 21, 3452. [Google Scholar] [CrossRef] [PubMed]
- Joerger, A.C.; Fersht, A.R. Structural Biology of the Tumor Suppressor p53. Annu. Rev. Biochem. 2008, 77, 557–582. [Google Scholar] [CrossRef]
- Teufel, D.P.; Freund, S.M.; Bycroft, M.; Fersht, A.R. Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53. Proc. Natl. Acad. Sci. USA 2007, 104, 7009–7014. [Google Scholar] [CrossRef]
- Gu, W.; Shi, X.-L.; Roeder, R.G. Synergistic activation of transcription by CBP and p53. Nature 1997, 387, 819–823. [Google Scholar] [CrossRef]
- Kussie, P.H.; Gorina, S.; Marechal, V.; Ellenbas, B.; Moreau, J.; Levine, A.J.; Pavletich, N.P. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 1996, 274, 948–953. [Google Scholar] [CrossRef]
- Marine, J.-C.; Jochemsen, A.G. Mdmx as an essential regulator of p53 activity. Biochem. Biophys. Res. Commun. 2005, 331, 750–760. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Levine, A.J. Human TAFII31 protein is a transcriptional coactivator of the p53 protein. Proc. Natl. Acad. Sci. USA 1995, 92, 5154–5158. [Google Scholar] [CrossRef]
- Di Lello, P.; Jenkins, L.M.; Jones, T.N.; Nguyen, B.D.; Hara, T.; Yamaguchi, H.; Dikeakos, J.D.; Appella, E.; Legault, P.; Omichinski, J.G. Structure of the Tfb1/p53 complex: Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol. Cell 2006, 22, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Marechal, V.; Levine, A.J. Mapping of the p53 and mdm-2 interaction domains. Mol. Cell. Biol. 1993, 13, 4107–4114. [Google Scholar] [CrossRef] [PubMed]
- Venot, C.; Maratrat, M.; Dureuil, C.; Conseiller, E.; Bracco, L.; Debussche, L. The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO J. 1998, 17, 4668–4679. [Google Scholar] [CrossRef]
- Zhao, K.; Chai, X.; Johnston, K.; Clements, A.; Marmorstein, R. Crystal Structure of the Mouse p53 Core DNA-binding Domain at 2.7 Å Resolution. J. Biol. Chem. 2001, 276, 12120–12127. [Google Scholar] [CrossRef] [PubMed]
- Balagurumoorthy, P.; Sakamoto, H.; Lewis, M.S.; Zambrano, N.; Clore, G.M.; Gronenborn, A.M.; Appella, E.; Harrington, R.E. Four p53 DNA-binding domain peptides bind natural p53-response elements and bend the DNA. Proc. Natl. Acad. Sci. USA 1995, 92, 8591–8595. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.; Klein, C.; Müller, L.; Hansen, S.; Buchner, J. p53 Contains Large Unstructured Regions in its Native State. J. Mol. Biol. 2002, 322, 917–927. [Google Scholar] [CrossRef]
- Toledo, F.; Wahl, G.M. Regulating the p53 pathway: In vitro hypotheses, in vivo veritas. Nat. Rev. Cancer 2006, 6, 909–923. [Google Scholar] [CrossRef] [PubMed]
- Lavin, M.F.; Gueven, N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006, 13, 941–950. [Google Scholar] [CrossRef]
- Bode, A.M.; Dong, Z. Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer 2004, 4, 793–805. [Google Scholar] [CrossRef]
- Feng, L.; Lin, T.; Uranishi, H.; Gu, W.; Xu, Y. Functional Analysis of the Roles of Posttranslational Modifications at the p53 C Terminus in Regulating p53 Stability and Activity. Mol. Cell. Biol. 2005, 25, 5389–5395. [Google Scholar] [CrossRef] [PubMed]
- Kastan, M.B.; Onyekwere, O.; Sidransky, D.; Vogelstein, B.; Craig, R.W. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991, 51, 6304–6311. [Google Scholar] [CrossRef]
- Harper, J.W.; Adami, G.R.; Wei, N.; Keyomarsi, K.; Elledge, S.J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 1993, 75, 805–816. [Google Scholar] [CrossRef]
- El-Deiry, W.S.; Tokino, T.; Velculescu, V.E.; Levy, D.B.; Parsons, R.; Trent, J.M.; Lin, D.; Mercer, W.E.; Kinzler, K.W.; Vogelstein, B. WAF1, a potential mediator of p53 tumor suppression. Cell 1993, 75, 817–825. [Google Scholar] [CrossRef]
- Miyashita, T.; Krajewski, S.; Krajewska, M.; Wang, H.G.; Lin, H.K.; A Liebermann, D.; Hoffman, B.; Reed, J.C. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene 1994, 9, 1799–1805. [Google Scholar]
- Nakano, K.; Vousden, K.H. PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell 2001, 3, 683–694. [Google Scholar] [CrossRef]
- Oda, E.; Ohki, R.; Murasawa, H.; Nemoto, J.; Shibue, T.; Yamashita, T.; Tokino, T.; Taniguchi, T.; Tanaka, N. Noxa, a BH3-Only Member of the Bcl-2 Family and Candidate Mediator of p53-Induced Apoptosis. Science 2000, 288, 1053–1058. [Google Scholar] [CrossRef]
- A Boehme, K.; Blattner, C. Regulation of p53—Insights into a complex process. Crit. Rev. Biochem. Mol. Biol. 2009, 44, 367–392. [Google Scholar] [CrossRef]
- Carr, M.I.; Jones, S.N. Regulation of the Mdm2-p53 signaling axis in the DNA damage response and tumorigenesis. Transl. Cancer Res. 2016, 5, 707–724. [Google Scholar] [CrossRef] [PubMed]
- Boehme, K.A.; Kulikov, R.; Blattner, C. p53 stabilization in response to DNA damage requires Akt/PKB and DNA-PK. Proc. Natl. Acad. Sci. USA 2008, 105, 7785–7790. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.K.; Allton, K.L.; Duncan, A.D.; Barton, M.C. TRIM24 Is a p53-Induced E3-Ubiquitin Ligase That Undergoes ATM-Mediated Phosphorylation and Autodegradation during DNA Damage. Mol. Cell. Biol. 2014, 34, 2695–2709. [Google Scholar] [CrossRef] [PubMed]
- Zindy, F.; Eischen, C.M.; Randle, D.H.; Kamijo, T.; Cleveland, J.L.; Sherr, C.J.; Roussel, M.F. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998, 12, 2424–2433. [Google Scholar] [CrossRef]
- Pomerantz, J.; Schreiber-Agus, N.; Liégeois, N.J.; Silverman, A.; Alland, L.; Chin, L.; Potes, J.; Chen, K.; Orlow, I.; Lee, H.-W.; et al. The Ink4a Tumor Suppressor Gene Product, p19Arf, Interacts with MDM2 and Neutralizes MDM2′s Inhibition of p53. Cell 1998, 92, 713–723. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiong, Y.; Yarbrough, W.G. ARF Promotes MDM2 Degradation and Stabilizes p53: ARF-INK4a Locus Deletion Impairs Both the Rb and p53 Tumor Suppression Pathways. Cell 1998, 92, 725–734. [Google Scholar] [CrossRef]
- Weber, J.D.; Taylor, L.J.; Roussel, M.F.; Sherr, C.J.; Bar-Sagi, D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1999, 1, 20–26. [Google Scholar] [CrossRef]
- Marcel, V.; Van Long, F.N.; Diaz, J.-J. 40 Years of Research Put p53 in Translation. Cancers 2018, 10, 152. [Google Scholar] [CrossRef]
- Aylon, Y.; Oren, M. The Paradox of p53: What, How, and Why? Cold Spring Harb. Perspect. Med. 2016, 6, a026328. [Google Scholar] [CrossRef] [PubMed]
- Kastenhuber, E.R.; Lowe, S.W. Putting p53 in Context. Cell 2017, 170, 1062–1078. [Google Scholar] [CrossRef]
- Glickman, M.H.; Ciechanover, A. The Ubiquitin-Proteasome Proteolytic Pathway: Destruction for the Sake of Construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef]
- Hoppe, T. Multiubiquitylation by E4 enzymes: ‘one size’ doesn’t fit all. Trends Biochem. Sci. 2005, 30, 183–187. [Google Scholar] [CrossRef]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.T.; Ciechanover, A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem. Sci. 2017, 42, 873–886. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Shabek, N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu. Rev. Biochem. 2017, 86, 129–157. [Google Scholar] [CrossRef] [PubMed]
- Spratt, D.E.; Walden, H.; Shaw, G.S. RBR E3 ubiquitin ligases: New structures, new insights, new questions. Biochem. J. 2014, 458, 421–437. [Google Scholar] [CrossRef]
- Li, W.; Bengtson, M.H.; Ulbrich, A.; Matsuda, A.; Reddy, V.A.; Orth, A.; Chanda, S.K.; Batalov, S.; Joazeiro, C.A.P. Genome-Wide and Functional Annotation of Human E3 Ubiquitin Ligases Identifies MULAN, a Mitochondrial E3 that Regulates the Organelle’s Dynamics and Signaling. PLoS ONE 2008, 3, e1487. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M. Mechanisms Underlying Ubiquitination. Annu. Rev. Biochem. 2001, 70, 503–533. [Google Scholar] [CrossRef]
- Hatakeyama, S.; Nakayama, K.I. U-box proteins as a new family of ubiquitin ligases. Biochem. Biophys. Res. Commun. 2003, 302, 635–645. [Google Scholar] [CrossRef]
- Haupt, Y.; Maya, R.; Kazaz, A.; Oren, M. Mdm2 promotes the rapid degradation of p53. Nature 1997, 387, 296–299. [Google Scholar] [CrossRef]
- Oliner, J.D.; Pietenpol, J.A.; Thiagalingam, S.; Gyuris, J.; Kinzler, K.W.; Vogelstein, B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 1993, 362, 857–860. [Google Scholar] [CrossRef]
- Honda, R.; Tanaka, H.; Yasuda, H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997, 420, 25–27. [Google Scholar] [CrossRef]
- Wu, X.; Bayle, J.H.; Olson, D.; Levine, A.J. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993, 7, 1126–1132. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.S.; Desterro, J.M.P.; Lain, S.; Lane, D.P.; Hay, R.T. Multiple C-Terminal Lysine Residues Target p53 for Ubiquitin-Proteasome-Mediated Degradation. Mol. Cell. Biol. 2000, 20, 8458–8467. [Google Scholar] [CrossRef]
- Lee, E.-W.; Lee, M.-S.; Camus, S.; Ghim, J.; Yang, M.-R.; Oh, W.; Ha, N.-C.; Lane, D.P.; Song, J. Differential regulation of p53 and p21 by MKRN1 E3 ligase controls cell cycle arrest and apoptosis. EMBO J. 2009, 28, 2100–2113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cui, N.; Zheng, G. Ubiquitination of P53 by E3 ligase MKRN2 promotes melanoma cell proliferation. Oncol. Lett. 2020, 19, 1975–1984. [Google Scholar] [CrossRef] [PubMed]
- Dornan, D.; Wertz, I.; Shimizu, H.; Arnott, D.; Frantz, G.D.; Dowd, P.; O’Rourke, K.; Koeppen, H.; Dixit, V.M. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 2004, 429, 86–92. [Google Scholar] [CrossRef]
- Leng, R.P.; Lin, Y.; Ma, W.; Wu, H.; Lemmers, B.; Chung, S.; Parant, J.M.; Lozano, G.; Hakem, R.; Benchimol, S. Pirh2, a p53-Induced Ubiquitin-Protein Ligase, Promotes p53 Degradation. Cell 2003, 112, 779–791. [Google Scholar] [CrossRef]
- Shloush, J.; Vlassov, J.E.; Engson, I.; Duan, S.; Saridakis, V.; Dhe-Paganon, S.; Raught, B.; Sheng, Y.; Arrowsmith, C.H. Structural and Functional Comparison of the RING Domains of Two p53 E3 Ligases, Mdm2 and Pirh2. J. Biol. Chem. 2011, 286, 4796–4808. [Google Scholar] [CrossRef]
- Duan, W.; Gao, L.; Wu, X.; Zhang, Y.; Otterson, G.A.; Villalona-Calero, M.A. Differential response between the p53 ubiquitin-protein ligases Pirh2 and MdM2 following DNA damage in human cancer cells. Exp. Cell Res. 2006, 312, 3370–3378. [Google Scholar] [CrossRef]
- Yamasaki, S.; Yagishita, N.; Sasaki, T.; Nakazawa, M.; Kato, Y.; Yamadera, T.; Bae, E.; Toriyama, S.; Ikeda, R.; Zhang, L.; et al. Cytoplasmic destruction of p53 by the endoplasmic reticulum-resident ubiquitin ligase ‘Synoviolin’. EMBO J. 2006, 26, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, S.; Nishioka, K.; Nakajima, T.; Yagishita, N. The Roles of Synoviolin in Crosstalk Between Endoplasmic Reticulum Stress-Induced Apoptosis and p53 Pathway. Cell Cycle 2007, 6, 1319–1323. [Google Scholar] [CrossRef]
- Zhou, R.; Wen, H.; Ao, S. Identification of a novel gene encoding a p53-associated protein. Gene 1999, 235, 93–101. [Google Scholar] [CrossRef]
- Rajendra, R.; Malegaonkar, D.; Pungaliya, P.; Henderson, M.; Rasheed, Z.; Brownell, J.; Liu, L.F.; Lutzker, S.; Saleem, A.; Rubin, E.H. Topors functions as an E3 ubiquitin ligase with specific E2 enzymes and ubiq-uitinates p53. J. Biol. Chem. 2004, 279, 36440–36444. [Google Scholar] [CrossRef]
- Yang, X.; Li, H.; Zhou, Z.; Wang, W.-H.; Deng, A.; Andrisani, O.; Liu, X. Plk1-mediated Phosphorylation of Topors Regulates p53 Stability. J. Biol. Chem. 2009, 284, 18588–18592. [Google Scholar] [CrossRef]
- Shen, J.; Li, P.; Shao, X.; Yang, Y.; Liu, X.-J.; Feng, M.; Pengyu, L.; Hu, R.; Wang, Z. The E3 Ligase RING1 Targets p53 for Degradation and Promotes Cancer Cell Proliferation and Survival. Cancer Res. 2018, 78, 359–371. [Google Scholar] [CrossRef]
- Wen, W.; Peng, C.; Kim, M.O.; Jeong, C.H.; Zhu, F.; Yao, K.; Zykova, T.; Ma, W.; Carper, A.; Langfald, A.; et al. Knockdown of RNF2 induces apoptosis by regulating MDM2 and p53 stability. Oncogene 2013, 33, 421–428. [Google Scholar] [CrossRef]
- Su, W.-J.; Fang, J.-S.; Cheng, F.; Liu, C.; Zhou, F.; Zhang, J. RNF2/Ring1b negatively regulates p53 expression in selective cancer cell types to promote tumor development. Proc. Natl. Acad. Sci. USA 2013, 110, 1720–1725. [Google Scholar] [CrossRef]
- Chen, Y.C.; Chan, J.Y.-H.; Chiu, Y.-L.; Liu, S.-T.; Lozano, G.; Wang, S.-L.; Ho, C.-L.; Huang, S.M. Grail as a molecular determinant for the functions of the tumor suppressor p53 in tumorigenesis. Cell Death Differ. 2013, 20, 732–743. [Google Scholar] [CrossRef]
- Allton, K.; Jain, A.K.; Herz, H.-M.; Tsai, W.-W.; Jung, S.Y.; Qin, J.; Bergmann, A.; Johnson, R.L.; Barton, M.C. Trim24 targets endogenous p53 for degradation. Proc. Natl. Acad. Sci. USA 2009, 106, 11612–11616. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Wang, X.L.; Ly, P.; Belyi, V.; Xumonette, Z.Y.; Young, K.H.; Hu, W.; Feng, Z. E3 ubiquitin ligase TRIM32 negatively regulates tumor suppressor p53 to promote tumorigenesis. Cell Death Differ. 2014, 21, 1792–1804. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, N.J.; Chen, C.; Tang, W.; Kornbluth, S. Ubiquitylation of p53 by the APC/C inhibitor Trim39. Proc. Natl. Acad. Sci. USA 2012, 109, 20931–20936. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Ji, Z.; Wang, Y.; Li, J.; Cao, H.; Zhu, H.H.; Gao, W.-Q. TRIM59 is up-regulated in gastric tumors, promoting ubiquitination and degradation of p53. Gastroenterology 2014, 147, 1043–1054. [Google Scholar] [CrossRef]
- Han, R.; Zhao, Q.; Zong, S.; Miao, S.; Song, W.; Wang, L. A novel TRIM family member, Trim69, regulates zebrafish development through p53-mediated apoptosis. Mol. Reprod. Dev. 2016, 83, 442–454. [Google Scholar] [CrossRef] [PubMed]
- Rong, X.; Rao, J.; Li, D.; Jing, Q.; Lu, Y.; Ji, Y. TRIM69 inhibits cataractogenesis by negatively regulating p53. Redox Biol. 2019, 22, 101157. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.T.T.; Richter, D.; Michel, G.; Mitsckha, S.; Kolanus, W.; Cuevas, E.; Wulczyn, F.G. The ubiquitin ligase LIN41/TRIM71 targets p53 to antagonize cell death and differ-entiation pathways during stem cell differentiation. Cell Death Differ. 2017, 24, 1063–1078. [Google Scholar] [CrossRef]
- Yang, W.; Rozan, L.M.; McDonald, E.R., III; Navaraj, A.; Liu, J.J.; Matthew, E.M.; Wang, W.; Dicker, D.T.; El-Deiry, W.S. CARPs are ubiquitin ligases that promote MDM2-independent p53 and phos-pho-p53ser20 degradation. J. Biol. Chem. 2007, 282, 3273–3281. [Google Scholar] [CrossRef]
- Scheffner, M.; Huibregtse, J.M.; Vierstra, R.D.; Howley, P.M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 1993, 75, 495–505. [Google Scholar] [CrossRef]
- Huibregtse, J.M.; Scheffner, M.; Howley, P.M. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol. Cell. Biol. 1993, 13, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Jana, N.R. Regulation of turnover of tumor suppressor p53 and cell growth by E6-AP, a ubiquitin protein ligase mutated in Angelman mental retardation syndrome. Cell. Mol. Life Sci. 2008, 65, 656–666. [Google Scholar] [CrossRef] [PubMed]
- Masuda, Y.; Saeki, Y.; Arai, N.; Kawai, H.; Kukimoto, I.; Tanaka, K.; Masutani, C. Stepwise multipolyubiquitination of p53 by the E6AP-E6 ubiquitin ligase complex. J. Biol. Chem. 2019, 294, 14860–14875. [Google Scholar] [CrossRef]
- Chen, D.; Kon, N.; Li, M.; Zhang, W.; Qin, J.; Gu, W. ARF-BP1/Mule Is a Critical Mediator of the ARF Tumor Suppressor. Cell 2005, 121, 1071–1083. [Google Scholar] [CrossRef] [PubMed]
- Kon, N.; Zhong, J.; Qiang, L.; Accili, D.; Gu, W. Inactivation of arf-bp1 Induces p53 Activation and Diabetic Phenotypes in Mice. J. Biol. Chem. 2012, 287, 5102–5111. [Google Scholar] [CrossRef]
- Maan, M.; Pati, U. CHIP promotes autophagy-mediated degradation of aggregating mutant p53 in hypoxic conditions. FEBS J. 2018, 285, 3197–3214. [Google Scholar] [CrossRef]
- Esser, C.; Scheffner, M.; Höhfeld, J. The chaperone-associated ubiquitin ligase CHIP is able to target p53 for proteasomal degra-dation. J. Biol. Chem. 2005, 280, 27443–27448. [Google Scholar] [CrossRef] [PubMed]
- Sisoula, C.; Trachana, V.; Patterson, C.; Gonos, E.S. CHIP-dependent p53 regulation occurs specifically during cellular senescence. Free Radic. Biol. Med. 2011, 50, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Pomeroy, S.L.; Ferreira, M.; Teider, N.; Mariani, J.; Nakayama, K.I.; Hatakeyama, S.; Tron, V.A.; Saltibus, L.F.; Spyracopoulos, L.; et al. P UBE4B promotes Hdm2-mediated degradation of the tumor suppressor p53. Nat. Med. 2011, 17, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Wu, H.; Leng, R.P. UBE4B targets phosphorylated p53 at serines 15 and 392 for degradation. Oncotarget 2015, 7, 2823–2836. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Joazeiro, C.A. RING Domain E3 Ubiquitin Ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef]
- De Rozieres, S.; Maya, R.; Oren, M.; Lozano, G. The loss of mdm2 induces p53-mediated apoptosis. Oncogene 2000, 19, 1691–1697. [Google Scholar] [CrossRef]
- Sigalas, I.; Calvert, A.H.; Anderson, J.J.; Neal, D.E.; Lunec, J. Alternatively spliced mdm2 transcripts with loss of p53 binding domain sequences: Transforming ability and frequent detection in human cancer. Nat. Med. 1996, 2, 912–917. [Google Scholar] [CrossRef] [PubMed]
- Bartel, F.; Taubert, H.; Harris, L.C. Alternative and aberrant splicing of MDM2 mRNA in human cancer. Cancer Cell 2002, 2, 9–15. [Google Scholar] [CrossRef]
- Volk, E.L.; Schuster, K.; Nemeth, K.M.; Fan, L.; Harris, L.C. MDM2-A, a common Mdm2 splice variant, causes perinatal lethality, reduced longevity and enhanced senescence. Dis. Model. Mech. 2009, 2, 47–55. [Google Scholar] [CrossRef]
- Evans, S.C.; Viswanathan, M.; Grier, J.D.; Narayana, M.; El-Naggar, A.K.; Lozano, G. An alternatively spliced HDM2 product increases p53 activity by inhibiting HDM2. Oncogene 2001, 20, 4041–4049. [Google Scholar] [CrossRef] [PubMed]
- Dang, J.; Kuo, M.-L.; Eischen, C.M.; Stepanova, L.; Sherr, C.J.; Roussel, M.F. The RING domain of Mdm2 can inhibit cell proliferation. Cancer Res. 2002, 62, 1222–1230. [Google Scholar]
- Kim, J.Y.; Lee, R.; Xiao, G.; Forbes, D.; Bargonetti, J. MDM2-C Functions as an E3 Ubiquitin Ligase. Cancer Manag. Res. 2020, 12, 7715–7724. [Google Scholar] [CrossRef]
- Savio, M.G.; Rotondo, G.; Maglie, S.; Rossetti, G.; Pardi, R. COP1D, an alternatively spliced constitutive photomorphogenic-1 (COP1) product, stabilizes UV stress-induced c-Jun through inhibition of full-length COP1. Oncogene 2008, 27, 2401–2411. [Google Scholar] [CrossRef]
- Dong, L.; Liu, Y.; Qiu, Y.; Peng, B.; Yin, J.; Liu, W.; He, X. Identification of Pirh2E and Pirh2F, two additional novel isoforms of Pirh2 ubiquitin ligase from human hepatocellular liver carcinoma cell line. Biomed. Mater. Eng. 2012, 22, 89–95. [Google Scholar] [CrossRef]
- Shi, J.; Huang, Y.; Sheikh, M.S. Identification of Pirh2D, an Additional Novel Isoform of Pirh2 Ubiquitin Ligase. Mol. Cell. Pharmacol. 2010, 2, 21–23. [Google Scholar] [PubMed]
- Corcoran, C.A.; Montalbano, J.; Sun, H.; He, Q.; Huang, Y.; Sheikh, M.S. Identification and Characterization of Two Novel Isoforms of Pirh2 Ubiquitin Ligase That Negatively Regulate p53 Independent of RING Finger Domains. J. Biol. Chem. 2009, 284, 21955–21970. [Google Scholar] [CrossRef]
- Lin, L.; Ozaki, T.; Takada, Y.; Kageyama, H.; Nakamura, Y.; Hata, A.; Zhang, J.; Simonds, W.F.; Nakagawara, A.; Koseki, H. topors, a p53 and topoisomerase I-binding RING finger protein, is a coactivator of p53 in growth suppression induced by DNA damage. Oncogene 2005, 24, 3385–3396. [Google Scholar] [CrossRef]
- Sen, N.; Satija, Y.K.; Das, S. PGC-1α, a Key Modulator of p53, Promotes Cell Survival upon Metabolic Stress. Mol. Cell 2011, 44, 621–634. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, I.; Hochner, H.; Levi, T.; Yelin, R.; Kahan, T.; Mitrani-Rosenbaum, S. Cloning and characterization of a novel human gene RNF38 encoding a conserved pu-tative protein with a RING finger domain. Biochem. Biophys. Res. Commun. 2002, 294, 1169–1176. [Google Scholar] [CrossRef]
- Lunardi, A.; Di Minin, G.; Provero, P.; Ferro, M.D.; Carotti, M.; Del Sal, G.; Collavin, L.L. A genome-scale protein interaction profile of Drosophila p53 uncovers additional nodes of the human p53 network. Proc. Natl. Acad. Sci. USA 2010, 107, 6322–6327. [Google Scholar] [CrossRef]
- Sheren, J.E.; Kassenbrock, C.K. RNF38 encodes a nuclear ubiquitin protein ligase that modifies p53. Biochem. Biophys. Res. Commun. 2013, 440, 473–478. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Seroogy, C.M.; Soares, L.; Ranheim, E.A.; Su, L.; Holness, C.; Bloom, D.; Fathman, C.G. The Gene Related to Anergy in Lymphocytes, an E3 Ubiquitin Ligase, Is Necessary for Anergy Induction in CD4 T Cells. J. Immunol. 2004, 173, 79–85. [Google Scholar] [CrossRef]
- Lee, Y.-Y.; Wang, C.-T.; Huang, S.K.-H.; Wu, W.-J.; Huang, C.-N.; Li, C.-C.; Chan, T.-C.; Liang, P.-I.; Hsing, C.-H.; Li, C.-F. Downregulation of RNF128 Predicts Progression and Poor Prognosis in Patients with Urothelial Carcinoma of the Upper Tract and Urinary Bladder. J. Cancer 2016, 7, 2187–2196. [Google Scholar] [CrossRef]
- Le Douarin, B.; Nielsen, A.L.; Garnier, J.M.; Ichinose, H.; Jeanmougin, F.; Losson, R.; Chambon, P. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996, 15, 6701–6715. [Google Scholar] [CrossRef]
- Khetchoumian, K.; Teletin, M.; Tisserand, J.; Mark, M.; Herquel, B.; Ignat, M.; Zucman-Rossi, J.; Cammas, F.; Lerouge, T.; Thibault, C.; et al. Loss of Trim24 (Tif1alpha) gene function confers oncogenic activity to retinoic acid receptor alpha. Nat. Genet. 2007, 39, 1500–1506. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.N.; Roe, A.E.; Donehower, L.A.; Bradley, A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 1995, 378, 206–208. [Google Scholar] [CrossRef]
- Nicklas, S.; Otto, A.; Wu, X.; Miller, P.; Stelzer, S.; Wen, Y.; Kuang, S.; Wrogemann, K.; Patel, K.; Ding, H.; et al. TRIM32 Regulates Skeletal Muscle Stem Cell Differentiation and Is Necessary for Normal Adult Muscle Regeneration. PLoS ONE 2012, 7, e30445. [Google Scholar] [CrossRef] [PubMed]
- Horn, E.J.; Albor, A.; Liu, Y.; El-Hizawi, S.; VanderBeek, G.E.; Babcock, M.; Bowden, G.T.; Hennings, H.; Lozano, G.; Weinberg, W.C.; et al. RING protein Trim32 associated with skin carcinogenesis has anti-apoptotic and E3-ubiquitin ligase properties. Carcinogenesis 2003, 25, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, Y.; Hu, W.; Feng, Z. TRIM32 is a novel negative regulator of p53. Mol. Cell. Oncol. 2015, 2, e970951. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.J.; Zhang, L.; Tang, W.; Chen, C.; Yang, C.-S.; Kornbluth, S. The Trim39 ubiquitin ligase inhibits APC/CCdh1-mediated degradation of the Bax activator MOAP-1. J. Cell Biol. 2012, 197, 361–367. [Google Scholar] [CrossRef]
- Han, Y.; Li, R.; Gao, J.; Miao, S.; Wang, L. Characterisation of human RING finger protein TRIM69, a novel testis E3 ubiquitin ligase and its subcellular localization. Biochem. Biophys. Res. Commun. 2012, 429, 6–11. [Google Scholar] [CrossRef]
- McDonald, E.R., III; El-Deiry, W.S. Suppression of caspase-8- and -10-associated RING proteins results in sensitization to death ligands and inhibition of tumor cell growth. Proc. Natl. Acad. Sci. USA 2004, 101, 6170–6175. [Google Scholar] [CrossRef]
- Yang, W.; Dicker, D.T.; Chen, J.; El-Deiry, W.S. CARPs enhance p53 turnover by degrading 14-3-3sigma and stabilizing MDM2. Cell Cycle 2008, 7, 670–682. [Google Scholar] [CrossRef]
- Lorenz, S. Structural mechanisms of HECT-type ubiquitin ligases. Biol. Chem. 2018, 399, 127–145. [Google Scholar] [CrossRef]
- Sluimer, J.; Distel, B. Regulating the human HECT E3 ligases. Cell. Mol. Life Sci. 2018, 75, 3121–3141. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Huibregtse, J.M.; Howley, P.M. The human E6-AP gene (UBE3A) encodes three potential protein isoforms generated by differential splicing. Genomics 1997, 41, 263–266. [Google Scholar] [CrossRef]
- Saporita, A.J.; Maggi, L.B.; Apicelli, A.J.; Weber, J.D. Therapeutic Targets in the ARF Tumor Suppressor Pathway. Curr. Med. Chem. 2007, 14, 1815–1827. [Google Scholar] [CrossRef]
- Aravind, L.; Koonin, E.V. The U box is a modified RING finger—A common domain in ubiquitination. Curr. Biol. 2000, 10, R132–R134. [Google Scholar] [CrossRef]
- Ballinger, C.A.; Connell, P.; Wu, Y.; Hu, Z.; Thompson, L.J.; Yin, L.-Y.; Patterson, C. Identification of CHIP, a Novel Tetratricopeptide Repeat-Containing Protein That Interacts with Heat Shock Proteins and Negatively Regulates Chaperone Functions. Mol. Cell. Biol. 1999, 19, 4535–4545. [Google Scholar] [CrossRef] [PubMed]
- Kuilman, T.; Michaloglou, C.; Mooi, W.J.; Peeper, D.S. The essence of senescence. Genes Dev. 2010, 24, 2463–2479. [Google Scholar] [CrossRef]
- Sisoula, C.; Gonos, E.S. CHIP E3 ligase regulates mammalian senescence by modulating the levels of oxidized proteins. Mech. Ageing Dev. 2011, 132, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Tonnessen-Murray, C.A.; Lozano, G.; Jackson, J.G. The Regulation of Cellular Functions by the p53 Protein: Cellular Senescence. Cold Spring Harb. Perspect. Med. 2016, 7, a026112. [Google Scholar] [CrossRef] [PubMed]
- Koegl, M.; Hoppe, T.; Schlenker, S.; Ulrich, H.D.; Mayer, T.U.; Jentsch, S. A Novel Ubiquitination Factor, E4, Is Involved in Multiubiquitin Chain Assembly. Cell 1999, 96, 635–644. [Google Scholar] [CrossRef]
- Kaneko-Oshikawa, C.; Nakagawa, T.; Yamada, M.; Yoshikawa, H.; Matsumoto, M.; Yada, M.; Hatakeyama, S.; Nakayama, K.; Nakayama, K.I. Mammalian E4 Is Required for Cardiac Development and Maintenance of the Nervous System. Mol. Cell. Biol. 2005, 25, 10953–10964. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Leng, R.P. UBE4B, a ubiquitin chain assembly factor, is required for MDM2-mediated p53 polyubiquitination and deg-radation. Cell Cycle 2011, 10, 1912–1915. [Google Scholar] [CrossRef]
- Cardozo, P.; Pagano, M. The SCF ubiquitin ligase: Insights into a molecular machine. Nat. Rev. Mol. Cell. Biol. 2004, 5, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Shi, L.; Li, W.; Yu, W.; Liang, J.; Zhang, H.; Yang, X.; Wang, Y.; Li, R.; Yao, X.; et al. JFK, a Kelch domain-containing F-box protein, links the SCF complex to p53 regulation. Proc. Natl. Acad. Sci. USA 2009, 106, 10195–10200. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Shi, L.; Wang, F.; Huangyang, P.; Si, W.; Yang, J.; Yao, Z.; Shang, Y. Substrate Phosphorylation and Feedback Regulation in JFK-promoted p53 Destabilization. J. Biol. Chem. 2011, 286, 4226–4235. [Google Scholar] [CrossRef]
- Li, M.; Brooks, C.L.; Wu-Baer, F.; Chen, D.; Baer, R.; Gu, W. Mono- versus polyubiquitination: Differential control of p53 fate by Mdm2. Science 2003, 302, 1972–1975. [Google Scholar] [CrossRef] [PubMed]
- Carter, S.; Bischof, O.; Dejean, A.; Vousden, K.H. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat. Cell. Biol. 2007, 9, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Kruse, J.P.; Gu, W. MSL2 promotes Mdm2-independent cytoplasmic localization of p53. J. Biol. Chem. 2009, 284, 3250–3263. [Google Scholar] [CrossRef]
- Laine, A.; Ronai, Z. Regulation of p53 localization and transcription by the HECT domain E3 ligase WWP1. Oncogene 2007, 26, 1477–1483. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Cao, X.; Yuan, F.; Li, G.; Tong, T. Knockdown of WWP1 inhibits growth and induces apoptosis in hepatoma carcinoma cells through the activation of caspase3 and p53. Biochem. Biophys. Res. Commun. 2014, 448, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Kasper, J.S.; Arai, T.; DeCaprio, J.A. A novel p53-binding domain in CUL7. Biochem. Biophys. Res. Commun. 2006, 348, 132–138. [Google Scholar] [CrossRef]
- Andrews, P.; He, Y.J.; Xiong, Y. Cytoplasmic localized ubiquitin ligase cullin 7 binds to p53 and promotes cell growth by antago-nizing p53 function. Oncogene 2006, 25, 4534–4548. [Google Scholar] [CrossRef]
- Le Cam, L.; Linares, L.K.; Paul, C.; Julien, E.; Lacroix, M.; Hatchi, D.; Triboulet, R.; Bossis, G.; Shmuel, A.; Rodriguez, M.S.; et al. E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell 2006, 127, 775–788. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Solozobova, V.; Zhang, P.; Armant, O.; Kuehl, B.; Brenner-Weiss, G.; Blattner, C. p53 is active in murine stem cells and alters the transcriptome in a manner that is remi-niscent of mutant p53. Cell Death Dis. 2015, 6, e1662. [Google Scholar] [CrossRef]
- Maki, C.G. p53 Localization. In p53. Molecular Biology Intelligence Unit; Springer: Boston, MA, USA, 2010; Volume 1, pp. 117–126. [Google Scholar]
- Mendjan, S.; Akhtar, A. The right dose for every sex. Chromosoma 2006, 116, 95–106. [Google Scholar] [CrossRef][Green Version]
- Lai, Z.; Moravcová, S.; Canitrot, Y.; Andrzejewski, L.P.; Walshe, D.M.; Rea, S. Msl2 Is a Novel Component of the Vertebrate DNA Damage Response. PLoS ONE 2013, 8, e68549. [Google Scholar] [CrossRef] [PubMed]
- Ingham, R.J.; Gish, G.; Pawson, T. The Nedd4 family of E3 ubiquitin ligases: Functional diversity within a common modular architecture. Oncogene 2004, 23, 1972–1984. [Google Scholar] [CrossRef]
- Zhi, X.; Chen, C. WWP1: A versatile ubiquitin E3 ligase in signaling and diseases. Cell. Mol. Life Sci. 2012, 69, 1425–1434. [Google Scholar] [CrossRef]
- Chen, J.; Shi, H.; Chen, Y.; Fan, S.; Liu, D.; Li, C. DNA damage induces expression of WWP1 to target ΔNp63α to degradation. PLoS ONE 2017, 12, e0176142. [Google Scholar] [CrossRef] [PubMed]
- Sarikas, A.; Hartmann, T.; Pan, Z.-Q. The cullin protein family. Genome Biol. 2011, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jung, P.; Verdoodt, B.; Bailey, A.; Yates, J.R.; Menssen, A.; Hermeking, H. Induction of Cullin 7 by DNA damage attenuates p53 function. Proc. Natl. Acad. Sci. USA 2007, 104, 11388–11393. [Google Scholar] [CrossRef]
- Sandy, P.; Gostissa, M.; Fogal, V.; De Cecco, L.; Szalay, K.; Rooney, R.J.; Schneider, C.; Del Sal, G. p53 is involved in the p120E4F-mediated growth arrest. Oncogene 2000, 19, 188–199. [Google Scholar] [CrossRef]
- Melchior, F.; Hengst, L. SUMO-1 and p53. Cell Cycle 2002, 1, 243–247. [Google Scholar] [CrossRef]
- Stindt, M.H.; Carter, S.; Vigneron, A.M.; Ryan, K.M.; Vousden, K.H. MDM2 promotes SUMO-2/3 modification of p53 to modulate transcriptional activity. Cell Cycle 2011, 10, 3176–3188. [Google Scholar] [CrossRef]
- Xirodimas, D.P.; Saville, M.K.; Bourdon, J.-C.; Hay, R.T.; Lane, D.P. Mdm2-Mediated NEDD8 Conjugation of p53 Inhibits Its Transcriptional Activity. Cell 2004, 118, 83–97. [Google Scholar] [CrossRef]
- Citro, S.; Chiocca, S. Sumo paralogs: Redundancy and divergencies. Front. Biosci. 2013, 5, 544–553. [Google Scholar] [CrossRef]
- Owerbach, D.; McKay, E.M.; Yeh, E.T.; Gabbay, K.H.; Bohren, K.M. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem. Biophys. Res. Commun. 2005, 337, 517–520. [Google Scholar] [CrossRef]
- Rabut, G.; Peter, M. Function and regulation of protein neddylation. ‘Protein modifications: Beyond the usual suspects’ review series. EMBO Rep. 2008, 9, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; He, Y.; Wang, X.; Liang, Z.; He, G.; Zhang, P.; Zhu, H.; Xu, N.; Liang, S. Protein SUMOylation modification and its associations with disease. Open Biol. 2017, 7, 7. [Google Scholar] [CrossRef]
- Gostissa, M.; Hengstermann, A.; Fogal, V.; Sandy, P.; Schwarz, S.E.; Scheffner, M.; Del Sal, G. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 1999, 18, 6462–6471. [Google Scholar] [CrossRef]
- Weger, S.; Hammer, E.; Heilbronn, R. Topors acts as a SUMO-1 E3 ligase for p53 in vitro and in vivo. FEBS Lett. 2005, 579, 5007–5012. [Google Scholar] [CrossRef] [PubMed]
- Stehmaier, P.; Muller, S. Regulation of p53 family members by the ubiquitin-like SUMO system. DNA Repair (Amsterdam) 2009, 8, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, D.; Müller, S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. USA 2002, 99, 2872–2877. [Google Scholar] [CrossRef]
- Bishof, O.; Schwamborn, K.; Martin, N.; Werner, A.; Sustmann, C.; Grosschedl, R.; Dejean, A. The E3 SUMO ligase PIASγ is a regulator of cellular senescence and apoptosis. Mol. Cell 2006, 22, 783–794. [Google Scholar] [CrossRef]
- Abida, W.M.; Nikolaev, A.; Zhao, W.; Zhang, W.; Gu, W. FBXO11 Promotes the Neddylation of p53 and Inhibits Its Transcriptional Activity. J. Biol. Chem. 2007, 282, 1797–1804. [Google Scholar] [CrossRef]
- Chen, L.; Chen, J. MDM2-ARF complex regulates p53 sumoylation. Oncogene 2003, 22, 5348–5357. [Google Scholar] [CrossRef]
- Stott, F.J.; Bates, S.; James, M.C.; McConnell, B.B.; Starborg, M.; Brookes, S.M.; Palmero, I.; Ryan, K.M.; Hara, E.; Vousden, K.H.; et al. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998, 17, 5001–5014. [Google Scholar] [CrossRef] [PubMed]
- Kwek, S.S.; Derry, J.; Tyner, A.L.; Shen, Z.; Gudkov, A.V. Functional analysis and intracellular localization of p53 modified by SUMO-1. Oncogene 2001, 20, 2587–2599. [Google Scholar] [CrossRef] [PubMed]
- Carter, S.A.; Vousden, K.H. p53-Ubl fusions as models of ubiquitination, sumoylation and neddylation of p53. Cell Cycle 2008, 7, 2519–2528. [Google Scholar] [CrossRef]
- Li, T.; Santockyte, R.; Shen, R.F.; Tekle, E.; Wang, G.; Yang, D.C.H.; Chock, P.B. Expression of SUMO-2/3 induced senescence through p53- and pRB-mediated pathways. J. Biol. Chem. 2006, 281, 36221–36227. [Google Scholar] [CrossRef]
- Batuello, C.N.; Hauck, P.M.; Gendron, J.M.; Lehman, J.A.; Mayo, L.D. Src phosphorylation converts Mdm2 from a ubiquitinating to a neddylating E3 ligase. Proc. Natl. Acad. Sci. USA 2015, 112, 1749–1754. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.H.; Megidish, T.; Xu, C.W. Activation of p53 by Protein Inhibitor of Activated Stat1 (PIAS1). J. Biol. Chem. 2002, 277, 8255–8259. [Google Scholar] [CrossRef] [PubMed]
- Nelson, V.; Davis, G.E.; Maxwell, S.A. A putative protein inhibitor of activated STAT (PIASy) interacts with p53 and inhibits p53-mediated transactivation but not apoptosis. Apoptosis 2001, 6, 221–234. [Google Scholar] [CrossRef]
- Urano, T.; Saito, T.; Tsukui, T.; Fujita, M.; Hosoi, T.; Muramatsu, M.; Ouchi, Y.; Inoue, S. Efp targets 14-3-3 sigma for proteolysis and promotes breast tumour growth. Nature 2002, 417, 871–875. [Google Scholar] [CrossRef]
- Zhang, P.; Elabd, S.; Hammer, S.; Solozobova, V.; Yan, H.; Bartel, F.; Inoue, S.; Henrich, T.; Wittbrodt, J.; Loosli, F.; et al. TRIM25 has a dual function in the p53/Mdm2 circuit. Oncogene 2015, 34, 5729–5738. [Google Scholar] [CrossRef]
- Nikolaev, A.Y.; Li, M.; Puskas, N.; Qin, J.; Gu, W. Parc: A cytoplasmic anchor for p53. Cell 2003, 112, 29–40. [Google Scholar] [CrossRef]
- Pei, X.-H.; Bai, F.; Li, Z.; Smith, M.D.; Whitewolf, G.; Jin, R.; Xiong, Y. Cytoplasmic CUL9/PARC Ubiquitin Ligase Is a Tumor Suppressor and Promotes p53-Dependent Apoptosis. Cancer Res. 2011, 71, 2969–2977. [Google Scholar] [CrossRef]
- Li, Z.; Xiong, Y. Cytoplasmic E3 ubiquitin ligase CUL9 controls cell proliferation, senescence, apoptosis and genome integrity through p53. Oncogene 2017, 36, 5212–5218. [Google Scholar] [CrossRef]
- Iaccarino, L.; Divona, M.; Ottone, T.; Cicconi, L.; Lavorgna, S.; Ciardi, C.; Alfonso, V.; Travaglini, S.; Facchini, L.; Cimino, G.; et al. Identification and monitoring of atypical PML/RARA fusion transcripts in acute pro-myelocytic leukemia. Genes Chromosomes Cancer 2019, 58, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Lallemand-Breitenbach, V.; de Thé, H. PML nuclear bodies. Cold Spring Harb. Perspect. Biol. 2010, 2, a000661. [Google Scholar] [CrossRef]
- Fogal, V.; Gostissa, M.; Sandy, P.; Zacchi, P.; Sternsdorf, T.; Jensen, K.; Pandolfi, P.P.; Will, H.; Schneider, C.; Del Sal, G. Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J. 2000, 19, 6185–6195. [Google Scholar] [CrossRef]
- de Stanchina, E.; Querido, E.; Narita, M.; Davuluri, R.V.; Pandolfi, P.P.; Ferbeyre, G.; Lowe, S.W. PML is a direct p53 target that modulates p53 effector functions. Mol. Cell 2004, 13, 523–535. [Google Scholar] [CrossRef]
- Zapatka, M.; Pathogens, P.; Borozan, I.; Brewer, D.S.; Iskar, M.; Grundhoff, A.; Alawi, M.; Desai, N.; Sültmann, H.; Moch, H.; et al. The landscape of viral associations in human cancers. Nat. Genet. 2020, 52, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Parkin, D.M. The global health burden of infection-associated cancers in the year. Int. J. Cancer 2006, 118, 3030–3044. [Google Scholar] [CrossRef]
- Scheffner, M.; Werness, B.A.; Huibregtse, J.M.; Levine, A.J.; Howley, P.M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 1990, 63, 1129–1136. [Google Scholar] [CrossRef]
- Schwarz, E.; Freese, U.K.; Gissmann, L.; Mayer, W.; Roggenbuck, B.; Stremlau, A.; zur Hausen, H. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature 1985, 314, 111–114. [Google Scholar] [CrossRef]
- Crook, T.; Tidy, J.A.; Vousden, K.F. Degradation of p53 can be targeted by HPV E6 sequences distinct from those required for p53 binding and trans-activation. Cell 1991, 67, 547–556. [Google Scholar] [CrossRef]
- Rowe, W.P.; Huebner, R.J.; Gilmore, L.K.; Parrott, R.H.; Ward, T.G. Isolation of a Cytopathogenic Agent from Human Adenoids Undergoing Spontaneous Degeneration in Tissue Culture. Exp. Biol. Med. 1953, 84, 570–573. [Google Scholar] [CrossRef]
- Lowe, S.W.; E Ruley, H. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 1993, 7, 535–545. [Google Scholar] [CrossRef] [PubMed]
- White, E.; Cipriani, R. Role of adenovirus E1B proteins in transformation: Altered organization of intermediate filaments in transformed cells that express the 19-kilodalton protein. Mol. Cell. Biol. 1990, 10, 120–130. [Google Scholar] [CrossRef]
- Martin, M.E.D.; Berk, A.J. Adenovirus E1B 55K Represses p53 Activation In Vitro. J. Virol. 1998, 72, 3146–3154. [Google Scholar] [CrossRef] [PubMed]
- Querido, E.; Marcellus, R.C.; Lai, A.; Charbonneau, R.; Teodoro, J.G.; Ketner, G.; Branton, P.E. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovi-rus-infected cells. J. Virol. 1997, 71, 3788–3798. [Google Scholar] [CrossRef]
- Moore, M.; Horikoshi, N.; Shenk, T. Oncogenic potential of the adenovirus E4orf6 protein. Proc. Natl. Acad. Sci. USA 1996, 93, 11295–11301. [Google Scholar] [CrossRef] [PubMed]
- Querido, E.; Morrison, M.R.; Chu-Pham-Dang, H.; Thirlwell, S.W.; Boivin, D.; Branton, P.E. Identification of three functions of the adenovirus e4orf6 protein that mediate p53 degradation by the E4orf6-E1B55K complex. J. Virol. 2001, 75, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Müller, S.; Dobner, T. The adenovirus E1B-55K oncoprotein induces SUMO modification of p53. Cell Cycle 2008, 7, 754–758. [Google Scholar] [CrossRef]
- Pennella, M.A.; Liu, Y.; Woo, J.L.; Kim, C.A.; Berk, A.J. Adenovirus E1B 55-Kilodalton Protein Is a p53-SUMO1 E3 Ligase That Represses p53 and Stimulates Its Nuclear Export through Interactions with Promyelocytic Leukemia Nuclear Bodies. J. Virol. 2010, 84, 12210–12225. [Google Scholar] [CrossRef]
- Hay, R.T. SUMO: A history of modification. Mol. Cell 2005, 18, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Raychaudhuri, P.; Rooney, R.; Nevins, J.R. Identification of an E1A-inducible cellular factor that interacts with regulatory se-quences within the adenovirus E4 promoter. EMBO J. 1987, 6, 4073–4081. [Google Scholar] [CrossRef]
- Boutell, C.; Everett, R.D. The Herpes Simplex Virus Type 1 (HSV-1) Regulatory Protein ICP0 Interacts with and Ubiquitinates p53. J. Biol. Chem. 2003, 278, 36596–36602. [Google Scholar] [CrossRef]
- Wilcock, D.; Lane, D.P. Localization of p53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature 1991, 349, 429–431. [Google Scholar] [CrossRef]
- Levrero, M.; Zucman-Rossi, J. Mechanisms of HBV-induced hepatocellular carcinoma. J. Hepatol. 2016, 64, S84–S101. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, K.; Das, P.; Chattopadhyay, N.R.; Mal, S.; Choudhuri, T. The interplay between Epstein-Bar virus (EBV) with the p53 and its homologs during EBV associated malignancies. Heliyon 2019, 5, e02624. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Abate, M.; Rice, P.W.; Cole, C.N. The ability of simian virus 40 large T antigen to immortalize primary mouse embryo fibroblasts cosegregates with its ability to bind to p53. J. Virol. 1991, 65, 6872–6880. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.L.; Gallo, C.V.D.M.; Costa, D.C.; Rangel, L.P. Prion-like aggregation of mutant p53 in cancer. Trends Biochem. Sci. 2014, 39, 260–267. [Google Scholar] [CrossRef]
- Muller, P.A.J.; Vousden, K.H. p53 mutations in cancer. Nat. Cell Biol. 2013, 15, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Bykov, V.J.N.; Eriksson, S.E.; Bianchi, J.; Wiman, K.G. Targeting mutant p53 for efficient cancer therapy. Nat. Rev. Cancer 2018, 18, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Vakifahmetoglu-Norberg, H.; Kim, M.; Xia, H.-G.; Iwanicki, M.P.; Ofengeim, D.; Coloff, J.L.; Pan, L.; Ince, T.A.; Kroemer, G.; Brugge, J.S.; et al. Chaperone-mediated autophagy degrades mutant p53. Genes Dev. 2013, 27, 1718–1730. [Google Scholar] [CrossRef] [PubMed]
- Lukashchuk, N.; Vousden, K.H. Ubiquitination and Degradation of Mutant p53. Mol. Cell. Biol. 2007, 27, 8284–8295. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Jung, Y.-S.; Zhang, Y.; Chen, X. Arsenic Trioxide Reactivates Proteasome-Dependent Degradation of Mutant p53 Protein in Cancer Cells in Part via Enhanced Expression of Pirh2 E3 Ligase. PLoS ONE 2014, 9, e103497. [Google Scholar] [CrossRef]
- Chen, Y.; Hao, Q.; Wang, J.; Li, J.; Huang, C.; Zhang, Y.; Wu, X.; Lu, H.; Zhou, X. Ubiquitin ligase TRIM71 suppresses ovarian tumorigenesis by degrading mutant p53. Cell Death Dis. 2019, 10, 1–14. [Google Scholar] [CrossRef]
- Buschmann, T.; Minamoto, T.; Wagle, N.; Fuchs, S.Y.; Adler, V.; Mai, M.; Ronai, Z. Analysis of JNK, Mdm2 and p14ARF contribution to the regulation of mutant p53 stability. J. Mol. Biol. 2000, 295, 1009–1021. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Marchenko, N.D.; Schulz, R.; Fischer, V.; Velasco-Hernandez, T.; Talos, F.; Moll, U.M. Functional Inactivation of Endogenous MDM2 and CHIP by HSP90 Causes Aberrant Stabilization of Mutant p53 in Human Cancer Cells. Mol. Cancer Res. 2011, 9, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Zhang, Y.; Zhang, J.; Liu, S.; Cho, S.J.; Chen, X. Mutant p53 Protein Is Targeted by Arsenic for Degradation and Plays a Role in Arsenic-mediated Growth Suppression. J. Biol. Chem. 2011, 286, 17478–17486. [Google Scholar] [CrossRef]
- Ray, D.; Ray, P.; Ferrer-Torres, D.; Wang, Z.; Nancarrow, D.; Yoon, H.-W.; Martinho, M.S.; Hinton, T.; Owens, S.; Thomas, D.; et al. Isoforms of RNF128 Regulate the Stability of Mutant P53 in Barrett’s Esophageal Cells. Gastroenterology 2020, 158, 583–597. [Google Scholar] [CrossRef]
- Scheffner, M.; Takahashi, T.; Huibregtse, J.M.; Minna, J.D.; Howley, P.M. Interaction of the human papillomavirus type 16 E6 oncoprotein with wild-type and mutant human p53 proteins. J. Virol. 1992, 66, 5100–5105. [Google Scholar] [CrossRef]
- Medcalf, E.A.; Milner, J. Targeting and degradation of p53 by E6 of human papillomavirus type 16 is preferential for the 1620+ p53 conformation. Oncogene 1993, 8, 2847–2851. [Google Scholar] [PubMed]
- Marcel, V.; Olivier, M.; Mollereau, B.; Hainaut, P.; Bourdon, J.-C. First International p53 Isoforms Meeting: ‘p53 isoforms through evolution: From identification to biological function’. Cell Death Differ. 2011, 18, 563–564. [Google Scholar] [CrossRef] [PubMed]
- Khoury, M.P.; Bourdon, J.-C. The Isoforms of the p53 Protein. Cold Spring Harb. Perspect. Biol. 2009, 2, a000927. [Google Scholar] [CrossRef]
- Camus, S.; Menéndez, S.; Fernandes, K.; Kua, N.; Liu, G.; Xirodimas, D.P.; Lane, D.P.; Bourdon, J.-C. The p53 isoforms are differentially modified by Mdm2. Cell Cycle 2012, 11, 1646–1655. [Google Scholar] [CrossRef][Green Version]
- Bourdon, J.-C.; Fernandes, K.; Murray-Zmijewski, F.; Liu, G.; Diot, A.; Xirodimas, D.P.; Saville, M.K.; Lane, D.P. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005, 19, 2122–2137. [Google Scholar] [CrossRef]
- Horikawa, I.; Fujita, K.; Jenkins, L.M.M.; Hiyoshi, Y.; Mondal, A.M.; Vojtesek, B.; Lane, D.P.; Appella, E.; Harris, C.C. Autophagic degradation of the inhibitory p53 isoform Δ133p53α as a regulatory mechanism for p53-mediated senescence. Nat. Commun. 2014, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Donehower, L.A.; Harvey, M.; Slagle, B.L.; McArthur, M.J.; Montgomery, C.A.M., Jr.; Butel, J.S.; Bradley, A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 1992, 356, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Parant, J.M.; Lang, G.; Chau, P.; Chavez-Reyes, A.; El-Naggar, A.K.; Multani, A.S.; Chang, S.; Lozano, G. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nat. Genet. 2003, 36, 63–68. [Google Scholar] [CrossRef]
- Lukas, J.; Gao, D.Q.; Keshmeshian, M.; Wen, W.H.; Tsao-Wei, D.; Rosenberg, S.; Press, M.F. Alternative and aberrant messenger RNA splicing of the mdm2 oncogene in invasive breast cancer. Cancer Res. 2001, 61, 3212–3219. [Google Scholar]
- Leite, K.R.M.; Franco, M.F.; Srougi, M.; Nesrallah, L.J.; Nesrallah, A.; Bevilacqua, R.G.; Darini, E.; Carvalho, C.M.; Meirelles, M.I.; Santana, I.; et al. Abnormal Expression of MDM2 in Prostate Carcinoma. Mod. Pathol. 2001, 14, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Fattah, G.; Yoffe, B.; Krishnan, B.; Khaoustov, V.; Itani, K. MDM2/p53 protein expression in the development of colorectal adenocarcinoma. J. Gastrointest. Surg. 2000, 4, 109–114. [Google Scholar] [CrossRef]
- Okamoto, H.; Fujishima, F.; Nakamura, Y.; Zuguchi, M.; Miyata, G.; Kamei, T.; Nakano, T.; Katsura, K.; Abe, S.; Taniyama, Y.; et al. Murine double minute 2 and its association with chemoradioresistance of esophageal squamous cell carcinoma. Anticancer Res. 2013, 33, 1463–1471. [Google Scholar]
- Ye, Y.; Li, X.; Yang, J.; Miao, S.; Wang, S.; Chen, Y.; Xia, X.; Wu, X.; Zhang, J.; Zhou, Y.; et al. MDM2 is a useful prognostic biomarker for resectable gastric cancer. Cancer Sci. 2013, 104, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Dworakowska, D.; Jassem, E.; Jassem, J.; Peters, B.; Dziadziuszko, R.; Żylicz, M.; Jakóbkiewicz-Banecka, J.; Kobierska-Gulida, G.; Szymanowska, A.; Skokowski, J.; et al. MDM2 gene amplification: A new independent factor of adverse prognosis in non-small cell lung cancer (NSCLC). Lung Cancer 2004, 43, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Onel, K.; Cordon-Cardo, C. MDM2 and prognosis. Mol. Cancer Res. 2004, 2, 1–8. [Google Scholar]
- Lee, Y.-H.; Andersen, J.B.; Song, H.-T.; Judge, A.D.; Seo, D.; Ishikawa, T.; Marquardt, J.U.; Kitade, M.; Durkin, M.E.; Raggi, C.; et al. Definition of Ubiquitination Modulator COP1 as a Novel Therapeutic Target in Human Hepatocellular Carcinoma. Cancer Res. 2010, 70, 8264–8269. [Google Scholar] [CrossRef]
- Dornan, D.; Bheddah, S.; Newton, K.; Ince, W.; Frantz, G.D.; Dowd, P.; Koeppen, H.; Dixit, V.M.; French, D.M. COP1, the negative regulator of p53, is overexpressed in breast and ovarian adeno-carcinomas. Cancer Res. 2004, 64, 7226–7230. [Google Scholar] [CrossRef]
- Su, C.H.; Zhao, R.; Zhang, F.; Qu, C.; Chen, B.; Feng, Y.-H.; Phan, L.; Chen, J.; Wang, H.; Wang, H.; et al. 14-3-3sigma exerts tumor-suppressor activity mediated by regulation of COP1 stability. Cancer Res. 2011, 71, 884–894. [Google Scholar] [CrossRef]
- Sawada, G.; Ueo, H.; Matsumura, T.; Uchi, R.; Ishibashi, M.; Mima, K.; Kurashige, J.; Takahashi, Y.; Akiyoshi, S.; Sudo, T.; et al. Loss of COP1 expression determines poor prognosis in patients with gastric cancer. Oncol. Rep. 2013, 30, 1971–1975. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-F.; Wang, D.-D.; Zhao, B.-W.; Wang, W.; Huang, C.-Y.; Chen, Y.-M.; Zheng, Y.; Keshari, R.P.; Xia, J.-C.; Zhou, Z.-W. High Level of COP1 Expression is Associated with Poor Prognosis in Primary Gastric Cancer. Int. J. Biol. Sci. 2012, 8, 1168–1177. [Google Scholar] [CrossRef]
- Song, Y.; Liu, Y.; Pan, S.; Xie, S.; Wang, Z.; Zhu, X. Role of the COP1 protein in cancer development and therapy. Semin. Cancer Biol. 2020, 67, 43–52. [Google Scholar] [CrossRef]
- Wang, X.-M.; Yang, L.-Y.; Guo, L.; Fan, C.; Wu, F. p53-induced RING-H2 protein, a novel marker for poor survival in hepatocellular carcinoma after hepatic resection. Cancer 2009, 115, 4554–4563. [Google Scholar] [CrossRef] [PubMed]
- Logan, I.R.; Gaughan, L.; McCracken, S.R.; Sapountzi, V.; Leung, H.Y.; Robson, C.N. Human PIRH2 enhances androgen receptor signaling through inhibition of histone deacetylase 1 and is overexpressed in prostate cancer. Mol. Cell. Biol. 2006, 26, 6502–6510. [Google Scholar] [CrossRef]
- Duan, W.; Gao, L.; Druhan, L.J.; Zhu, W.-G.; Morrison, C.; Otterson, G.A.; Villalona-Calero, M.A. Expression of Pirh2, a Newly Identified Ubiquitin Protein Ligase, in Lung Cancer. J. Natl. Cancer Inst. 2004, 96, 1718–1721. [Google Scholar] [CrossRef]
- Shimada, M.; Kitagawa, K.; Dobashi, Y.; Isobe, T.; Hattori, T.; Uchida, C.; Abe, K.; Kotake, Y.; Oda, T.; Suzuki, H.; et al. High expression of Pirh2, an E3 ligase for p27, is associated with low expression of p27 and poor prognosis in head and neck cancers. Cancer Sci. 2009, 100, 866–872. [Google Scholar] [CrossRef]
- Cooper, C.S.; Foster, C.S. Concepts of epigenetics in prostate cancer development. Br. J. Cancer 2008, 100, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wan, C.; Liu, Y.; Lv, L.; Chen, B.; Ni, R.; Huang, Y.; Li, Y.; Zheng, X.; Yang, D.; et al. Polycomb group oncogene RING1 is over-expressed in non-small cell lung cancer. Pathol. Oncol. Res. 2014, 20, 549–556. [Google Scholar] [CrossRef]
- Sánchez-Beato, M.; Sánchez, E.; González-Carreró, J.; Morente, M.M.; Díez, A.; Sánchez-Verde, L.; Martín, M.C.; Cigudosa, J.C.; Vidal, M.; Piris, M. Ángel Variability in the expression of polycomb proteins in different normal and tumoral tissues. A pilot study using tissue microarrays. Mod. Pathol. 2006, 19, 684–694. [Google Scholar] [CrossRef] [PubMed]
- Rai, K.; Akdemir, K.C.; Kwong, L.N.; Fiziev, P.; Wu, C.-J.; Keung, E.Z.; Sharma, S.; Samant, N.S.; Williams, M.; Axelrad, J.B.; et al. Dual Roles of RNF2 in Melanoma Progression. Cancer Discov. 2015, 5, 1314–1327. [Google Scholar] [CrossRef]
- Saleem, A.; Dutta, J.; Malegaonkar, D.; Rasheed, F.; Rasheed, Z.; Rajendra, R.; Marshall, H.; Luo, M.; Li, H.; Rubin, E.H. The topoisomerase I- and p53-binding protein topors is differentially expressed in normal and malignant human tissues and may function as a tumor suppressor. Oncogene 2004, 23, 5293–5300. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, X.; Huang, Y.; Yang, D.; Li, X.; Liang, J.; Lin, L.; Zhang, M.; Zhong, K.; Liang, B.; Li, J. Overexpression of TRIM24 Is Associated with the Onset and Progress of Human Hepatocellular Carcinoma. PLoS ONE 2014, 9, e85462. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sun, L.; Tang, Z.; Fu, L.; Xu, Y.; Li, Z.; Luo, W.; Qiu, X.; Wang, E. Overexpression of TRIM24 Correlates with Tumor Progression in Non-Small Cell Lung Cancer. PLoS ONE 2012, 7, e37657. [Google Scholar] [CrossRef]
- Chambon, M.; Orsetti, B.; Berthe, M.-L.; Bascoul-Mollevi, C.; Rodriguez, C.; Duong, V.; Gleizes, M.; Thénot, S.; Bibeau, F.; Theillet, C.; et al. Prognostic Significance of TRIM24/TIF-1α Gene Expression in Breast Cancer. Am. J. Pathol. 2011, 178, 1461–1469. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Cao, W.; Li, J.; Song, X.; Mao, L.; Chen, W. TRIM24 Overexpression Is Common in Locally Advanced Head and Neck Squamous Cell Carcinoma and Correlates with Aggressive Malignant Phenotypes. PLoS ONE 2013, 8, e63887. [Google Scholar] [CrossRef]
- Zhang, L.-H.; Yin, A.-A.; Cheng, J.-X.; Huang, H.-Y.; Li, X.-M.; Zhang, Y.-Q.; Han, N.; Zhang, X. TRIM24 promotes glioma progression and enhances chemoresistance through activation of the PI3K/Akt signaling pathway. Oncogene 2014, 34, 600–610. [Google Scholar] [CrossRef]
- Miao, Z.-F.; Wang, Z.; Zhao, T.-T.; Xu, Y.; Wu, J.-H.; Liu, X.-Y.; Xu, H.; You, Y.; Xu, H.-M. TRIM24 is upregulated in human gastric cancer and promotes gastric cancer cell growth and chemoresistance. Virchows Archiv 2015, 466, 525–532. [Google Scholar] [CrossRef]
- Xue, D.; Zhang, X.; Zhang, X.; Liu, J.; Li, N.; Liu, C.; Liu, Y.; Wang, P. Clinical significance and biological roles of TRIM24 in human bladder carcinoma. Tumour Biol. 2015, 36, 6849–6855. [Google Scholar] [CrossRef]
- Ikeda, K.; Orimo, A.; Higashi, Y.; Muramatsu, M.; Inoue, S. Efp as a primary estrogen-responsive gene in human breast cancer. FEBS Lett. 2000, 472, 9–13. [Google Scholar] [CrossRef]
- Suzuki, T.; Urano, T.; Tsukui, T.; Horie-Inoue, K.; Moriya, T.; Ishida, T.; Muramatsu, M.; Ouchi, Y.; Sasano, H.; Inoue, S. Estrogen-Responsive Finger Protein as a New Potential Biomarker for Breast Cancer. Clin. Cancer Res. 2005, 11, 6148–6154. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Kollipara, R.K.; Humphries, C.G.; Ma, S.-H.; Hutchinson, R.; Li, R.; Siddiqui, J.; Tomlins, S.A.; Raj, G.V.; Kittler, R. The ubiquitin ligase TRIM25 targets ERG for degradation in prostate cancer. Oncotarget 2016, 7, 64921–64931. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, M.; Akahira, J.-I.; Suzuki, T.; Inoue, S.; Ito, K.; Moriya, T.; Sasano, H.; Okamura, K.; Yaegashi, N. Expression of estrogen-responsive finger protein (Efp) is associated with advanced disease in human epithelial ovarian cancer. Gynecol. Oncol. 2005, 99, 664–670. [Google Scholar] [CrossRef]
- Sato, W.; Ikeda, K.; Urano, T.; Abe, Y.; Nakasato, N.; Horie-Inoue, K.; Takeda, S.; Inoue, S. Efp promotes in vitro and in vivo growth of endometrial cancer cells along with the activation of nuclear factor-κB signaling. PLoS ONE 2018, 13, e0208351. [Google Scholar]
- Yokoe, T.; Toiyama, Y.; Okugawa, Y.; Tanaka, K.; Ohi, M.; Inoue, Y.; Mohri, Y.; Miki, C.; Kusunoki, M. KAP1 Is Associated With Peritoneal Carcinomatosis in Gastric Cancer. Ann. Surg. Oncol. 2009, 17, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Fu, X.; Cui, Y.; Xu, S.; Xu, Y.; Dong, Q.; Sun, L. Expression of KAP1 in epithelial ovarian cancer and its correlation with drug-resistance. Int. J. Clin. Exp. Med. 2015, 8, 17308–17320. [Google Scholar]
- Qi, Z.-X.; Cai, J.-J.; Chen, L.-C.; Yue, Q.; Gong, Y.; Yao, Y.; Mao, Y. TRIM28 as an independent prognostic marker plays critical roles in glioma progression. J. Neurooncol. 2015, 126, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jiang, J.; Li, Q.; Ma, H.; Xu, Z.; Gao, Y. KAP1 is overexpressed in hepatocellular carcinoma and its clinical significance. Int. J. Clin. Oncol. 2016, 21, 927–933. [Google Scholar] [CrossRef]
- Addison, J.B.; Koontz, C.; Fugett, J.H.; Creighton, C.J.; Chen, N.; Farrugia, M.K.; Padon, R.R.; Voronkova, M.A.; McLaughlin, S.L.; Livengood, R.H.; et al. KAP1 promotes proliferation and metastatic progression of breast cancer cells. Cancer Res. 2014, 75, 344–355. [Google Scholar] [CrossRef]
- Kano, S.; Miyajima, N.; Fukuda, S.; Hatakeyama, S. Tripartite motif protein 32 facilitates cell growth and migration via degradation of Abl-interactor 2. Cancer Res. 2008, 68, 5572–5580. [Google Scholar] [CrossRef]
- Zhao, T.-T.; Jin, F.; Li, J.-G.; Xu, Y.-Y.; Dong, H.-T.; Liu, Q.; Xing, P.; Zhu, G.-L.; Xu, H.; Yin, S.-C.; et al. F. TRIM32 promotes proliferation and confers chemoresistance to breast cancer cells through activation of the NF-κB pathway. J. Cancer 2018, 9, 1349–1356. [Google Scholar] [CrossRef]
- Du, Y.; Zhang, W.; Du, B.; Zang, S.; Wang, X.; Mao, X.; Hu, Z. TRIM32 overexpression improves chemoresistance through regulation of mitochondrial function in non-small-cell lung cancers. OncoTargets Ther. 2018, 11, 7841–7852. [Google Scholar] [CrossRef]
- Liu, Y.; Dong, Y.; Zhao, L.; Su, L.; Diao, K.; Mi, X. TRIM59 overexpression correlates with poor prognosis and contributes to breast cancer progression through AKT signaling pathway. Mol. Carcinog. 2018, 57, 1792–1802. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Weng, L.; Liu, B.; Zhu, L.; Zhang, W.; Tian, G.; Hu, L.; Li, Q.; Jiang, S.; Shang, M. TRIM59 predicts poor prognosis and promotes pancreatic cancer progression via the PI3K/AKT/mTOR-glycolysis signaling axis. J. Cell. Biochem. 2020, 121, 1986–1997. [Google Scholar] [CrossRef]
- Sun, Y.; Ji, B.; Feng, Y.; Zhang, Y.; Ji, D.; Zhu, C.; Wang, S.; Zhang, C.; Zhang, D.; Sun, Y. TRIM59 facilitates the proliferation of colorectal cancer and promotes metastasis via the PI3K/AKT pathway. Oncol. Rep. 2017, 38, 43–52. [Google Scholar] [CrossRef]
- Tan, P.; Ye, Y.; He, L.; Xie, J.; Jing, J.; Ma, G.; Pan, H.; Han, L.; Han, W.; Zhou, Y. TRIM59 promotes breast cancer motility by suppressing p62-selective autophagic degradation of PDCD10. PLoS Biol. 2018, 16, e3000051. [Google Scholar] [CrossRef] [PubMed]
- Beer-Romero, P.; Glass, S.; Rolfe, M. Antisense targeting of E6AP elevates p53 in HPV-infected cells but not in normal cells. Oncogene 1997, 14, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Wolyniec, K.; Shortt, J.; de Stanchina, E.; Levav-Cohen, Y.; Alsheich-Bartok, O.; Louria-Haynon, I.; Corneille, V.; Kumar, B.; Woods, S.J.; Opat, S.; et al. E6AP ubiquitin ligase regulates PML-induced senescence in Myc-driven lymphomagenesis. Blood 2012, 120, 822–832. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Nawaz, Z. E3 ubiquitin protein ligase, E6-associated protein (E6-AP) regulates PI3K-Akt signaling and prostate cell growth. Biochim. Biophys. Acta 2011, 1809, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Gamell, C.; Gulati, T.; Levav-Cohen, Y.; Young, R.J.; Do, H.; Pilling, P.; Takano, E.; Watkins, N.; Fox, S.B.; Russell, P.; et al. Reduced abundance of the E3 ubiquitin ligase E6AP contributes to decreased expression of theINK4/ARFlocus in non–small cell lung cancer. Sci. Signal. 2017, 10, eaaf8223. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, S.; Tufail, R.; El Hokayem, J.; Jorda, M.; Zhao, W.; Reis, Z.; Nawaz, Z. Overexpression of ligase defective E6-associated protein, E6-AP, results in mammary tumorigenesis. Breast Cancer Res. Treat. 2011, 132, 97–108. [Google Scholar] [CrossRef]
- Zhang, X.F.; Pan, Q.Z.; Pan, K.; Weng, D.-S.; Wang, Q.-J.; Zhao, J.-J.; He, J.; Liu, Q.; Wang, D.-D.; Jiang, S.-S.; et al. Expression and prognostic role of ubiquitination factor E4B in primary hepatocellular carci-noma. Mol. Carcinog. 2016, 55, 64–76. [Google Scholar] [CrossRef]
- Xu, J.; Wang, H.; Li, W.; Liu, K.; Zhang, T.; He, Z.; Guo, F. E3 ubiquitin ligase CHIP attenuates cellular proliferation and invasion abilities in triple-negative breast cancer cells. Clin. Exp. Med. 2019, 20, 109–119. [Google Scholar] [CrossRef]
- Xu, J.; Zhou, J.; Dai, H.; Liu, F.; Li, W.; Wang, W.; Guo, F. CHIP functions as an oncogene by promoting colorectal cancer metastasis via activation of MAPK and AKT signaling and suppression of E-cadherin. J. Transl. Med. 2018, 16, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhou, J.; Zhou, P.; Chen, W.; Guo, F. The ubiquitin ligase CHIP inactivates NF-κB signaling and impairs the ability of migration and invasion in gastric cancer cells. Int. J. Oncol. 2015, 46, 2096–2106. [Google Scholar] [CrossRef] [PubMed]
- Canfield, K.; Wells, W.; Geradts, J.; Kinlaw, W.B.; Cheng, C.; Kurokawa, M. Inverse association between MDM2 and HUWE1 protein expression levels in human breast cancer and liposarcoma. Int. J. Clin. Exp. Pathol. 2016, 9, 6342–6349. [Google Scholar] [PubMed]
- Hakem, A.; Bohgaki, M.; Lemmers, B.; Tai, E.; Salmena, L.; Matysiak-Zablocki, E.; Jung, Y.-S.; Karaskova, J.; Kaustov, L.; Duan, S.; et al. Role of Pirh2 in Mediating the Regulation of p53 and c-Myc. PLoS Genet. 2011, 7, e1002360. [Google Scholar] [CrossRef]
- Berchuck, A.; Iversen, E.S.; Lancaster, J.M.; Pittman, J.; Luo, J.; Lee, P.; Murphy, S.K.; Dressman, H.K.; Febbo, P.G.; West, M.; et al. Patterns of Gene Expression That Characterize Long-term Survival in Advanced Stage Serous Ovarian Cancers. Clin. Cancer Res. 2005, 11, 3686–3696. [Google Scholar] [CrossRef] [PubMed]
- Chin, K.; Devries, S.; Fridlyand, J.; Spellman, P.T.; Roydasgupta, R.; Kuo, W.-L.; Lapuk, A.; Neve, R.M.; Qian, Z.; Ryder, T.; et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 2006, 10, 529–541. [Google Scholar] [CrossRef]
- Zhu, K.; Li, J.; Li, J.; Sun, J.; Guo, Y.; Tian, H.; Li, L.; Zhang, C.; Shi, M.; Kong, G.; et al. Ring1 promotes the transformation of hepatic progenitor cells into cancer stem cells through the Wnt/β-catenin signaling pathway. J. Cell. Biochem. 2019, 121, 3941–3951. [Google Scholar] [CrossRef]
- Pierce, S.B.; Stewart, M.D.; Gulsuner, S.; Walsh, T.; Dhall, A.; McClellan, J.M.; Klevit, R.E.; King, M.-C. De novo mutation in RING1 with epigenetic effects on neurodevelopment. Proc. Natl. Acad. Sci. USA 2018, 115, 1558–1563. [Google Scholar] [CrossRef]
- Marshall, H.; Bhaumik, M.; Aviv, H.; Moore, D.; Yao, M.; Dutta, J.; Rahim, H.; Gounder, M.; Ganesan, S.; Saleem, A.; et al. Deficiency of the dual ubiqutin/SUMO ligase Topors results in genetic instability and an increased rate of malignancy in mice. BMC Mol. Biol. 2010, 11, 31. [Google Scholar] [CrossRef]
- Jiang, S.; Minter, L.C.; Stratton, S.A.; Yang, P.; Abbas, A.A.; Akdemir, Z.C.; Pant, V.; Post, S.; Gagea, M.; Lee, R.G.; et al. TRIM24 suppresses development of spontaneous hepatic lipid accumulation and hepa-tocellular carcinoma in mice. J. Hepatol. 2015, 62, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Feldman, I.; Rauscher, F.J., III. Hetero-oligomerization among the TIF family of RBCC/TRIM domain-containing nuclear cofactors: A potential mechanism for regulating the switch between coactivation and corepression. J. Mol. Biol. 2002, 320, 629–644. [Google Scholar] [CrossRef]
- Qin, Y.; Cui, H.; Zhang, H. Overexpression of TRIM25 in Lung Cancer Regulates Tumor Cell Progression. Technol. Cancer Res. Treat. 2016, 15, 707–715. [Google Scholar] [CrossRef]
- Ueyama, K.; Ikeda, K.; Sato, W.; Nakasato, N.; Horie-Inoue, K.; Takeda, S.; Inoue, S. Knockdown of Efp by DNA-modified small interfering RNA inhibits breast cancer cell pro-liferation and in vivo tumor growth. Cancer Gene Ther. 2010, 17, 624–632. [Google Scholar] [CrossRef]
- Zhu, Z.; Wang, Y.; Zhang, C.; Yu, S.; Zhu, Q.; Hou, K.; Yan, B. TRIM25 blockade by RNA interference inhibited migration and invasion of gastric cancer cells through TGF-β signaling. Sci. Rep. 2016, 6, 19070. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, D.-T.; Kurtyka, C.; Rawal, B.; Fulp, W.J.; Haura, E.B.; Cress, W.D. Tripartite Motif Containing 28 (Trim28) Can Regulate Cell Proliferation by Bridging HDAC1/E2F Interactions. J. Biol. Chem. 2012, 287, 40106–40118. [Google Scholar] [CrossRef] [PubMed]
- Bojkowska, K.; Aloisio, F.; Cassano, M.; Kapopoulou, A.; De Sio, F.S.; Zangger, N.; Offner, S.; Cartoni, C.; Thomas, C.; Quenneville, S.; et al. Liver-specific ablation of Krüppel-associated box-associated protein 1 in mice leads to male-predominant hepatosteatosis and development of liver adenoma. Hepatology 2012, 56, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Lazzari, E.; Meroni, G. TRIM32 ubiquitin E3 ligase, one enzyme for several pathologies: From muscular dystrophy to tumours. Int. J. Biochem. Cell Biol. 2016, 79, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Ryu, Y.S.; Lee, Y.; Lee, K.W.; Hwang, C.Y.; Maeng, J.-S.; Kim, J.-H.; Seo, Y.-S.; You, K.-H.; Song, B.; Kwon, K.-S. TRIM32 Protein Sensitizes Cells to Tumor Necrosis Factor (TNFα)-induced Apoptosis via Its RING Domain-dependent E3 Ligase Activity against X-linked Inhibitor of Apoptosis (XIAP). J. Biol. Chem. 2011, 286, 25729–25738. [Google Scholar] [CrossRef] [PubMed]
- Izumi, H.; Kaneko, Y.; Veenstra, J.J.; Gibson, H.M.; Littrup, P.J.; Reyes, J.D.; Cher, M.L.; Takashima, A.; Wei, W.-Z. Trim32 Facilitates Degradation of MYCN on Spindle Poles and Induces Asymmetric Cell Division in Human Neuroblastoma Cells. Cancer Res. 2014, 74, 5620–5630. [Google Scholar] [CrossRef]
- Louria-Hayon, I.; Alsheich-Bartok, O.; Levav-Cohen, Y.; Silberman, I.; Berger, M.; Grossman, T.; Matentzoglu, K.; Jiang, Y.-H.; Muller, S.; Scheffner, M.; et al. E6AP promotes the degradation of the PML tumor suppressor. Cell Death Differ. 2009, 16, 1156–1166. [Google Scholar] [CrossRef]
- Raghu, D.; Paul, P.J.; Gulati, T.; Deb, S.; Khoo, C.; Russo, A.; Gallo, E.; Blandino, G.; Chan, A.-L.; Takano, E.; et al. E6AP promotes prostate cancer by reducing p27 expression. Oncotarget 2017, 8, 42939–42948. [Google Scholar] [CrossRef]
- Nawaz, Z.; Lonard, D.M.; Smith, C.L.; Lev-Lehman, E.; Tsai, S.Y.; Tsai, M.-J.; O’Malley, B.W. The Angelman Syndrome-Associated Protein, E6-AP, Is a Coactivator for the Nuclear Hormone Receptor Superfamily. Mol. Cell. Biol. 1999, 19, 1182–1189. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Luo, K.-J.; Hu, Y.; Yang, H.; Fu, J.-H. Metastatic Lymph Node CHIP Expression is a Potential Prognostic Marker for Resected Esophageal Squamous Cell Carcinoma Patients. Ann. Surg. Oncol. 2013, 20, 1668–1675. [Google Scholar] [CrossRef]
- Wang, Y.; Ren, F.; Wang, Y.; Feng, Y.; Wang, D.; Jia, B.; Qiu, Y.; Wang, S.; Yu, J.; Sung, J.J.; et al. CHIP/Stub1 functions as a tumor suppressor and represses NF-κB-mediated signaling in colo-rectal cancer. Carcinogenesis 2014, 35, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, X.; Zhang, J.; Chen, Y.; Xu, J.; Xia, X.; He, S.; Qiang, F.; Li, A.; Shu, Y.; et al. CHIP functions as a novel suppressor of tumour angiogenesis with prognostic significance in human gastric cancer. Gut 2012, 62, 496–508. [Google Scholar] [CrossRef]
- Mokbel, K.; Patani, N.; Jiang, W.G.; Newbold, R. Prognostic implications of carboxyl-terminus of Hsc70 interacting protein and lysyl-oxidase expression in human breast cancer. J. Carcinog. 2010, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Brautigan, D.L.; Parsons, S.J.; Larner, J.M. Androgen receptor degradation by the E3 ligase CHIP modulates mitotic arrest in prostate cancer cells. Oncogene 2014, 33, 26–33. [Google Scholar] [CrossRef]
- Sun, Y. Targeting E3 Ubiquitin Ligases for Cancer Therapy. Cancer Biol. Ther. 2003, 2, 621–627. [Google Scholar] [CrossRef]
- Tortora, G.; Caputo, R.; Damiano, V.; Bianco, R.; Chen, J.; Agrawal, S.; Bianco, A.R.; Ciardiello, F. A novel MDM2 anti-sense oligonucleotide has anti-tumor activity and potentiates cytotoxic drugs acting by different mechanisms in human colon cancer. Int. J. Cancer 2000, 88, 804–809. [Google Scholar] [CrossRef]
- Yang, Y.; Ludwig, R.L.; Jensen, J.P.; Pierre, S.A.; Medaglia, M.V.; Davydov, I.V.; Safiran, Y.J.; Oberoi, P.; Kenten, J.H.; Phillips, A.C.; et al. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell 2005, 7, 547–559. [Google Scholar] [CrossRef] [PubMed]
- Vassilev, L.T.; Vu, B.T.; Graves, B.; Carvajal, D.; Podlaski, F.; Filipovic, Z.; Kong, N.; Kammlott, U.; Lukacs, C.; Klein, C.; et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004, 303, 844–848. [Google Scholar] [CrossRef]
- Grasberger, B.L.; Lu, T.; Schubert, C.; Parks, D.J.; Carver, T.E.; Koblish, H.K.; Cummings, M.D.; Lafrance, L.V.; Milkiewicz, K.L.; Calvo, R.R.; et al. Discovery and Cocrystal Structure of Benzodiazepinedione HDM2 Antagonists That Activate p53 in Cells. J. Med. Chem. 2005, 48, 909–912. [Google Scholar] [CrossRef]
- Ding, K.; Lu, Y.; Nikolovska-Coleska, Z.; Wang, G.; Qiu, S.; Shangary, S.; Gao, W.; Qin, D.; Stuckey, J.; Krajewski, K.; et al. Structure-Based Design of Spiro-oxindoles as Potent, Specific Small-Molecule Inhibitors of the MDM2−p53 Interaction. J. Med. Chem. 2006, 49, 3432–3435. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Nikolovska-Coleska, Z.; Fang, X.; Gao, W.; Shangary, S.; Qiu, S.; Qin, D.; Wang, S. Discovery of a Nanomolar Inhibitor of the Human Murine Double Minute 2 (MDM2)−p53 Interaction through an Integrated, Virtual Database Screening Strategy. J. Med. Chem. 2006, 49, 3759–3762. [Google Scholar] [CrossRef]
- Issaeva, N.; Bozko, P.; Enge, M.; Protopopova, M.; Verhoef, L.G.G.C.; Masucci, M.G.; Pramanik, A.; Selivanova, G. Small molecule RITA binds to p53, blocks p53–HDM-2 interaction and activates p53 function in tumors. Nat. Med. 2004, 10, 1321–1328. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.-J.; Nag, S.; Voruganti, S.; Wang, W.; Zhang, R. Natural Product MDM2 Inhibitors: Anticancer Activity and Mechanisms of Action. Curr. Med. Chem. 2012, 19, 5705–5725. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, Z.; Hill, D.L.; Chen, X.; Wang, H.; Zhang, R. Genistein, a Dietary Isoflavone, Down-Regulates the MDM2 Oncogene at Both Transcriptional and Posttranslational Levels. Cancer Res. 2005, 65, 8200–8208. [Google Scholar] [CrossRef]
- Fang, J.; Xia, C.; Cao, Z.; Zheng, J.Z.; Reed, E.; Jiang, B.-H. Apigenin inhibits VEGF and HIF-1 expression via PI3K/AKT/p70S6K1 and HDM2/p53 pathways. FASEB J. 2004, 19, 342–353. [Google Scholar] [CrossRef]
- Mu, R.; Qi, Q.; Gu, H.; Wang, J.; Yang, Y.; Rong, J.; Liu, W.; Lu, N.; You, Q.; Guo, Q. Involvement of p53 in oroxylin A-induced apoptosis in cancer cells. Mol. Carcinog. 2009, 48, 1159–1169. [Google Scholar] [CrossRef]
- Nag, S.A.; Qin, J.-J.; Wang, W.; Wang, M.-H.; Wang, H.; Zhang, R. Ginsenosides as anticancer agents: In vitro and in vivo activities, structure–activity relationships, and molecular mechanisms of action. Front. Pharmacol. 2012, 3, 25. [Google Scholar] [CrossRef]
- Zhang, X.; Gu, L.; Li, J.; Shah, N.; He, J.; Yang, L.; Hu, Q.; Zhou, M. Degradation of MDM2 by the Interaction between Berberine and DAXX Leads to Potent Apoptosis in MDM2-Overexpressing Cancer Cells. Cancer Res. 2010, 70, 9895–9904. [Google Scholar] [CrossRef]
- Gu, H.; Wang, X.; Rao, S.; Wang, J.; Zhao, J.; Ren, F.L.; Mu, R.; Yang, Y.; Qi, Q.; Liu, W.; et al. Gambogic acid mediates apoptosis as a p53 inducer through down-regulation of mdm2 in wild-type p53-expressing cancer cells. Mol. Cancer Ther. 2008, 7, 3298–3305. [Google Scholar] [CrossRef]
- Li, M.; Zhang, Z.; Hill, D.L.; Wang, H.; Zhang, R. Curcumin, a Dietary Component, Has Anticancer, Chemosensitization, and Radiosensitization Effects by Down-regulating the MDM2 Oncogene through the PI3K/mTOR/ETS2 Pathway. Cancer Res. 2007, 67, 1988–1996. [Google Scholar] [CrossRef] [PubMed]
- Gopal, Y.V.; Chanchorn, E.; Van Dyke, M.W. Parthenolide promotes the ubiquitination of MDM2 and activates p53 cellular functions. Mol. Cancer Ther. 2009, 8, 552–562. [Google Scholar] [CrossRef] [PubMed]
- Sasiela, C.A.; Stewart, D.H.; Kitagaki, J.; Safiran, Y.J.; Yang, Y.; Weissman, A.M.; Oberoi, P.; Davydov, I.V.; Goncharova, E.; Beutler, J.A.; et al. Identification of Inhibitors for MDM2 Ubiquitin Ligase Activity from Natural Product Extracts by a Novel High-Throughput Electrochemiluminescent Screen. J. Biomol. Screen. 2008, 13, 229–237. [Google Scholar] [CrossRef]
- Clement, J.A.; Kitagaki, J.; Yang, Y.; Saucedo, C.J.; O’Keefe, B.R.; Weissman, A.M.; McKee, T.C.; McMahon, J.B. Discovery of new pyridoacridine alkaloids from Lissoclinum cf. badium that inhibit the ubiquitin ligase activity of Hdm2 and stabilize p53. Bioorg. Med. Chem. 2008, 16, 10022–10028. [Google Scholar] [CrossRef] [PubMed]
- Stoll, R.; Renner, C.; Hansen, S.; Palme, S.; Klein, C.; Belling, A.; Zeslawski, W.; Kamionka, M.; Rehm, T.; Mühlhahn, P.; et al. Chalcone derivatives antagonize interactions between the human oncoprotein MDM2 and p53. Biochemistry 2001, 40, 336–344. [Google Scholar] [CrossRef]
- Tsukamoto, S.; Yoshida, T.; Hosono, H.; Ohta, T.; Yokosawa, H. Hexylitaconic acid: A new inhibitor of p53-HDM2 interaction isolated from a marine-derived fungus, Arthrinium sp. Bioorg. Med. Chem. Lett. 2006, 16, 69–71. [Google Scholar] [CrossRef]
- Duncan, S.J.; Grüschow, S.; Williams, D.H.; McNicholas, C.; Purewal, R.; Hajek, M.; Gerlitz, M.; Martin, S.; Wrigley, A.S.K.; Moore, M. Isolation and Structure Elucidation of Chlorofusin, a Novel p53-MDM2 Antagonist from a Fusarium sp. J. Am. Chem. Soc. 2001, 123, 554–560. [Google Scholar] [CrossRef]
- Malloy, K.L.; Choi, H.; Fiorilla, C.; Valeriote, F.A.; Matainaho, T.; Gerwick, W.H. Hoiamide D, a marine cyanobacteria-derived inhibitor of p53/MDM2 interaction. Bioorg. Med. Chem. Lett. 2012, 22, 683–688. [Google Scholar] [CrossRef]
- Huang, L.; Kinnucan, E.; Wang, G.; Beaudenon, S.; Howley, P.M.; Huibregtse, J.M.; Pavletich, N.P. Structure of an E6AP-UbcH7 complex: Insights into ubiquitination by the E2-E3 enzyme cascade. Science 1999, 286, 1321–1326. [Google Scholar] [CrossRef]
- Zhang, W.; Wu, K.-P.; Sartori, M.A.; Kamadurai, H.B.; Ordureau, A.; Jiang, C.; Mercredi, P.Y.; Murchie, R.; Hu, J.; Persaud, A.; et al. System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes. Mol. Cell 2016, 62, 121–136. [Google Scholar] [CrossRef]
- Mund, T.; Lewis, M.J.; Maslen, S.; Pelham, H.R. Peptide and small molecule inhibitors of HECT-type ubiquitin ligases. Proc. Natl. Acad. Sci. USA 2014, 111, 16736–16741. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Marchenko, N.; Moll, U.M. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ. 2011, 18, 1904–1913. [Google Scholar] [CrossRef] [PubMed]
- Léveillard, T.; Gorry, P.; Niederreither, K.; Wasylyk, B. MDM2 expression during mouse embryogenesis and the requirement of p53. Mech. Dev. 1998, 74, 189–193. [Google Scholar] [CrossRef]
- Rolfo, A.; Garcia, J.; Todros, T.; Post, M.; Caniggia, I. The double life of MULE in preeclamptic and IUGR placentae. Cell Death Dis. 2012, 3, e305. [Google Scholar] [CrossRef]
- Zeinab, R.A.; Wu, H.; Sergi, C.M.; Leng, R.P. UBE4B: A Promising Regulatory Molecule in Neuronal Death and Survival. Int. J. Mol. Sci. 2012, 13, 16865–16879. [Google Scholar] [CrossRef]
- Dornan, D.; Shimizu, H.; Mah, A.; Dudhela, T.; Eby, M.; O’Rourke, K.; Seshagiri, S.; Dixit, V.M. ATM Engages Autodegradation of the E3 Ubiquitin Ligase COP1 After DNA Damage. Science 2006, 313, 1122–1126. [Google Scholar] [CrossRef] [PubMed]

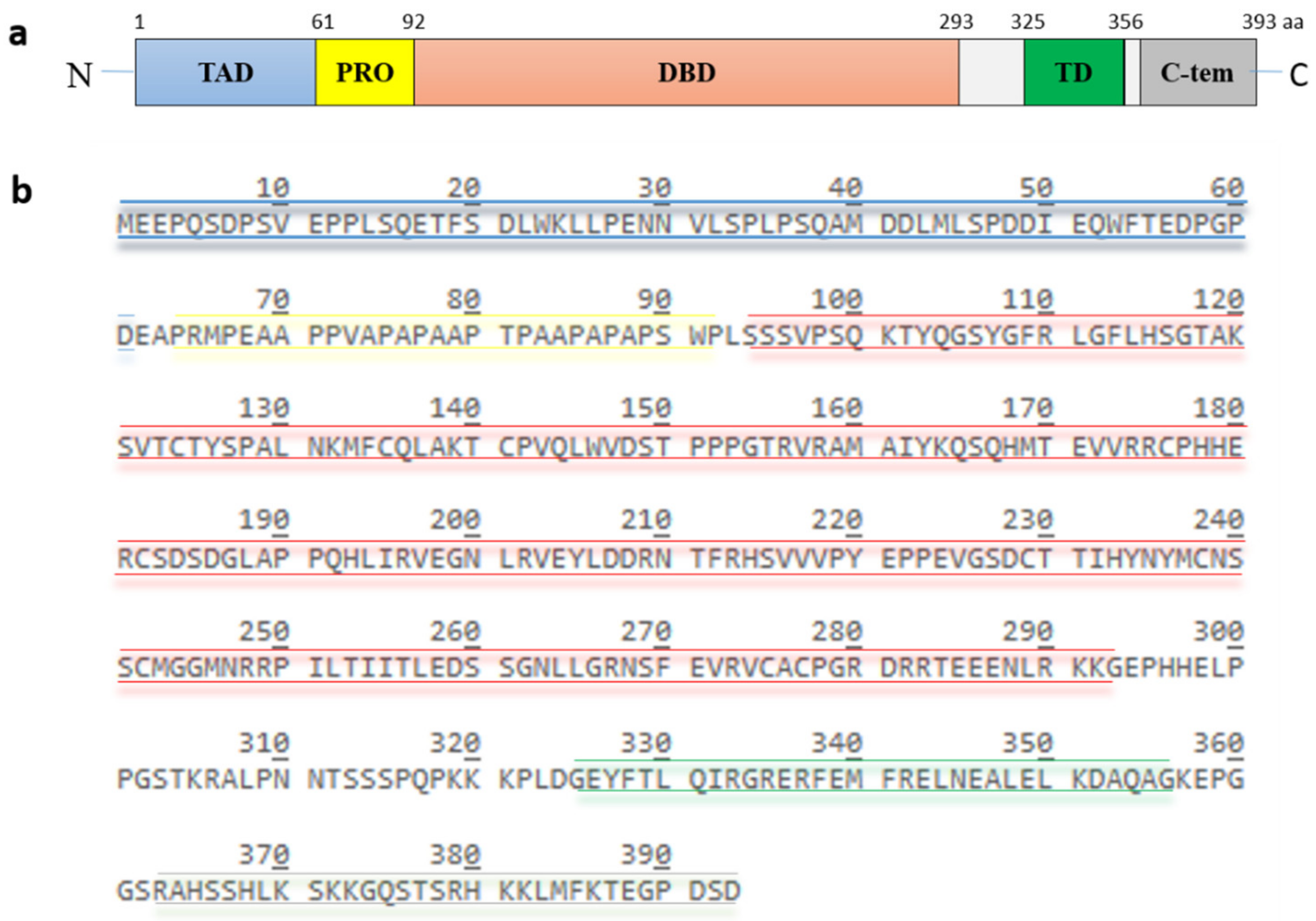



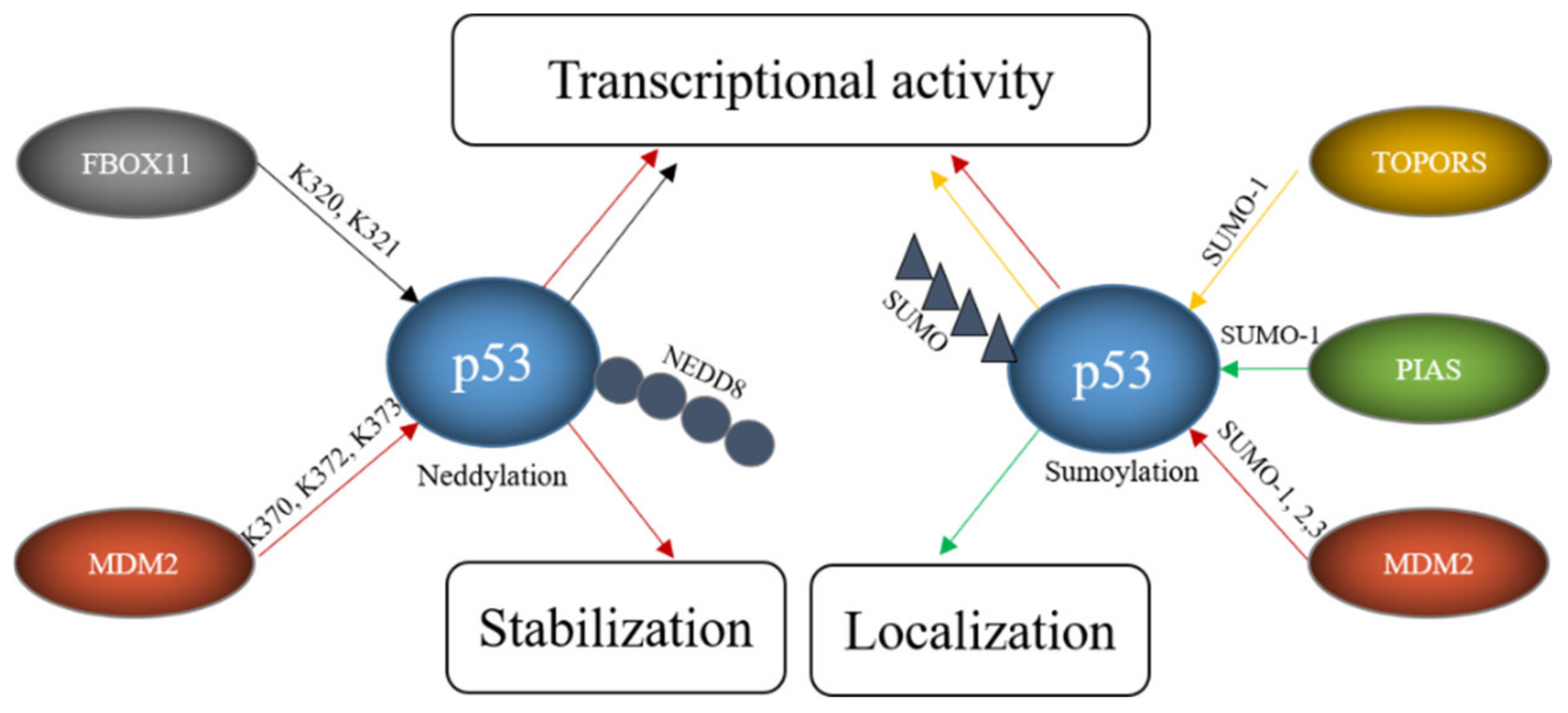
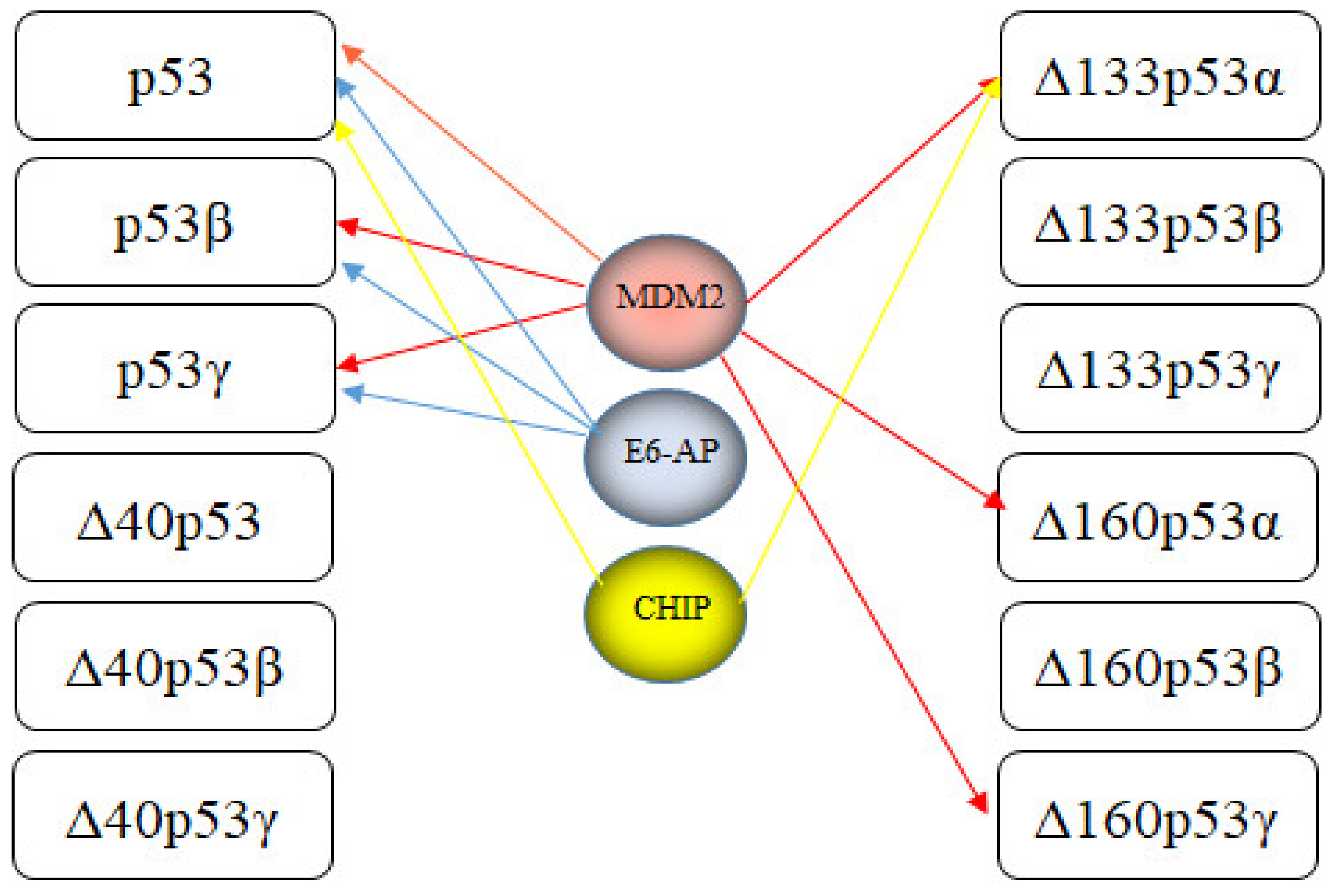
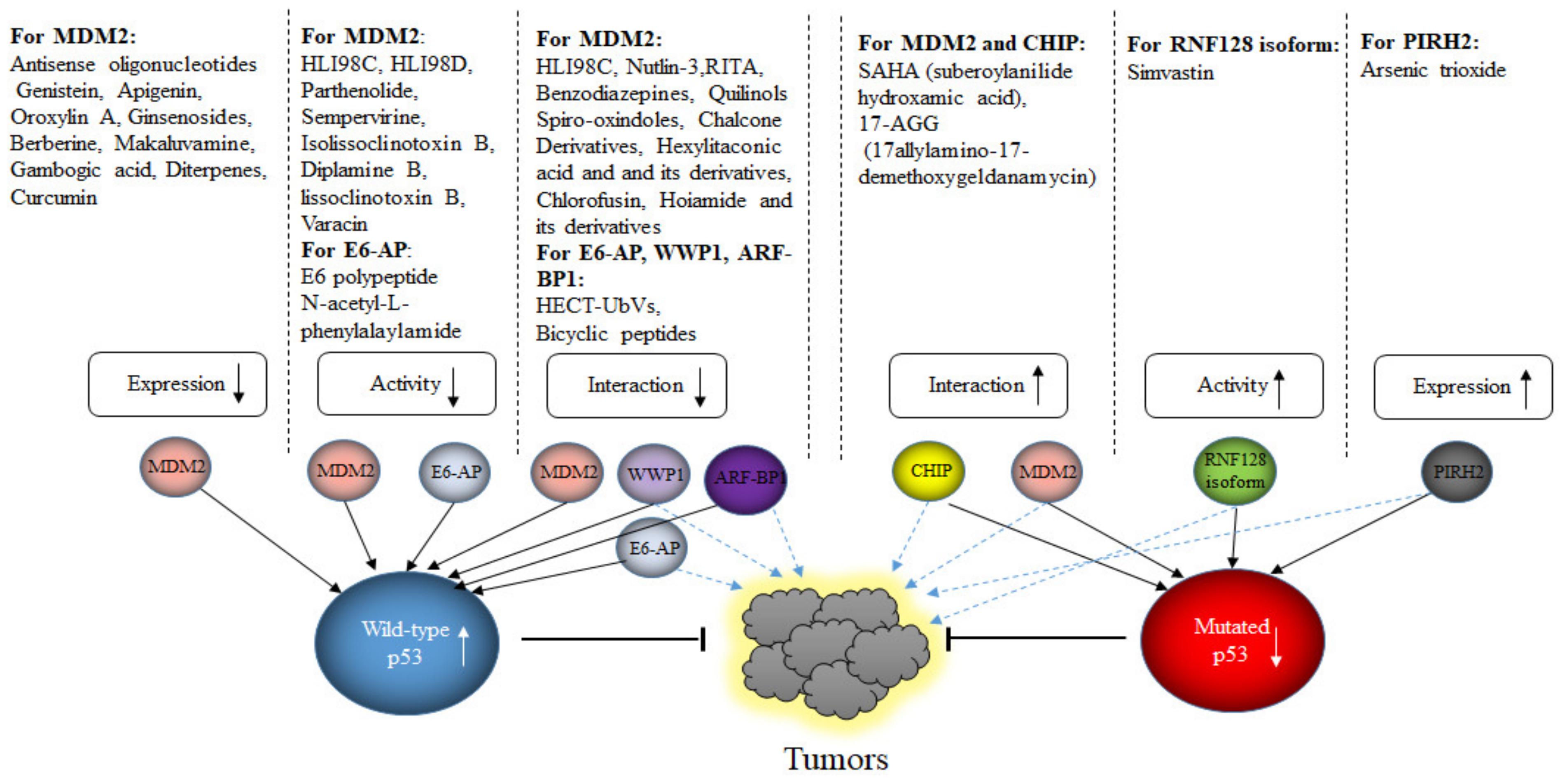
| E3 | Alias | Type | References |
|---|---|---|---|
| MDM2 | Hdm2 | RING | [55,56,57,58,59] |
| MKRN1 | RNF61 | RING | [60] |
| MKRN2 | RNF62 | RING | [61] |
| COP1 | RNF200 | RING | [62] |
| PIRH2 | ZN363 | RING | [63,64,65] |
| Synoviolin | HRD1 | RING | [66,67] |
| TOPORS | P53BP3 | RING | [68,69,70] |
| RNF1 | RING1 | RING | [71] |
| RNF2 | RING1B | RING | [72,73] |
| RNF128 | Grail | RING | [74] |
| TRIM24 | TIF1A | RING | [75] |
| TRIM32 | HT2A | RING | [38,76] |
| TRIM39 | RNF23 | RING | [77] |
| TRIM59 | RNF104 | RING | [78] |
| TRIM 69 | RNF36 | RING | [79,80] |
| TRIM71 | Lin41 | RING | [81] |
| CARP1/2 | RNF34/RNF189 | RING | [82] |
| UBE3A | E6-AP | HECT | [83,84,85,86] |
| ARF-BP1 | HUWE1 | HECT | [87,88] |
| CHIP | UBOX1 | U-box | [89,90,91] |
| UBE4B | UBOX3 | U-box | [92,93] |
| E3 | Alias | Type | Impact on p53 | References | ||
|---|---|---|---|---|---|---|
| Nuclear export | Transcriptional activity | Protein stabilization | ||||
| MDM2 | Hdm2 | RING | √ | √ | [138,139] | |
| MSL2 | RNF184 | RING | √ | [140] | ||
| WWP1 | AIP5 | HECT | √ | √ | √ | [141,142] |
| CUL7 | KIAA0076 | - | √ | [143,144] | ||
| E4F1 | E4F | RING | √ | [145] | ||
| E3s | Type | Sumoylation | Neddylation | Impact on p53 | References | ||
|---|---|---|---|---|---|---|---|
| Protein stabilization | Subcellular Localization | Transcriptional activity | |||||
| MDM2 | RING | √ | √ | √ | √ | [157,158,163], | |
| TOPORS | RING | √ | √ | √ | [164] | ||
| PIAS-4 | RING | √ | √ | √ | [165,166,167] | ||
| FBXO11 | F-Box | √ | √ | √ | [168] | ||
| E3 | Modulation in Cancers | References |
|---|---|---|
| Mdm2 | breast cancer, prostate cancer, colorectal adenocarcinoma, esophageal squamous cell carcinoma, gastric cancer, non-small cell lung cancer | [228,229,230,231,232,233] |
| COP1 | breast adenocarcinomas, ovarian adenocarcinomas, pancreatic cancer, hepatocellular carcinoma, gastric cancer | [235,236,237] |
| PIRH2 | hepatocellular carcinoma, head and neck cancer, prostate cancer, lung cancer, prostate cancer, ovarian cancer, breast cancer | [241,242,243,244] |
| RNF1 | non-small cell lung cancer, hepatocellular cancer, prostate cancer | [245,246] |
| RNF2 | colon cancer, gastric cancer B-cell lymphoma, Burkitt’s lymphoma, Hodgkin’s lymphomas, Melanoma | [247,248] |
| TOPORS | colon adenocarcinoma, breast cancer, | [249] |
| TRIM24 | non-small cell lung cancer, chronic myeloid leukemia, head and neck cancer squamous cell carcinoma, glioma, gastric cancer, bladder cancer, hepatocellular carcinoma | [250,251,252,253,254,255,256] |
| TRIM25 | endometrial cancer, ovary cancer, prostate cancer, lung cancer, breast cancer | [257,258,259,260,261] |
| TRIM28 | gastric cancer, ovarian cancer, glioma, hepatocellular carcinoma, breast cancer | [262,263,264,265,266] |
| TRIM32 | breast cancer, skin cancer, head and neck cancer, non-small cell lung cancer | [267,268,269,270] |
| TRIM59 | colorectal cancer, gastric cancer, pancreatic cancer | [78,270,271,272,273] |
| UBE3A | HPV-associated cervical cancer, Burkitt’s lymphoma, prostate cancer, non-small cell lung cancer, breast cancer | [121,274,275,276,277,278] |
| UBE4B | hepatocellular carcinoma, brain tumors | [132,279] |
| CHIP | breast cancer, gastric cancer, colorectal cancer, esophageal squamous cell carcinoma | [280,281,282] |
| ARF-BP1 | breast cancer, liposarcoma | [283] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, M.; Blattner, C. Regulation of p53 by E3s. Cancers 2021, 13, 745. https://doi.org/10.3390/cancers13040745
Pan M, Blattner C. Regulation of p53 by E3s. Cancers. 2021; 13(4):745. https://doi.org/10.3390/cancers13040745
Chicago/Turabian StylePan, Mengwu, and Christine Blattner. 2021. "Regulation of p53 by E3s" Cancers 13, no. 4: 745. https://doi.org/10.3390/cancers13040745
APA StylePan, M., & Blattner, C. (2021). Regulation of p53 by E3s. Cancers, 13(4), 745. https://doi.org/10.3390/cancers13040745






