Genetic Variations and Health-Related Quality of Life (HRQOL): A Genome-Wide Study Approach
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
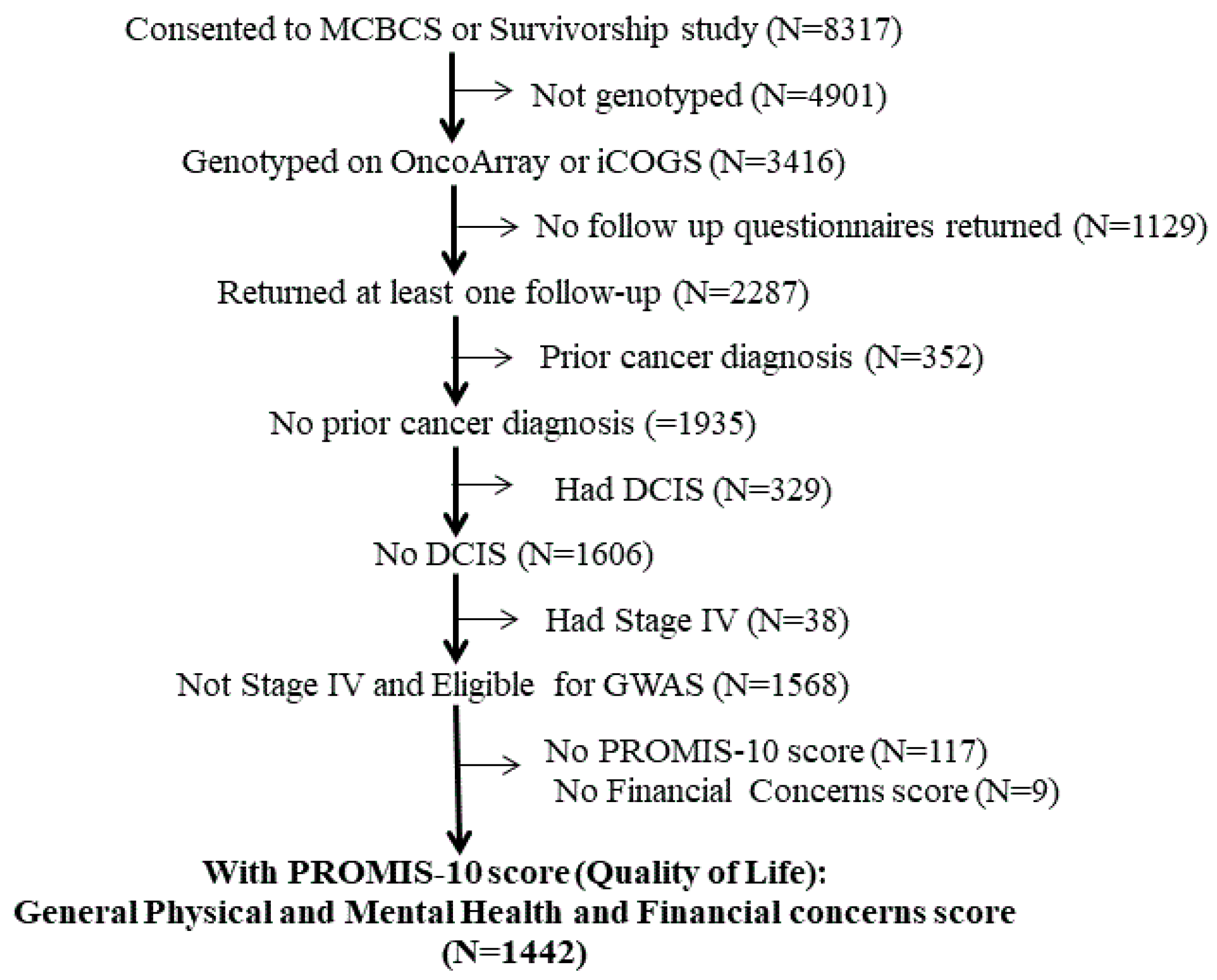
2.1. Patient Characteristics
2.2. GWAS Results
3. Discussion
4. Materials and Methods
4.1. Patient Cohort
4.2. Quality of Life (QOL) Assessments
4.3. Genotyping, Quality Control, and Imputation
4.4. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schoormans, D.; Li, J.; Darabi, H.; Brandberg, Y.; Sprangers, M.A.; Eriksson, M.; Zwinderman, K.H.; Hall, P. The genetic basis of quality of life in healthy Swedish women: A candidate gene approach. PLoS ONE 2015, 10, e0118292. [Google Scholar] [CrossRef]
- Sprangers, M.A.; Thong, M.S.; Bartels, M.; Barsevick, A.; Ordoñana, J.; Shi, Q.; Wang, X.S.; Klepstad, P.; Wierenga, E.A.; Singh, J.A.; et al. Biological pathways, candidate genes, and molecular markers associated with quality-of-life domains: An update. Qual. Life Res. 2014, 23, 1997–2013. [Google Scholar] [CrossRef]
- Kullowatz, A.; Kanniess, F.; Dahme, B.; Magnussen, H.; Ritz, T. Association of depression and anxiety with health care use and quality of life in asthma patients. Respir. Med. 2007, 101, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Pelle, A.J.; Pedersen, S.S.; Szabó, B.M.; Denollet, J. Beyond Type D personality: Reduced positive affect (anhedonia) predicts impaired health status in chronic heart failure. Qual. Life Res. 2009, 18, 689–698. [Google Scholar] [CrossRef]
- Wald, A.; Sigurdsson, L. Quality of life in children and adults with constipation. Best Pract. Res. Clin. Gastroenterol. 2011, 25, 19–27. [Google Scholar] [CrossRef]
- Schoormans, D.; Mulder, B.J.; van Melle, J.P.; Pieper, P.G.; van Dijk, A.P.; Sieswerda, G.T.; Hulsbergen-Zwarts, M.S.; Plokker, T.H.; Brunninkhuis, L.G.; Vliegen, H.W.; et al. Illness perceptions of adults with congenital heart disease and their predictive value for quality of life two years later. Eur. J. Cardiovasc. Nurs. 2014, 13, 86–94. [Google Scholar] [CrossRef]
- Lewandowska, A.; Rudzki, G.; Lewandowski, T.; Prochnicki, M.; Rudzki, S.; Laskowska, B.; Brudniak, J. Quality of Life of Cancer Patients Treated with Chemotherapy. Int. J. Environ. Res. Public Health 2020, 17, 6938. [Google Scholar] [CrossRef]
- Hamilton, J.G.; Wu, L.M.; Austin, J.E.; Valdimarsdottir, H.; Basmajian, K.; Vu, A.; Rowley, S.D.; Isola, L.; Redd, W.H.; Rini, C. Economic survivorship stress is associated with poor health-related quality of life among distressed survivors of hematopoietic stem cell transplantation. Psychooncology 2013, 22, 911–921. [Google Scholar] [CrossRef]
- Hastert, T.A.; Kyko, J.M.; Reed, A.R.; Harper, F.W.K.; Beebe-Dimmer, J.L.; Baird, T.E.; Schwartz, A.G. Financial Hardship and Quality of Life among African American and White Cancer Survivors: The Role of Limiting Care Due to Cost. Cancer Epidemiol. Biomark. Prev. 2019, 28, 1202–1211. [Google Scholar] [CrossRef]
- Brandolini, L.; d’Angelo, M.; Antonosante, A.; Allegretti, M.; Cimini, A. Chemokine Signaling in Chemotherapy-Induced Neuropathic Pain. Int. J. Mol. Sci. 2019, 20, 2904. [Google Scholar] [CrossRef]
- Nicholl, B.I.; Holliday, K.L.; Macfarlane, G.J.; Thomson, W.; Davies, K.A.; O’Neill, T.W.; Bartfai, G.; Boonen, S.; Casanueva, F.; Finn, J.D.; et al. No evidence for a role of the catechol-O-methyltransferase pain sensitivity haplotypes in chronic widespread pain. Ann. Rheum. Dis. 2010, 69, 2009–2012. [Google Scholar] [CrossRef]
- Tander, B.; Gunes, S.; Boke, O.; Alayli, G.; Kara, N.; Bagci, H.; Canturk, F. Polymorphisms of the serotonin-2A receptor and catechol-O-methyltransferase genes: A study on fibromyalgia susceptibility. Rheumatol. Int. 2008, 28, 685–691. [Google Scholar] [CrossRef] [PubMed]
- van Meurs, J.B.; Uitterlinden, A.G.; Stolk, L.; Kerkhof, H.J.; Hofman, A.; Pols, H.A.; Bierma-Zeinstra, S.M. A functional polymorphism in the catechol-O-methyltransferase gene is associated with osteoarthritis-related pain. Arthritis Rheum. 2009, 60, 628–629. [Google Scholar] [CrossRef] [PubMed]
- Cohen, H.; Neumann, L.; Glazer, Y.; Ebstein, R.P.; Buskila, D. The relationship between a common catechol-O-methyltransferase (COMT) polymorphism val(158) met and fibromyalgia. Clin. Exp. Rheumatol. 2009, 27 (Suppl. 56), 51–56. [Google Scholar]
- Sprangers, M.A.; Sloan, J.A.; Veenhoven, R.; Cleeland, C.S.; Halyard, M.Y.; Abertnethy, A.P.; Baas, F.; Barsevick, A.M.; Bartels, M.; Boomsma, D.I.; et al. The establishment of the GENEQOL consortium to investigate the genetic disposition of patient-reported quality-of-life outcomes. Twin Res. Hum. Genet. 2009, 12, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Bartels, M. Genetics of wellbeing and its components satisfaction with life, happiness, and quality of life: A review and meta-analysis of heritability studies. Behav. Genet. 2015, 45, 137–156. [Google Scholar] [CrossRef] [PubMed]
- Schoormans, D.; Darabi, H.; Li, J.; Brandberg, Y.; Eriksson, M.; Zwinderman, K.H.; Sprangers, M.A.; Hall, P. In Search for the Genetic Basis of Quality of Life in Healthy Swedish Women—A GWAS Study Using the iCOGS Custom Genotyping Array. PLoS ONE 2015, 10, e0140563. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rausch, S.M.; Clark, M.M.; Patten, C.; Liu, H.; Felten, S.; Li, Y.; Sloan, J.; Yang, P. Relationship between cytokine gene single nucleotide polymorphisms and symptom burden and quality of life in lung cancer survivors. Cancer 2010, 116, 4103–4113. [Google Scholar] [CrossRef] [PubMed]
- Schoormans, D.; Radonic, T.; de Witte, P.; Groenink, M.; Azim, D.; Lutter, R.; Mulder, B.J.; Sprangers, M.A.; Zwinderman, A.H. Mental quality of life is related to a cytokine genetic pathway. PLoS ONE 2012, 7, e45126. [Google Scholar] [CrossRef] [PubMed]
- Sloan, J.A.; de Andrade, M.; Decker, P.; Wampfler, J.; Oswold, C.; Clark, M.; Yang, P. Genetic variations and patient-reported quality of life among patients with lung cancer. J. Clin. Oncol. 2012, 30, 1699–1704. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sprangers, M.A.; Sloan, J.A.; Barsevick, A.; Chauhan, C.; Dueck, A.C.; Raat, H.; Shi, Q.; Van Noorden, C.J. Scientific imperatives, clinical implications, and theoretical underpinnings for the investigation of the relationship between genetic variables and patient-reported quality-of-life outcomes. Qual. Life Res. 2010, 19, 1395–1403. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rausch, S.M.; Gonzalez, B.D.; Clark, M.M.; Patten, C.; Felten, S.; Liu, H.; Li, Y.; Sloan, J.; Yang, P. SNPs in PTGS2 and LTA predict pain and quality of life in long term lung cancer survivors. Lung Cancer 2012, 77, 217–223. [Google Scholar] [CrossRef]
- Alexander, K.; Cooper, B.; Paul, S.M.; West, C.; Yates, P.; Kober, K.M.; Aouizerat, B.E.; Miaskowski, C. Evidence of associations between cytokine gene polymorphisms and quality of life in patients with cancer and their family caregivers. Oncol. Nurs. Forum 2014, 41, 267–281. [Google Scholar] [CrossRef]
- Reyes-Gibby, C.C.; Shete, S.; Yennurajalingam, S.; Frazier, M.; Bruera, E.; Kurzrock, R.; Crane, C.H.; Abbruzzese, J.; Evans, D.; Spitz, M.R. Genetic and nongenetic covariates of pain severity in patients with adenocarcinoma of the pancreas: Assessing the influence of cytokine genes. J. Pain Symptom. Manag. 2009, 38, 894–902. [Google Scholar] [CrossRef]
- Reyes-Gibby, C.C.; Spitz, M.R.; Yennurajalingam, S.; Swartz, M.; Gu, J.; Wu, X.; Bruera, E.; Shete, S. Role of inflammation gene polymorphisms on pain severity in lung cancer patients. Cancer Epidemiol. Biomark. Prev. 2009, 18, 2636–2642. [Google Scholar] [CrossRef] [PubMed]
- Pierzynski, J.A.; Ye, Y.; Lippman, S.M.; Rodriguez, M.A.; Wu, X.; Hildebrandt, M.A.T. Socio-demographic, Clinical, and Genetic Determinants of Quality of Life in Lung Cancer Patients. Sci. Rep. 2018, 8, 10640. [Google Scholar] [CrossRef]
- Kapoor, M.; Wang, J.C.; Wetherill, L.; Le, N.; Bertelsen, S.; Hinrichs, A.L.; Budde, J.; Agrawal, A.; Bucholz, K.; Dick, D.; et al. A meta-analysis of two genome-wide association studies to identify novel loci for maximum number of alcoholic drinks. Hum. Genet. 2013, 132, 1141–1151. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Sun, H.; Lu, X.; He, C.; Xiao, L.; He, J.; Yang, Y.; Tang, Y. Genetic polymorphisms and haplotypes of BRCA1 gene associated with quality of life and survival among patients with non-small-cell lung cancer. Qual. Life Res. 2020, 29, 2631–2640. [Google Scholar] [CrossRef]
- Alexander, K.; Conley, Y.P.; Levine, J.D.; Cooper, B.A.; Paul, S.M.; Mastick, J.; West, C.; Miaskowski, C. Cytokine Gene Polymorphisms Associated with Various Domains of Quality of Life in Women With Breast Cancer. J. Pain Symptom. Manag. 2018, 55, 334–350. [Google Scholar] [CrossRef] [PubMed]
- Alexander, K.E.; Chambers, S.; Spurdle, A.B.; Batra, J.; Lose, F.; O’Mara, T.A.; Gardiner, R.A.; Aitken, J.F.; Clements, J.A.; Kedda, M.A.; et al. Association between single-nucleotide polymorphisms in growth factor genes and quality of life in men with prostate cancer and the general population. Qual. Life Res. 2015, 24, 2183–2193. [Google Scholar] [CrossRef] [PubMed]
- Peters, C.A.; Stock, R.G.; Cesaretti, J.A.; Atencio, D.P.; Peters, S.; Burri, R.J.; Stone, N.N.; Ostrer, H.; Rosenstein, B.S. TGFB1 single nucleotide polymorphisms are associated with adverse quality of life in prostate cancer patients treated with radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2008, 70, 752–759. [Google Scholar] [CrossRef]
- Zorina-Lichtenwalter, K.; Parisien, M.; Diatchenko, L. Genetic studies of human neuropathic pain conditions: A review. Pain 2018, 159, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Derks, M.G.; de Glas, N.A.; Bastiaannet, E.; de Craen, A.J.; Portielje, J.E.; van de Velde, C.J.; van Leeuwen, F.E.; Liefers, G.J. Physical Functioning in Older Patients With Breast Cancer: A Prospective Cohort Study in the TEAM Trial. Oncologist 2016, 21, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Ecclestone, C.; Chow, R.; Pulenzas, N.; Zhang, L.; Leahey, A.; Hamer, J.; DeAngelis, C.; Bedard, G.; McDonald, R.; Bhatia, A.; et al. Quality of life and symptom burden in patients with metastatic breast cancer. Support Care Cancer 2016, 24, 4035–4043. [Google Scholar] [CrossRef] [PubMed]
- Hamer, J.; McDonald, R.; Zhang, L.; Verma, S.; Leahey, A.; Ecclestone, C.; Bedard, G.; Pulenzas, N.; Bhatia, A.; Chow, R.; et al. Quality of life (QOL) and symptom burden (SB) in patients with breast cancer. Support Care Cancer 2017, 25, 409–419. [Google Scholar] [CrossRef]
- Bouskill, K.; Kramer, M. The impact of cancer and quality of life among long-term survivors of breast cancer in Austria. Support Care Cancer 2016, 24, 4705–4712. [Google Scholar] [CrossRef]
- Shin, J.A.; El-Jawahri, A.; Parkes, A.; Schleicher, S.M.; Knight, H.P.; Temel, J.S. Quality of Life, Mood, and Prognostic Understanding in Patients with Metastatic Breast Cancer. J. Palliat. Med. 2016, 19, 863–869. [Google Scholar] [CrossRef]
- Barsevick, A.; Frost, M.; Zwinderman, A.; Hall, P.; Halyard, M. I’m so tired: Biological and genetic mechanisms of cancer-related fatigue. Qual. Life Res. 2010, 19, 1419–1427. [Google Scholar] [CrossRef]
- Bower, J.E.; Ganz, P.A.; Irwin, M.R.; Castellon, S.; Arevalo, J.; Cole, S.W. Cytokine genetic variations and fatigue among patients with breast cancer. J. Clin. Oncol. 2013, 31, 1656–1661. [Google Scholar] [CrossRef]
- Illi, J.; Miaskowski, C.; Cooper, B.; Levine, J.D.; Dunn, L.; West, C.; Dodd, M.; Dhruva, A.; Paul, S.M.; Baggott, C.; et al. Association between pro- and anti-inflammatory cytokine genes and a symptom cluster of pain, fatigue, sleep disturbance, and depression. Cytokine 2012, 58, 437–447. [Google Scholar] [CrossRef]
- Kao, C.F.; Jia, P.; Zhao, Z.; Kuo, P.H. Enriched pathways for major depressive disorder identified from a genome-wide association study. Int. J. Neuropsychopharmacol. 2012, 15, 1401–1411. [Google Scholar] [CrossRef] [PubMed]
- Shi, Q.; Cleeland, C.S.; Klepstad, P.; Miaskowski, C.; Pedersen, N.L. Biological pathways and genetic variables involved in pain. Qual. Life Res. 2010, 19, 1407–1417. [Google Scholar] [CrossRef] [PubMed]
- Rabert, D.K.; Koch, B.D.; Ilnicka, M.; Obernolte, R.A.; Naylor, S.L.; Herman, R.C.; Eglen, R.M.; Hunter, J.C.; Sangameswaran, L. A tetrodotoxin-resistant voltage-gated sodium channel from human dorsal root ganglia, hPN3/SCN10A. Pain 1998, 78, 107–114. [Google Scholar] [CrossRef]
- Liu, X.D.; Yang, J.J.; Fang, D.; Cai, J.; Wan, Y.; Xing, G.G. Functional upregulation of nav1.8 sodium channels on the membrane of dorsal root Ganglia neurons contributes to the development of cancer-induced bone pain. PLoS ONE 2014, 9, e114623. [Google Scholar] [CrossRef] [PubMed]
- Klein-Weigel, P.F.; Volz, T.S.; Richter, J.G. Erythromelalgia. Vasa 2018, 47, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Reimann, F.; Cox, J.J.; Belfer, I.; Diatchenko, L.; Zaykin, D.V.; McHale, D.P.; Drenth, J.P.; Dai, F.; Wheeler, J.; Sanders, F.; et al. Pain perception is altered by a nucleotide polymorphism in SCN9A. Proc. Natl. Acad. Sci. USA 2010, 107, 5148–5153. [Google Scholar] [CrossRef]
- Macri, V.; Brody, J.A.; Arking, D.E.; Hucker, W.J.; Yin, X.; Lin, H.; Mills, R.W.; Sinner, M.F.; Lubitz, S.A.; Liu, C.T.; et al. Common Coding Variants in SCN10A Are Associated With the Nav1.8 Late Current and Cardiac Conduction. Circ. Genom. Precis Med. 2018, 11, e001663. [Google Scholar] [CrossRef]
- Iio, C.; Ogimoto, A.; Nagai, T.; Suzuki, J.; Inoue, K.; Nishimura, K.; Uetani, T.; Okayama, H.; Okura, T.; Shigematsu, Y.; et al. Association between Genetic Variation in the SCN10A Gene and Cardiac Conduction Abnormalities in Patients with Hypertrophic Cardiomyopathy. Int. Heart J. 2015, 56, 421–427. [Google Scholar] [CrossRef]
- He, Y.J.; Winham, S.J.; Hoskins, J.M.; Glass, S.; Paul, J.; Brown, R.; Motsinger-Reif, A.; McLeod, H.L. Carboplatin/taxane-induced gastrointestinal toxicity: A pharmacogenomics study on the SCOTROC1 trial. Pharm. J. 2016, 16, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Trabelsi, M.; Kavian, N.; Daoud, F.; Commere, V.; Deburgrave, N.; Beugnet, C.; Llense, S.; Barbot, J.C.; Vasson, A.; Kaplan, J.C.; et al. Revised spectrum of mutations in sarcoglycanopathies. Eur. J. Hum. Genet. 2008, 16, 793–803. [Google Scholar] [CrossRef]
- Moreira, E.S.; Vainzof, M.; Marie, S.K.; Nigro, V.; Zatz, M.; Passos-Bueno, M.R. A first missense mutation in the delta sarcoglycan gene associated with a severe phenotype and frequency of limb-girdle muscular dystrophy type 2F (LGMD2F) in Brazilian sarcoglycanopathies. J. Med. Genet. 1998, 35, 951–953. [Google Scholar] [CrossRef][Green Version]
- Pegoraro, E.; Hoffman, E.P. Limb-Girdle Muscular Dystrophy Overview—RETIRED CHAPTER, FOR HISTORICAL REFERENCE ONLY. In GeneReviews(®); Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Stephens, K., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Tsubata, S.; Bowles, K.R.; Vatta, M.; Zintz, C.; Titus, J.; Muhonen, L.; Bowles, N.E.; Towbin, J.A. Mutations in the human delta-sarcoglycan gene in familial and sporadic dilated cardiomyopathy. J. Clin. Investig. 2000, 106, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Carinelli, S.; Blanco, O.A.; Perdomo-Ramirez, A.; Claverie-Martin, F. Nail-Patella syndrome with early onset end-stage renal disease in a child with a novel heterozygous missense mutation in the LMX1B homeodomain: A case report. Biomed. Rep. 2020, 13, 49. [Google Scholar] [CrossRef] [PubMed]
- Bech, S.; Lokkegaard, A.; Nielsen, T.T.; Norremolle, A.; Gronborg, S.; Hasholt, L.; Steffensen, G.K.; Graehn, G.; Olesen, J.H.; Tommerup, N.; et al. Paroxysmal Cranial Dyskinesia and Nail-Patella Syndrome Caused by a Novel Variant in the LMX1B Gene. Mov. Disord. 2020, 35, 2343–2347. [Google Scholar] [CrossRef] [PubMed]
- Bongers, E.M.; Huysmans, F.T.; Levtchenko, E.; de Rooy, J.W.; Blickman, J.G.; Admiraal, R.J.; Huygen, P.L.; Cruysberg, J.R.; Toolens, P.A.; Prins, J.B.; et al. Genotype-phenotype studies in nail-patella syndrome show that LMX1B mutation location is involved in the risk of developing nephropathy. Eur. J. Hum. Genet. 2005, 13, 935–946. [Google Scholar] [CrossRef]
- Locke, A.E.; Kahali, B.; Berndt, S.I.; Justice, A.E.; Pers, T.H.; Day, F.R.; Powell, C.; Vedantam, S.; Buchkovich, M.L.; Yang, J.; et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015, 518, 197–206. [Google Scholar] [CrossRef]
- Zhu, Q.; Xue, K.; Guo, H.W.; Yang, Y.H. LMX1B rs10733682 Polymorphism Interacts with Macronutrients, Dietary Patterns on the Risk of Obesity in Han Chinese Girls. Nutrients 2020, 12, 1227. [Google Scholar] [CrossRef] [PubMed]
- Hilario, J.D.; Rodino-Klapac, L.R.; Wang, C.; Beattie, C.E. Semaphorin 5A is a bifunctional axon guidance cue for axial motoneurons in vivo. Dev. Biol. 2009, 326, 190–200. [Google Scholar] [CrossRef]
- Melin, M.; Carlsson, B.; Anckarsater, H.; Rastam, M.; Betancur, C.; Isaksson, A.; Gillberg, C.; Dahl, N. Constitutional downregulation of SEMA5A expression in autism. Neuropsychobiology 2006, 54, 64–69. [Google Scholar] [CrossRef]
- Mosca-Boidron, A.L.; Gueneau, L.; Huguet, G.; Goldenberg, A.; Henry, C.; Gigot, N.; Pallesi-Pocachard, E.; Falace, A.; Duplomb, L.; Thevenon, J.; et al. A de novo microdeletion of SEMA5A in a boy with autism spectrum disorder and intellectual disability. Eur. J. Hum. Genet. 2016, 24, 838–843. [Google Scholar] [CrossRef]
- Lin, L.; Lesnick, T.G.; Maraganore, D.M.; Isacson, O. Axon guidance and synaptic maintenance: Preclinical markers for neurodegenerative disease and therapeutics. Trends Neurosci. 2009, 32, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Willing, M.; Grange, D.K.; Shinawi, M.; Manwaring, L.; Vineyard, M.; Kulkarni, S.; Cottrell, C.E. Multigenerational autosomal dominant inheritance of 5p chromosomal deletions. Am. J. Med. Genet. A 2016, 170, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Purohit, A.; Sadanandam, A.; Myneni, P.; Singh, R.K. Semaphorin 5A mediated cellular navigation: Connecting nervous system and cancer. Biochim. Biophys. Acta 2014, 1846, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Gras, C.; Eiz-Vesper, B.; Jaimes, Y.; Immenschuh, S.; Jacobs, R.; Witte, T.; Blasczyk, R.; Figueiredo, C. Secreted semaphorin 5A activates immune effector cells and is a biomarker for rheumatoid arthritis. Arthritis Rheumatol. 2014, 66, 1461–1471. [Google Scholar] [CrossRef]
- Hoyer, H.; Braathen, G.J.; Eek, A.K.; Nordang, G.B.; Skjelbred, C.F.; Russell, M.B. Copy number variations in a population-based study of Charcot-Marie-Tooth disease. Biomed. Res. Int. 2015, 2015, 960404. [Google Scholar] [CrossRef]
- Lu, Q.; Zhu, L. The Role of Semaphorins in Metabolic Disorders. Int. J. Mol. Sci. 2020, 21, 5641. [Google Scholar] [CrossRef]
- Vyas, S.; Matic, I.; Uchima, L.; Rood, J.; Zaja, R.; Hay, R.T.; Ahel, I.; Chang, P. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat. Commun. 2014, 5, 4426. [Google Scholar] [CrossRef]
- Shao, C.; Qiu, Y.; Liu, J.; Feng, H.; Shen, S.; Saiyin, H.; Yu, W.; Wei, Y.; Yu, L.; Su, W.; et al. PARP12 (ARTD12) suppresses hepatocellular carcinoma metastasis through interacting with FHL2 and regulating its stability. Cell Death Dis. 2018, 9, 856. [Google Scholar] [CrossRef]
- van der Harst, P.; Verweij, N. Identification of 64 Novel Genetic Loci Provides an Expanded View on the Genetic Architecture of Coronary Artery Disease. Circ. Res. 2018, 122, 433–443. [Google Scholar] [CrossRef]
- Jin, Y.; Andersen, G.; Yorgov, D.; Ferrara, T.M.; Ben, S.; Brownson, K.M.; Holland, P.J.; Birlea, S.A.; Siebert, J.; Hartmann, A.; et al. Genome-wide association studies of autoimmune vitiligo identify 23 new risk loci and highlight key pathways and regulatory variants. Nat. Genet. 2016, 48, 1418–1424. [Google Scholar] [CrossRef]
- Hays, R.D.; Bjorner, J.B.; Revicki, D.A.; Spritzer, K.L.; Cella, D. Development of physical and mental health summary scores from the patient-reported outcomes measurement information system (PROMIS) global items. Qual. Life Res. 2009, 18, 873–880. [Google Scholar] [CrossRef]
- Cook, K.F.; Jensen, S.E.; Schalet, B.D.; Beaumont, J.L.; Amtmann, D.; Czajkowski, S.; Dewalt, D.A.; Fries, J.F.; Pilkonis, P.A.; Reeve, B.B.; et al. PROMIS measures of pain, fatigue, negative affect, physical function, and social function demonstrated clinical validity across a range of chronic conditions. J. Clin. Epidemiol. 2016, 73, 89–102. [Google Scholar] [CrossRef]
- Schalet, B.D.; Hays, R.D.; Jensen, S.E.; Beaumont, J.L.; Fries, J.F.; Cella, D. Validity of PROMIS physical function measured in diverse clinical samples. J. Clin. Epidemiol. 2016, 73, 112–118. [Google Scholar] [CrossRef]
- Cella, D.; Riley, W.; Stone, A.; Rothrock, N.; Reeve, B.; Yount, S.; Amtmann, D.; Bode, R.; Buysse, D.; Choi, S.; et al. The Patient-Reported Outcomes Measurement Information System (PROMIS) developed and tested its first wave of adult self-reported health outcome item banks: 2005–-2008. J. Clin. Epidemiol. 2010, 63, 1179–1194. [Google Scholar] [CrossRef]
- Rothrock, N.E.; Hays, R.D.; Spritzer, K.; Yount, S.E.; Riley, W.; Cella, D. Relative to the general US population, chronic diseases are associated with poorer health-related quality of life as measured by the Patient-Reported Outcomes Measurement Information System (PROMIS). J. Clin. Epidemiol. 2010, 63, 1195–1204. [Google Scholar] [CrossRef]
- Rose, M.; Bjorner, J.B.; Gandek, B.; Bruce, B.; Fries, J.F.; Ware, J.E., Jr. The PROMIS Physical Function item bank was calibrated to a standardized metric and shown to improve measurement efficiency. J. Clin. Epidemiol. 2014, 67, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Sloan, J.A.; Dueck, A. Issues for statisticians in conducting analyses and translating results for quality of life end points in clinical trials. J. Biopharm. Stat. 2004, 14, 73–96. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.A.; Satele, D.; Pattabasavaiah, S.; Buckner, J.C.; Sloan, J.A. Normative data and clinically significant effect sizes for single-item numerical linear analogue self-assessment (LASA) scales. Health Qual. Life Outcomes 2014, 12, 187. [Google Scholar] [CrossRef]
- Garcia-Closas, M.; Couch, F.J.; Lindstrom, S.; Michailidou, K.; Schmidt, M.K.; Brook, M.N.; Orr, N.; Rhie, S.K.; Riboli, E.; Feigelson, H.S.; et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat. Genet. 2013, 45, 392–398, 398e1–2. [Google Scholar] [CrossRef] [PubMed]
- Michailidou, K.; Hall, P.; Gonzalez-Neira, A.; Ghoussaini, M.; Dennis, J.; Milne, R.L.; Schmidt, M.K.; Chang-Claude, J.; Bojesen, S.E.; Bolla, M.K.; et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 2013, 45, 353–361, 361e1–2. [Google Scholar] [CrossRef]
- Manichaikul, A.; Mychaleckyj, J.C.; Rich, S.S.; Daly, K.; Sale, M.; Chen, W.M. Robust relationship inference in genome-wide association studies. Bioinformatics 2010, 26, 2867–2873. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Das, S.; Forer, L.; Schonherr, S.; Sidore, C.; Locke, A.E.; Kwong, A.; Vrieze, S.I.; Chew, E.Y.; Levy, S.; McGue, M.; et al. Next-generation genotype imputation service and methods. Nat. Genet. 2016, 48, 1284–1287. [Google Scholar] [CrossRef] [PubMed]
- Manolio, T.A. Genomewide association studies and assessment of the risk of disease. N. Engl. J. Med. 2010, 363, 166–176. [Google Scholar] [CrossRef]
- Pe’er, I.; Yelensky, R.; Altshuler, D.; Daly, M.J. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 2008, 32, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Pruim, R.J.; Welch, R.P.; Sanna, S.; Teslovich, T.M.; Chines, P.S.; Gliedt, T.P.; Boehnke, M.; Abecasis, G.R.; Willer, C.J. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics 2010, 26, 2336–2337. [Google Scholar] [CrossRef] [PubMed]
- Alexander, T.A.; Machiela, M.J. LDpop: An interactive online tool to calculate and visualize geographic LD patterns. BMC Bioinform. 2020, 21, 14. [Google Scholar] [CrossRef]
- Myers, T.A.; Chanock, S.J.; Machiela, M.J. LDlinkR: An R Package for Rapidly Calculating Linkage Disequilibrium Statistics in Diverse Populations. Front. Genet. 2020, 11, 157. [Google Scholar] [CrossRef]


| Demographic Factors | n (%) | |
|---|---|---|
| Race | Non-white, unknown, or undisclosed | 56 (3.9%) |
| White | 1386 (96.1%) | |
| Age at diagnosis in years | Mean (SD) | 53.4 (11.5) |
| Median (Q1, Q3) | 51.5 (45.6, 62.4) | |
| Range | 22.7–90.0 | |
| Age at time of QOL assessment in years | Mean (SD) | 61.6 (12.8) |
| Median (Q1, Q3) | 61.0 (52.5, 71.4) | |
| Range | 26.2–95.5 | |
| Years between cancer diagnosis and QOL assessment | Mean (SD) | 8.2 (3.8) |
| Median (Q1, Q3) | 8.3 (5.0, 10.9) | |
| Range | 0.9–16.7 | |
| Gender | Male | 3 (0.2%) |
| Female | 1439 (99.8%) | |
| Financial concerns rating (patient-reported) | Mean (SD) | 2.1 (2.7) |
| Median (Q1, Q3) | 1.0 (0.0, 3.0) | |
| Range | 0.0–10.0 | |
| Treatment received any time before QOL assessment (Yes/No) | Mastectomy | 782 (54.2%)/660 (45.8%) |
| Axillary lymph node dissection (ALND) | 530 (36.8%)/912 (63.2%) | |
| Chemotherapy | 608 (42.2%)/834 (57.8%) | |
| Radiation | 689 (47.8%)/753 (52.2%) | |
| Endocrine therapy | 997 (69.1%)/445 (30.9%) | |
| HRQOL (PROMIS-10 Measure) | Covariate | Coefficient | b p-Value | ||
|---|---|---|---|---|---|
| Estimate | SE | ap-Value | |||
| Global Physical Health | Genotyped with iCOGS | 0.63 | 0.83 | 0.45 | - |
| Age c z-score | −4.00 | 0.45 | 2.0 × 10−18 | 1.6 × 10−23 | |
| Age z-score, squared | −1.67 | 0.32 | 3.0 × 10−7 | ||
| Financial concerns z-score | −6.93 | 0.64 | 6.0 × 10−26 | 4.8 × 10−40 | |
| Financial concerns z-score, squared | 1.12 | 0.37 | 0.002 | ||
| Chemotherapy treatment | −0.59 | 0.83 | 0.48 | - | |
| Global Mental Health | Genotyped with iCOGS | −0.05 | 0.99 | 0.96 | - |
| Age z-score | −1.27 | 0.54 | 0.02 | 3.5 × 10−7 | |
| Age z-score, squared | −1.88 | 0.39 | 1.0 × 10−6 | ||
| Financial concerns z-score | −9.83 | 0.77 | 2.0 × 10−35 | 2.0 × 10−69 | |
| Financial concerns z-score, squared | 0.67 | 0.44 | 0.13 | ||
| Chemotherapy treatment | −2.47 | 1.00 | 0.01 | - | |
| QOL Domain | Gene | SNP rsID | Chr | BP | Ref Allele | Alt Allele | Alt Allele Freq | Coefficient (β) Estimate | SE | p-Value |
|---|---|---|---|---|---|---|---|---|---|---|
| Global Physical Health | SCN10A | rs112718371 | 3 | 38750460 | C | T | 0.011 | −17.79 | 3.25 | 5.21 × 10−8 |
| - | rs79198292 | 14 | 57649431 | G | A | 0.022 | −10.46 | 2.01 | 2.38 × 10−7 | |
| rs1954548 | 14 | 57644518 | G | A | 0.023 | −10.33 | 1.99 | 2.62 × 10−7 | ||
| rs1954547 | 14 | 57644605 | C | T | 0.023 | −10.27 | 1.99 | 2.91 × 10−7 | ||
| rs2211582 | 14 | 57643873 | C | T | 0.022 | −10.40 | 2.02 | 2.97 × 10−7 | ||
| rs73300594 | 14 | 57641616 | C | T | 0.023 | −10.31 | 2.01 | 3.50 × 10−7 | ||
| rs60372931 | 14 | 57642708 | A | G | 0.023 | −10.29 | 2.01 | 3.56 × 10−7 | ||
| rs7159115 | 14 | 57645169 | A | G | 0.023 | −10.09 | 1.99 | 4.58 × 10−7 | ||
| rs11848373 | 14 | 57649554 | G | A | 0.024 | −9.86 | 1.96 | 5.51 × 10−7 | ||
| rs7144304 | 14 | 57655035 | A | T | 0.034 | −8.08 | 1.61 | 5.57 × 10−7 | ||
| Global Mental Health | SGCD | rs73813229 | 5 | 155504517 | T | C | 0.185 | −5.10 | 0.99 | 2.84 × 10−7 |
| rs73298688 | 5 | 155508728 | C | T | 0.184 | −5.10 | 0.99 | 2.99 × 10−7 | ||
| LMX1B | rs71497626 | 9 | 129405863 | G | A | 0.011 | −17.16 | 3.34 | 3.17 × 10−7 | |
| SGCD | rs113472609 | 5 | 155501582 | T | C | 0.186 | −5.07 | 0.99 | 3.17 × 10−7 | |
| rs80138336 | 5 | 155501438 | A | G | 0.185 | −5.06 | 0.99 | 3.27 × 10−7 | ||
| rs73813228 | 5 | 155501356 | A | G | 0.185 | −5.06 | 0.99 | 3.29 × 10−7 | ||
| rs4704970 | 5 | 155500992 | G | A | 0.185 | −5.05 | 0.99 | 3.42 × 10−7 | ||
| rs73813227 | 5 | 155500542 | T | C | 0.185 | −5.04 | 0.99 | 3.59 × 10−7 | ||
| PARP12 | rs1544460 | 7 | 139731709 | G | A | 0.430 | −3.41 | 0.67 | 4.28 × 10−7 | |
| SGCD | rs10476276 | 5 | 155500647 | A | G | 0.188 | −4.96 | 0.98 | 4.50 × 10−7 | |
| SEMA5A | rs76677754 | 5 | 9084176 | C | T | 0.021 | −14.26 | 2.81 | 4.58 × 10−7 | |
| rs11741186 | 5 | 9044674 | G | A | 0.020 | −13.26 | 2.65 | 6.13 × 10−7 | ||
| SGCD | rs73812917 | 5 | 155464275 | G | A | 0.186 | −4.84 | 0.97 | 6.15 × 10−7 | |
| SEMA5A | rs78456783 | 5 | 9037340 | A | C | 0.020 | −13.21 | 2.65 | 6.76 × 10−7 | |
| - | rs9899933 | 17 | 48032244 | A | G | 0.060 | −8.35 | 1.69 | 7.99 × 10−7 | |
| SGCD | rs75174473 | 5 | 155462439 | A | G | 0.203 | −4.61 | 0.93 | 8.84 × 10−7 | |
| SEMA5A | rs11741172 | 5 | 9044647 | G | A | 0.020 | −13.24 | 2.68 | 9.02 × 10−7 | |
| SGCD | rs58327079 | 5 | 155463960 | T | C | 0.203 | −4.60 | 0.93 | 9.93 × 10−7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adjei, A.A.; Lopez, C.L.; Schaid, D.J.; Sloan, J.A.; Le-Rademacher, J.G.; Loprinzi, C.L.; Norman, A.D.; Olson, J.E.; Couch, F.J.; Beutler, A.S.; et al. Genetic Variations and Health-Related Quality of Life (HRQOL): A Genome-Wide Study Approach. Cancers 2021, 13, 716. https://doi.org/10.3390/cancers13040716
Adjei AA, Lopez CL, Schaid DJ, Sloan JA, Le-Rademacher JG, Loprinzi CL, Norman AD, Olson JE, Couch FJ, Beutler AS, et al. Genetic Variations and Health-Related Quality of Life (HRQOL): A Genome-Wide Study Approach. Cancers. 2021; 13(4):716. https://doi.org/10.3390/cancers13040716
Chicago/Turabian StyleAdjei, Araba A., Camden L. Lopez, Daniel J. Schaid, Jeff A. Sloan, Jennifer G. Le-Rademacher, Charles L. Loprinzi, Aaron D. Norman, Janet E. Olson, Fergus J. Couch, Andreas S. Beutler, and et al. 2021. "Genetic Variations and Health-Related Quality of Life (HRQOL): A Genome-Wide Study Approach" Cancers 13, no. 4: 716. https://doi.org/10.3390/cancers13040716
APA StyleAdjei, A. A., Lopez, C. L., Schaid, D. J., Sloan, J. A., Le-Rademacher, J. G., Loprinzi, C. L., Norman, A. D., Olson, J. E., Couch, F. J., Beutler, A. S., Vachon, C. M., & Ruddy, K. J. (2021). Genetic Variations and Health-Related Quality of Life (HRQOL): A Genome-Wide Study Approach. Cancers, 13(4), 716. https://doi.org/10.3390/cancers13040716






