Bioinformatic Approaches to Validation and Functional Analysis of 3D Lung Cancer Models
Abstract
:Simple Summary
Abstract
1. Introduction
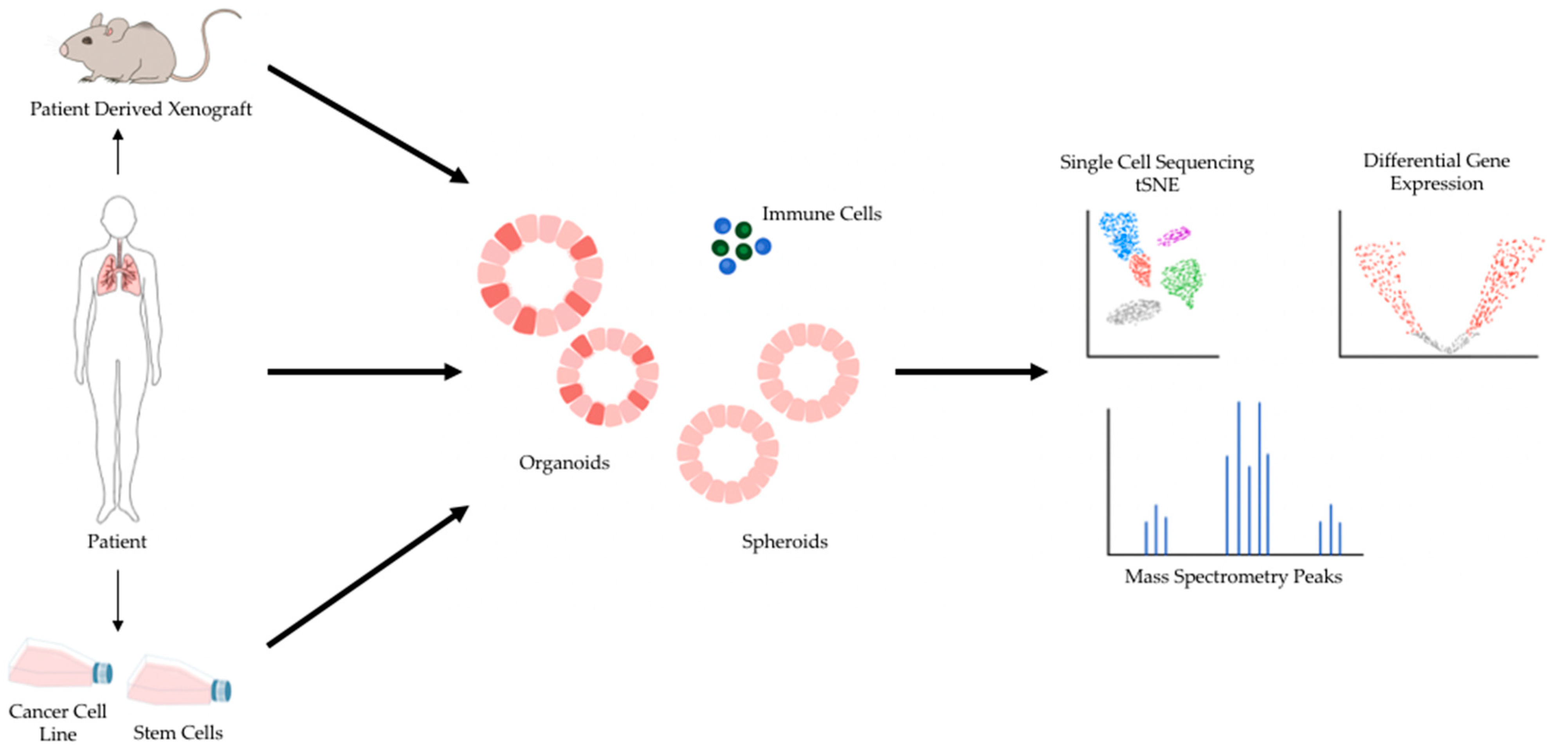
2. 3D Preclinical Models for Cancer Research
3. Lethality of Lung Cancer and the Need for Improved Models
4. Sequencing Validation of Patient-Derived 3D Lung Cancer Models for Drug Efficacy Screens
5. 3D Models of Cancer, Single Cell Profiling, and Genomic Screens as Scalable Discovery Platforms
6. Single-Cell Profiling of Tumor-Microenvironments in 3D Lung Cancer Models
7. 3D Lung Cancer Models for Neoantigen Discovery
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Rosenthal, R.; Cadieux, E.L.; Salgado, R.; Bakir, M.A.; Moore, D.A.; Hiley, C.T.; Lund, T.; Tanić, M.; Reading, J.L.; Joshi, K.; et al. Neoantigen-Directed Immune Escape in Lung Cancer Evolution. Nature 2019, 567, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Valdes-Mora, F.; Handler, K.; Law, A.M.K.; Salomon, R.; Oakes, S.R.; Ormandy, C.J.; Gallego-Ortega, D. Single-Cell Transcriptomics in Cancer Immunobiology: The Future of Precision Oncology. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Maynard, A.; McCoach, C.E.; Rotow, J.K.; Harris, L.; Haderk, F.; Kerr, D.L.; Yu, E.A.; Schenk, E.L.; Tan, W.; Zee, A.; et al. Therapy-Induced Evolution of Human Lung Cancer Revealed by Single-Cell RNA Sequencing. Cell 2020, 182, 1232–1251.e22. [Google Scholar] [CrossRef]
- Langhans, S.A. Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front. Pharmacol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DiMasi, J.A.; Reichert, J.M.; Feldman, L.; Malins, A. Clinical Approval Success Rates for Investigational Cancer Drugs. Front. Mol. Biosci. 2013, 94, 329–335. [Google Scholar] [CrossRef]
- Nass, S.J.; Rothenberg, M.L.; Pentz, R.; Hricak, H.; Abernethy, A.; Anderson, K.; Gee, A.W.; Harvey, R.D.; Piantadosi, S.; Bertagnolli, M.M.; et al. Accelerating anticancer drug development—opportunities and trade-offs. Nat. Rev. Clin. Oncol. 2018, 15, 777–786. [Google Scholar] [CrossRef]
- Garner, H.; de Visser, K.E. Immune Crosstalk in Cancer Progression and Metastatic Spread: A Complex Conversation. Nat. Rev. Clin. Oncol. 2020, 20, 483–497. [Google Scholar] [CrossRef]
- Couzin-Frankel, J. Cancer Immunotherapy. Science 2013, 342, 1432–1433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hegde, P.S.; Chen, D.S. Top 10 Challenges in Cancer Immunotherapy. Immunity 2020, 52, 17–35. [Google Scholar] [CrossRef]
- Chen, D.S.; Mellman, I. Elements of Cancer Immunity and the Cancer–Immune Set Point. Nature 2017, 541, 321–330. [Google Scholar] [CrossRef]
- Yu, J.X.; Hubbard-Lucey, V.M.; Tang, J. Immuno-Oncology Drug Development Goes Global. Nat. Rev. Drug Discov. 2019, 18, 899–900. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.X.; Upadhaya, S.; Tatake, R.; Barkalow, F.; Hubbard-Lucey, V.M. Cancer Cell Therapies: The Clinical Trial Landscape. Nat. Rev. Drug Discov. 2020, 19, 583–584. [Google Scholar] [CrossRef]
- Riedl, A.; Schlederer, M.; Pudelko, K.; Stadler, M.; Walter, S.; Unterleuthner, D.; Unger, C.; Kramer, N.; Hengstschläger, M.; Kenner, L.; et al. Comparison of Cancer Cells in 2D vs 3D Culture Reveals Differences in AKT–MTOR–S6K Signaling and Drug Responses. J. Cell Sci. 2017, 130, 203–218. [Google Scholar] [CrossRef] [Green Version]
- Melissaridou, S.; Wiechec, E.; Magan, M.; Jain, M.V.; Chung, M.K.; Farnebo, L.; Roberg, K. The Effect of 2D and 3D Cell Cultures on Treatment Response, EMT Profile and Stem Cell Features in Head and Neck Cancer. Cancer Cell Int. 2019, 19, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ekert, J.E.; Johnson, K.; Strake, B.; Pardinas, J.; Jarantow, S.; Perkinson, R.; Colter, D.C. Three-Dimensional Lung Tumor Microenvironment Modulates Therapeutic Compound Responsiveness In Vitro—Implication for Drug Development. PLoS ONE 2014, 9, e92248. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, M.; Amant, F.; Biankin, A.V.; Budinská, E.; Byrne, A.T.; Caldas, C.; Clarke, R.B.; de Jong, S.; Jonkers, J.; Mælandsmo, G.M.; et al. Patient-Derived Xenograft Models: An Emerging Platform for Translational Cancer Research. Cancer Discov. 2014, 4, 998–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weeber, F.; Ooft, S.N.; Dijkstra, K.K.; Voest, E.E. Tumor Organoids as a Pre-Clinical Cancer Model for Drug Discovery. Cell Chem. Biol. 2017, 24, 1092–1100. [Google Scholar] [CrossRef]
- Mebarki, M.; Bennaceur, A.; Bonhomme-Faivre, L. Human-Cell-Derived Organoids as a New Ex Vivo Model for Drug Assays in Oncology. Drug Discov. Today 2018, 23, 857–863. [Google Scholar] [CrossRef]
- Simian, M.; Bissell, M.J. Organoids: A Historical Perspective of Thinking in Three Dimensions. J. Cell Biol. 2017, 216, 31–40. [Google Scholar] [CrossRef]
- Costa, E.C.; Moreira, A.F.; de Melo-Diogo, D.; Gaspar, V.M.; Carvalho, M.P.; Correia, I.J. 3D Tumor Spheroids: An Overview on the Tools and Techniques Used for Their Analysis. Biotechnol. Adv. 2016, 34, 1427–1441. [Google Scholar] [CrossRef] [PubMed]
- Zanoni, M.; Cortesi, M.; Zamagni, A.; Arienti, C.; Pignatta, S.; Tesei, A. Modeling Neoplastic Disease with Spheroids and Organoids. J. Hematol. Oncol. 2020, 13, 97. [Google Scholar] [CrossRef] [PubMed]
- Fatehullah, A.; Tan, S.H.; Barker, N. Organoids as an in Vitro Model of Human Development and Disease. Nat. Cell Biol. 2016, 18, 246–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a Dish: Modeling Development and Disease Using Organoid Technologies. Science 2014, 345. [Google Scholar] [CrossRef] [PubMed]
- Schutgens, F.; Clevers, H. Human Organoids: Tools for Understanding Biology and Treating Diseases. Annu. Rev. Pathol. Mech. Dis. 2020, 15, 211–234. [Google Scholar] [CrossRef] [Green Version]
- Powley, I.R.; Patel, M.; Miles, G.; Pringle, H.; Howells, L.; Thomas, A.; Kettleborough, C.; Bryans, J.; Hammonds, T.; MacFarlane, M.; et al. Patient-Derived Explants (PDEs) as a Powerful Preclinical Platform for Anti-Cancer Drug and Biomarker Discovery. Br. J. Cancer 2020, 122, 735–744. [Google Scholar] [CrossRef] [Green Version]
- Gendoo, D.M.A. Bioinformatics and Computational Approaches for Analyzing Patient-Derived Disease Models in Cancer Research. Comput. Struct. Biotechnol. J. 2020, 18, 375–380. [Google Scholar] [CrossRef]
- Bruun, J.; Kryeziu, K.; Eide, P.W.; Moosavi, S.H.; Eilertsen, I.A.; Langerud, J.; Røsok, B.; Totland, M.Z.; Brunsell, T.H.; Pellinen, T.; et al. Patient-Derived Organoids from Multiple Colorectal Cancer Liver Metastases Reveal Moderate Intra-Patient Pharmacotranscriptomic Heterogeneity. Clin. Cancer Res. 2020, 26, 4107–4119. [Google Scholar] [CrossRef] [Green Version]
- Neal, J.T.; Li, X.; Zhu, J.; Giangarra, V.; Grzeskowiak, C.L.; Ju, J.; Liu, I.H.; Chiou, S.-H.; Salahudeen, A.A.; Smith, A.R.; et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 2018, 175, 1972–1988.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuki, K.; Cheng, N.; Nakano, M.; Kuo, C.J. Organoid Models of Tumor Immunology. Trends Immunol. 2020, 41, 652–664. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, K.K.; Cattaneo, C.M.; Weeber, F.; Chalabi, M.; van de Haar, J.; Fanchi, L.F.; Slagter, M.; van der Velden, D.L.; Kaing, S.; Kelderman, S.; et al. Generation of Tumor-Reactive T Cells by Co-Culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 2018, 174, 1586–1598.e12. [Google Scholar] [CrossRef] [Green Version]
- Schreurs, R.R.C.E.; Baumdick, M.E.; Sagebiel, A.F.; Kaufmann, M.; Mokry, M.; Klarenbeek, P.L.; Schaltenberg, N.; Steinert, F.L.; van Rijn, J.M.; Drewniak, A.; et al. Human Fetal TNF-α-Cytokine-Producing CD4+ Effector Memory T Cells Promote Intestinal Development and Mediate Inflammation Early in Life. Immunity 2019, 50, 462–476.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finnberg, N.K.; Gokare, P.; Lev, A.; Grivennikov, S.I.; MacFarlane, A.W.; Campbell, K.S.; Winters, R.M.; Kaputa, K.; Farma, J.M.; Abbas, A.E.-S.; et al. Application of 3D Tumoroid Systems to Define Immune and Cytotoxic Therapeutic Responses Based on Tumoroid and Tissue Slice Culture Molecular Signatures. Oncotarget 2017, 8, 66747–66757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cattaneo, C.M.; Dijkstra, K.K.; Fanchi, L.F.; Kelderman, S.; Kaing, S.; van Rooij, N.; van den Brink, S.; Schumacher, T.N.; Voest, E.E. Tumor Organoid–T-Cell Coculture Systems. Nat. Protoc. 2020, 15, 15–39. [Google Scholar] [CrossRef]
- Tsai, S.; McOlash, L.; Palen, K.; Johnson, B.; Duris, C.; Yang, Q.; Dwinell, M.B.; Hunt, B.; Evans, D.B.; Gershan, J.; et al. Development of Primary Human Pancreatic Cancer Organoids, Matched Stromal and Immune Cells and 3D Tumor Microenvironment Models. BMC Cancer 2018, 18, 335. [Google Scholar] [CrossRef]
- Schnalzger, T.E.; de Groot, M.H.; Zhang, C.; Mosa, M.H.; Michels, B.E.; Röder, J.; Darvishi, T.; Wels, W.S.; Farin, H.F. 3D Model for CAR-Mediated Cytotoxicity Using Patient-Derived Colorectal Cancer Organoids. EMBO J. 2019, 38, e100928. [Google Scholar] [CrossRef]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The Biology and Management of Non-Small Cell Lung Cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Woodard, G.A.; Wang, S.X.; Kratz, J.R.; Zoon-Besselink, C.T.; Chiang, C.-Y.; Gubens, M.A.; Jahan, T.M.; Blakely, C.M.; Jones, K.D.; Mann, M.J.; et al. Adjuvant Chemotherapy Guided by Molecular Profiling and Improved Outcomes in Early Stage, Non–Small-Cell Lung Cancer. Clin. Lung Cancer 2018, 19, 58–64. [Google Scholar] [CrossRef]
- Sachs, N.; Papaspyropoulos, A.; Zomer-van Ommen, D.D.; Heo, I.; Böttinger, L.; Klay, D.; Weeber, F.; Huelsz-Prince, G.; Iakobachvili, N.; Amatngalim, G.D.; et al. Long-Term Expanding Human Airway Organoids for Disease Modeling. EMBO J. 2019, 38, e100300. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Mun, H.; Sung, C.O.; Cho, E.J.; Jeon, H.-J.; Chun, S.-M.; Jung, D.J.; Shin, T.H.; Jeong, G.S.; Kim, D.K.; et al. Patient-Derived Lung Cancer Organoids as in Vitro Cancer Models for Therapeutic Screening. Nat. Commun. 2019, 10, 3991. [Google Scholar] [CrossRef]
- Dye, B.R.; Hill, D.R.; Ferguson, M.A.; Tsai, Y.-H.; Nagy, M.S.; Dyal, R.; Wells, J.M.; Mayhew, C.N.; Nattiv, R.; Klein, O.D.; et al. In Vitro Generation of Human Pluripotent Stem Cell Derived Lung Organoids. eLife 2015, 4, e05098. [Google Scholar] [CrossRef]
- Lehmann, R.; Lee, C.M.; Shugart, E.C.; Benedetti, M.; Charo, R.A.; Gartner, Z.; Hogan, B.; Knoblich, J.; Nelson, C.M.; Wilson, K.M. Human Organoids: A New Dimension in Cell Biology. Mol. Biol. Cell 2019, 30, 1129–1137. [Google Scholar] [CrossRef]
- Izumchenko, E.; Paz, K.; Ciznadija, D.; Sloma, I.; Katz, A.; Vasquez-Dunddel, D.; Ben-Zvi, I.; Stebbing, J.; McGuire, W.; Harris, W.; et al. Patient-Derived Xenografts Effectively Capture Responses to Oncology Therapy in a Heterogeneous Cohort of Patients with Solid Tumors. Ann. Oncol. 2017, 28, 2595–2605. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-H.; Chu, X.-P.; Zhang, J.-T.; Nie, Q.; Tang, W.-F.; Su, J.; Yan, H.-H.; Zheng, H.-P.; Chen, Z.-X.; Chen, X.; et al. Genomic Characteristics and Drug Screening among Organoids Derived from Non-Small Cell Lung Cancer Patients. Thoracic. Cancer 2020, 11, 2279–2290. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Radulovich, N.; Ng, C.; Liu, N.; Notsuda, H.; Cabanero, M.; Martins-Filho, S.N.; Raghavan, V.; Li, Q.; Mer, A.S.; et al. Organoid Cultures as Preclinical Models of Non–Small Cell Lung Cancer. Clin. Cancer Res. 2020, 26, 1162–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dost, A.F.M.; Moye, A.L.; Vedaie, M.; Tran, L.M.; Fung, E.; Heinze, D.; Villacorta-Martin, C.; Huang, J.; Hekman, R.; Kwan, J.H.; et al. Organoids Model Transcriptional Hallmarks of Oncogenic KRAS Activation in Lung Epithelial Progenitor Cells. Cell Stem Cell 2020, 27, 663–678.e8. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Pierce, S.E.; Li, A.; Spees, K.; Anderson, G.R.; Seoane, J.A.; Lo, Y.-H.; Dubreuil, M.; Olivas, M.; Kamber, R.A.; et al. CRISPR Screens in Cancer Spheroids Identify 3D Growth-Specific Vulnerabilities. Nature 2020, 580, 136–141. [Google Scholar] [CrossRef]
- Burdett, N.; Desai, J. New Biomarkers for Checkpoint Inhibitor Therapy. ESMO Open 2020, 5, e000597. [Google Scholar] [CrossRef]
- Alcantara, M.; Rusquec, P.D.; Romano, E. Current Clinical Evidence and Potential Solutions to Increase Benefit of CAR T-Cell Therapy for Patients with Solid Tumors. OncoImmunology 2020, 9, 1777064. [Google Scholar] [CrossRef]
- Shifrut, E.; Carnevale, J.; Tobin, V.; Roth, T.L.; Woo, J.M.; Bui, C.T.; Li, P.J.; Diolaiti, M.E.; Ashworth, A.; Marson, A. Genome-Wide CRISPR Screens in Primary Human T Cells Reveal Key Regulators of Immune Function. Cell 2018, 175, 1958–1971.e15. [Google Scholar] [CrossRef] [Green Version]
- Roth, T.L.; Li, P.J.; Blaeschke, F.; Nies, J.F.; Apathy, R.; Mowery, C.; Yu, R.; Nguyen, M.L.T.; Lee, Y.; Truong, A.; et al. Pooled Knockin Targeting for Genome Engineering of Cellular Immunotherapies. Cell 2020, 181, 728–744.e21. [Google Scholar] [CrossRef] [PubMed]
- Roybal, K.T.; Lim, W.A. Synthetic Immunology: Hacking Immune Cells to Expand Their Therapeutic Capabilities. Annu. Rev. Immunol. 2017, 35, 229–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashima, Y.; Sakamoto, Y.; Kaneko, K.; Seki, M.; Suzuki, Y.; Suzuki, A. Single-Cell Sequencing Techniques from Individual to Multiomics Analyses. Exp. Mol. Med. 2020, 52, 1419–1427. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, Y.; Zheng, L.; Zheng, C.; Song, J.; Zhang, Q.; Kang, B.; Liu, Z.; Jin, L.; Xing, R.; et al. Global Characterization of T Cells in Non-Small-Cell Lung Cancer by Single-Cell Sequencing. Nat. Med. 2018, 24, 978–985. [Google Scholar] [CrossRef]
- Wu, S.-H. (Sam); Lee, J.-H.; Koo, B.-K. Lineage Tracing: Computational Reconstruction Goes Beyond the Limit of Imaging. Mol. Cells 2019, 42, 104. [Google Scholar] [CrossRef]
- Kunz, D.J.; Gomes, T.; James, K.R. Immune Cell Dynamics Unfolded by Single-Cell Technologies. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.N.; Kishton, R.J.; Restifo, N.P. Developing Neoantigen-Targeted T Cell–Based Treatments for Solid Tumors. Nat. Med. 2019, 25, 1488–1499. [Google Scholar] [CrossRef] [PubMed]
- Richman, L.P.; Vonderheide, R.H.; Rech, A.J. Neoantigen Dissimilarity to the Self-Proteome Predicts Immunogenicity and Response to Immune Checkpoint Blockade. Cell Syst. 2019, 9, 375–382.e4. [Google Scholar] [CrossRef]
- Łuksza, M.; Riaz, N.; Makarov, V.; Balachandran, V.P.; Hellmann, M.D.; Solovyov, A.; Rizvi, N.A.; Merghoub, T.; Levine, A.J.; Chan, T.A.; et al. A Neoantigen Fitness Model Predicts Tumour Response to Checkpoint Blockade Immunotherapy. Nature 2017, 551, 517–520. [Google Scholar] [CrossRef]
- McGranahan, N.; Rosenthal, R.; Hiley, C.T.; Rowan, A.J.; Watkins, T.B.K.; Wilson, G.A.; Birkbak, N.J.; Veeriah, S.; Van Loo, P.; Herrero, J.; et al. Allele-Specific HLA Loss and Immune Escape in Lung Cancer Evolution. Cell 2017, 171, 1259–1271.e11. [Google Scholar] [CrossRef] [PubMed]
- Garrido, F.; Cabrera, T.; Aptsiauri, N. “Hard” and “Soft” Lesions Underlying the HLA Class I Alterations in Cancer Cells: Implications for Immunotherapy. Int. J. Cancer 2010, 127, 249–256. [Google Scholar] [CrossRef]
- Newey, A.; Griffiths, B.; Michaux, J.; Pak, H.S.; Stevenson, B.J.; Woolston, A.; Semiannikova, M.; Spain, G.; Barber, L.J.; Matthews, N.; et al. Immunopeptidomics of Colorectal Cancer Organoids Reveals a Sparse HLA Class I Neoantigen Landscape and No Increase in Neoantigens with Interferon or MEK-Inhibitor Treatment. J. Immunother. Cancer 2019, 7, 309. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhou, L.; Xie, N.; Nice, E.C.; Zhang, T.; Cui, Y.; Huang, C. Overcoming Cancer Therapeutic Bottleneck by Drug Repurposing. Signal. Transduct. Target. Ther. 2020, 5, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Muramatsu, T.; Kanai, Y.; Ojima, H.; Sukeda, A.; Hiraoka, N.; Arai, E.; Sugiyama, Y.; Matsuzaki, J.; Uchida, R.; et al. Establishment of Patient-Derived Organoids and Drug Screening for Biliary Tract Carcinoma. Cell Rep. 2019, 27, 1265–1276.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Key Technology | Use Case | Therapeutic Implications | Advantages/Disadvantages |
|---|---|---|---|
| Whole Exome Sequencing | Assess 3D cancer model’s ability to recapitulate genomic composition of tumor tissue derivative | Personalized Drug(s) Trialing and Repurposing | Enables characterization and comparison of mutation profile but does not capture gene expression levels. |
| RNA Sequencing | Assess 3D cancer model’s ability to recapitulate transcriptome of tumor tissue derivative | Personalized Drug(s) Trialing and Repurposing | Enables comparison of relative gene expression but does not guarantee function at the protein level. |
| Single Cell RNA Sequencing | Identify critical cancer cell subpopulations in 3D lung cancer models for the study of cancer stem cells and cancer evolution | Novel therapeutic regimens that target newly identified driving pathways | Enables identification of rare cell populations with functional significance. Limited throughput currently. |
| Single Cell RNA Sequencing with Paired TCR/BCR Sequencing | Characterize immune repertoire and immune cell states in 3D cancer and immune cell co-culture models | Development of novel immune modulating drugs and cell therapies | Enables correlation of immune cell specificity and function. Limited throughput currently. |
| Mass Spectrometry | Identify neoantigen burden and targets for immune-based therapies from 3D cancer models | Development of Autologous (Engineered) immune cell therapies | Enables definitive characterization of cancer cell protein expression. Requires significant sample input. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, P.J.; Roose, J.P.; Jablons, D.M.; Kratz, J.R. Bioinformatic Approaches to Validation and Functional Analysis of 3D Lung Cancer Models. Cancers 2021, 13, 701. https://doi.org/10.3390/cancers13040701
Li PJ, Roose JP, Jablons DM, Kratz JR. Bioinformatic Approaches to Validation and Functional Analysis of 3D Lung Cancer Models. Cancers. 2021; 13(4):701. https://doi.org/10.3390/cancers13040701
Chicago/Turabian StyleLi, P. Jonathan, Jeroen P. Roose, David M. Jablons, and Johannes R. Kratz. 2021. "Bioinformatic Approaches to Validation and Functional Analysis of 3D Lung Cancer Models" Cancers 13, no. 4: 701. https://doi.org/10.3390/cancers13040701






