Genomic Landscape of Hodgkin Lymphoma
Abstract
Simple Summary
Abstract
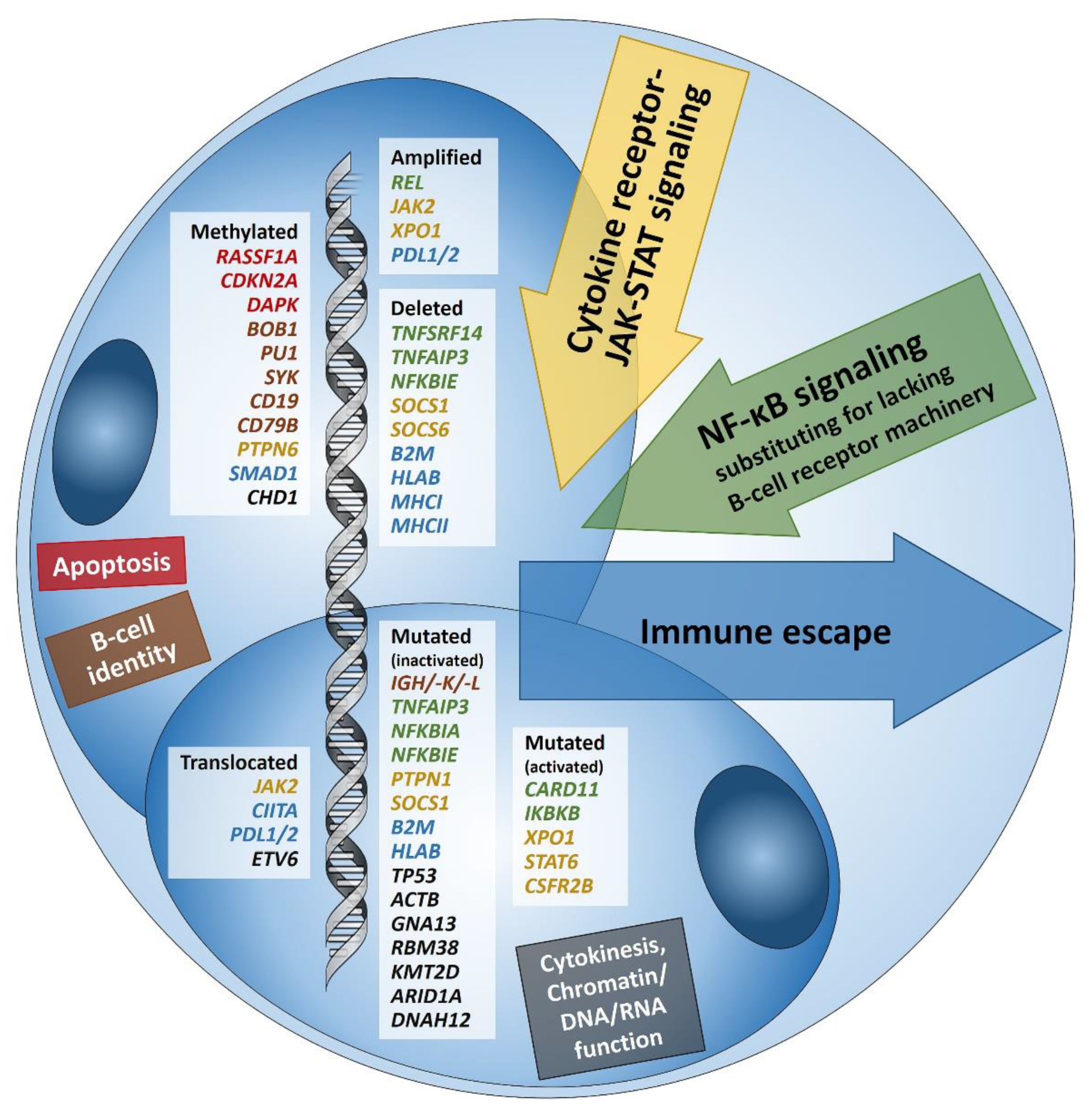
1. Introduction: Searching and Finding the Needle in the Haystack
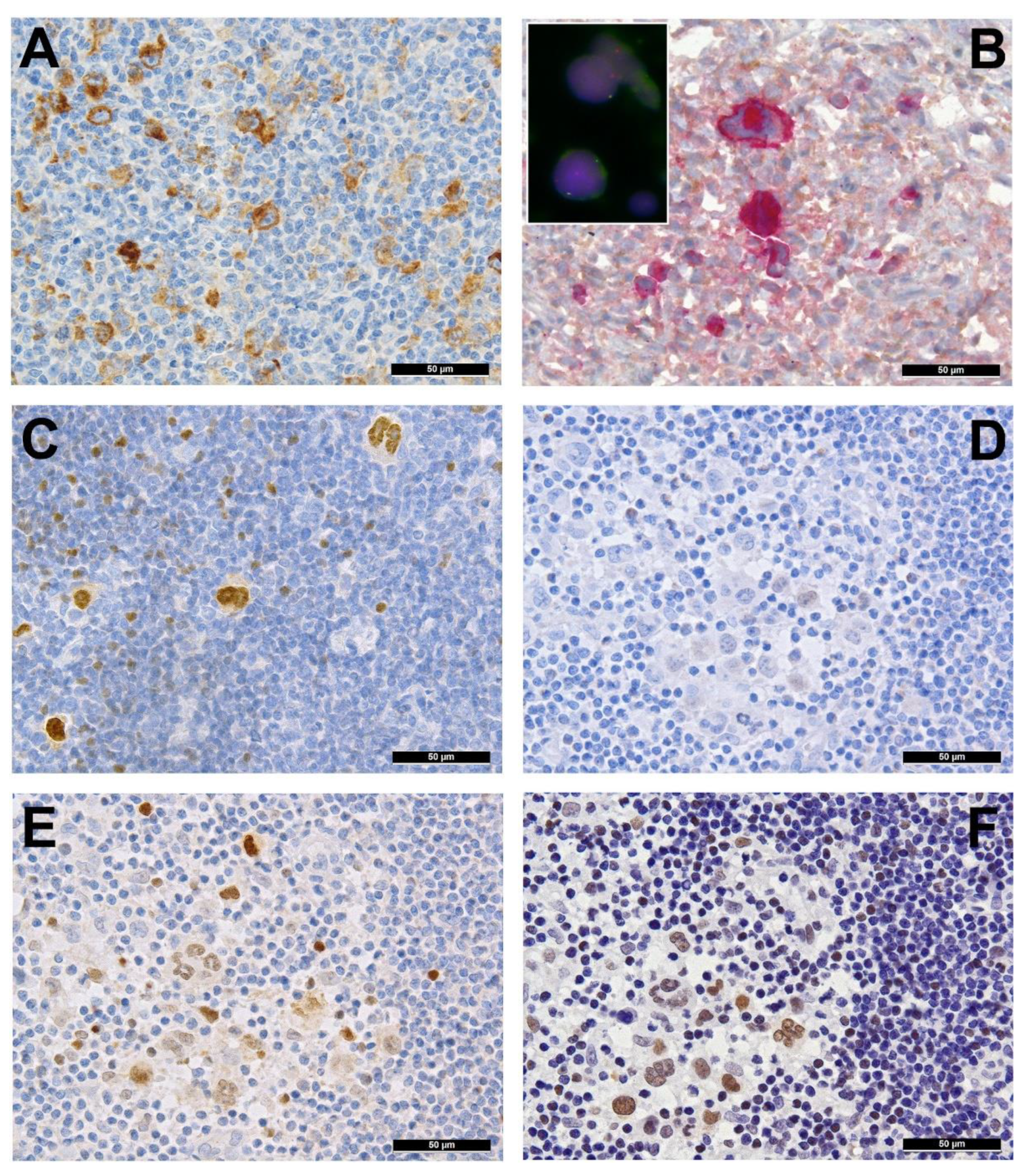
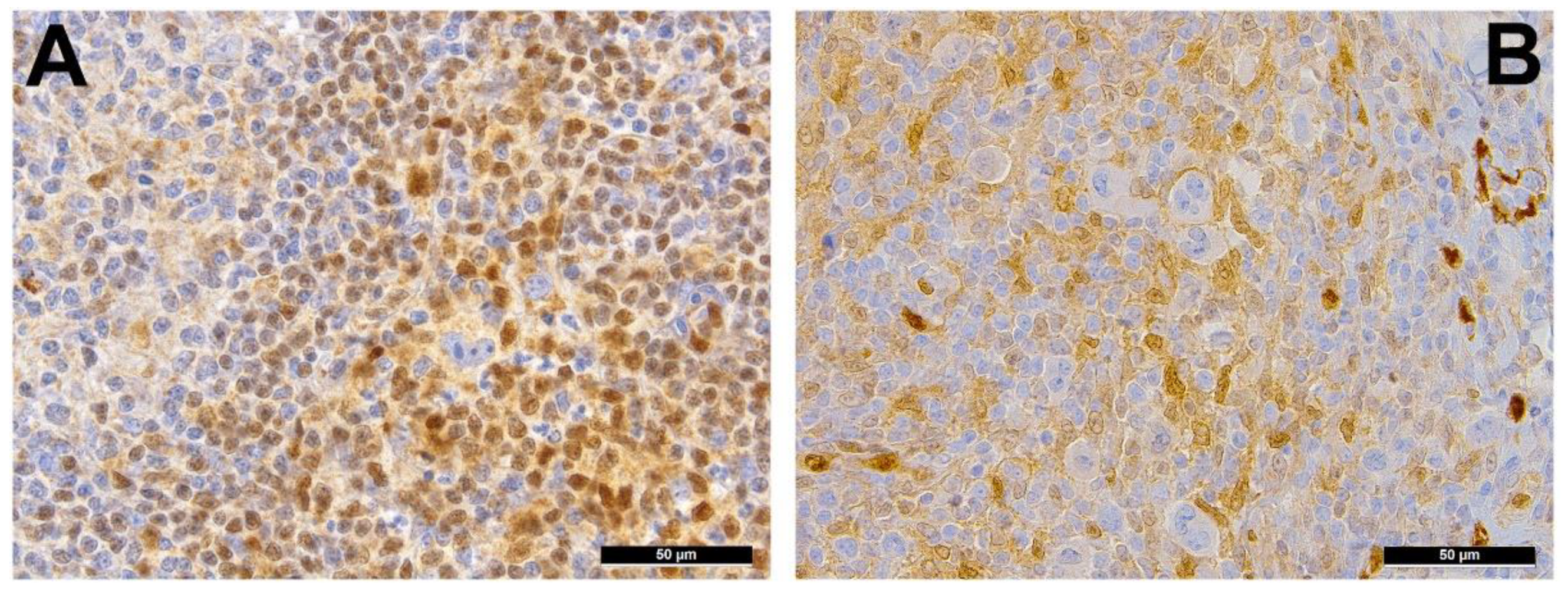
2. Immune Evasion
3. Pervasive JAK/STAT Signaling
4. Disruption of the NF-κB Pathway
5. Alterations in PI3K/AKT/mTOR Pathway and Impairment of Cytokinesis
6. Other Genetic Aberrations
7. Epstein-Barr Virus
8. Epigenetics in cHL
9. NLPHL
10. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, Y.; Abdul Razak, F.R.; Terpstra, M.; Chan, F.C.; Saber, A.; Nijland, M.; van Imhoff, G.; Visser, L.; Gascoyne, R.; Steidl, C.; et al. The mutational landscape of Hodgkin lymphoma cell lines determined by whole-exome sequencing. Leukemia 2014, 28, 2248–2251. [Google Scholar] [CrossRef]
- Drexler, H.G.; Pommerenke, C.; Eberth, S.; Nagel, S. Hodgkin lymphoma cell lines: To separate the wheat from the chaff. Biol. Chem. 2018, 399, 511–523. [Google Scholar] [CrossRef]
- Atayar, C.; Kok, K.; Kluiver, J.; Bosga, A.; van den Berg, E.; van der Vlies, P.; Blokzijl, T.; Harms, G.; Davelaar, I.; Sikkema-Raddatz, B.; et al. BCL6 alternative breakpoint region break and homozygous deletion of 17q24 in the nodular lymphocyte predominance type of Hodgkin’s lymphoma-derived cell line DEV. Hum. Pathol. 2006, 37, 675–683. [Google Scholar] [CrossRef]
- Drexler, H.G.; Dirks, W.G.; Matsuo, Y.; MacLeod, R.A. False leukemia-lymphoma cell lines: An update on over 500 cell lines. Leukemia 2003, 17, 416–426. [Google Scholar] [CrossRef]
- Hartmann, S.; Martin-Subero, J.I.; Gesk, S.; Hüsken, J.; Giefing, M.; Nagel, I.; Riemke, J.; Chott, A.; Klapper, W.; Parrens, M.; et al. Detection of genomic imbalances in microdissected Hodgkin and Reed-Sternberg cells of classical Hodgkin’s lymphoma by array-based comparative genomic hybridization. Haematologica 2008, 93, 1318–1326. [Google Scholar] [CrossRef] [PubMed]
- Weniger, M.A.; Barth, T.F.; Möller, P. Genomic alterations in Hodgkin’s lymphoma. Int. J. Hematol. 2006, 83, 379–384. [Google Scholar] [CrossRef]
- Juskevicius, D.; Jucker, D.; Dietsche, T.; Perrina, V.; Rufle, A.; Ruiz, C.; Dirnhofer, S.; Tzankov, A. Novel cell enrichment technique for robust genetic analysis of archival classical Hodgkin lymphoma tissues. Lab. Investig. 2018, 98, 1487–1499. [Google Scholar] [CrossRef]
- Fromm, J.R.; Kussick, S.J.; Wood, B.L. Identification and purification of classical Hodgkin cells from lymph nodes by flow cytometry and flow cytometric cell sorting. Am. J. Clin. Pathol. 2006, 126, 764–780. [Google Scholar] [CrossRef]
- Reichel, J.; Chadburn, A.; Rubinstein, P.G.; Giulino-Roth, L.; Tam, W.; Liu, Y.; Gaiolla, R.; Eng, K.; Brody, J.; Inghirami, G.; et al. Flow sorting and exome sequencing reveal the oncogenome of primary Hodgkin and Reed-Sternberg cells. Blood 2015, 125, 1061–1072. [Google Scholar] [CrossRef]
- Juskevicius, D.; Lorber, T.; Gsponer, J.; Perrina, V.; Ruiz, C.; Stenner-Liewen, F.; Dirnhofer, S.; Tzankov, A. Distinct genetic evolution patterns of relapsing diffuse large B-cell lymphoma revealed by genome-wide copy number aberration and targeted sequencing analysis. Leukemia 2016, 30, 2385–2395. [Google Scholar] [CrossRef]
- Wienand, K.; Chapuy, B.; Stewart, C.; Dunford, A.J.; Wu, D.; Kim, J.; Kamburov, A.; Wood, T.R.; Cader, F.Z.; Ducar, M.D.; et al. Genomic analyses of flow-sorted Hodgkin Reed-Sternberg cells reveal complementary mechanisms of immune evasion. Blood Adv. 2019, 3, 4065–4080. [Google Scholar] [CrossRef]
- Spina, V.; Bruscaggin, A.; Cuccaro, A.; Martini, M.; Di Trani, M.; Forestieri, G.; Manzoni, M.; Condoluci, A.; Arribas, A.; Terzi-Di-Bergamo, L.; et al. Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood 2018, 131, 2413–2425. [Google Scholar] [CrossRef] [PubMed]
- Desch, A.K.; Hartung, K.; Botzen, A.; Brobeil, A.; Rummel, M.; Kurch, L.; Georgi, T.; Jox, T.; Bielack, S.; Burdach, S. Genotyping circulating tumor DNA of pediatric Hodgkin lymphoma. Leukemia 2020, 34, 151–166. [Google Scholar] [CrossRef]
- Küppers, R.; Rajewsky, K.; Zhao, M.; Simons, G.; Laumann, R.; Fischer, R.; Hansmann, M.L. Hodgkin disease: Hodgkin and Reed-Sternberg cells picked from histological sections show clonal immunoglobulin gene rearrangements and appear to be derived from B cells at various stages of development. Proc. Natl. Acad. Sci. USA 1994, 91, 10962–10966. [Google Scholar] [CrossRef]
- Roemer, M.G.; Advani, R.H.; Ligon, A.H.; Natkunam, Y.; Redd, R.A.; Homer, H.; Connelly, C.F.; Sun, H.H.; Daadi, S.E.; Freeman, G.J.; et al. PD-L1 and PD-L2 Genetic Alterations Define Classical Hodgkin Lymphoma and Predict Outcome. J. Clin. Oncol. 2016, 34, 2690–2697. [Google Scholar] [CrossRef] [PubMed]
- Tiacci, E.; Ladewig, E.M.; Schiavoni, G.; Penson, A.; Fortini, E.; Pettirossi, V.; Wang, Y.; Rosseto, A.; Venanzi, A.; Vlasevska, S.; et al. Pervasive mutations of JAK-STAT pathway genes in classical Hodgkin lymphoma. Blood 2018, 131, 2454–2465. [Google Scholar] [CrossRef] [PubMed]
- Steidl, C.; Shah, S.P.; Woolcock, B.W.; Rui, L.; Kawahara, M.; Farinha, P.; Johnson, N.A.; Zhao, Y.; Telenius, A.; Neriah, S.B.; et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature 2011, 471, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Kilger, E.; Kieser, A.; Baumann, M.; Hammerschmidt, W. Epstein-Barr virus-mediated B-cell proliferation is dependent upon latent membrane protein 1, which simulates an activated CD40 receptor. EMBO J. 1998, 17, 1700–1709. [Google Scholar] [CrossRef]
- Gerlach, M.M.; Stelling-Germani, A.; Wu, C.T.; Newrzela, S.; Döring, C.; Vela, V.; Müller, A.; Hartmann, S.; Tzankov, A. SMAD1 promoter hypermethylation and lack of SMAD1 expression in Hodgkin lymphoma: A potential target for hypomethylating drug therapy. Haematologica 2021, 106, 619–621. [Google Scholar] [CrossRef]
- Menter, T.; Tzankov, A. Genetic alterations of 9p24 in lymphomas and their impact for cancer (immuno-)therapy. Virchows Arch. 2019, 474, 497–509. [Google Scholar] [CrossRef]
- Francisco, L.M.; Salinas, V.H.; Brown, K.E.; Vanguri, V.K.; Freeman, G.J.; Kuchroo, V.K.; Sharpe, A.H. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J. Exp. Med. 2009, 206, 3015–3029. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Monti, S.; Rodig, S.J.; Juszczynski, P.; Currie, T.; O’Donnell, E.; Chapuy, B.; Takeyama, K.; Neuberg, D.; Golub, T.R.; et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood 2010, 116, 3268–3277. [Google Scholar] [CrossRef]
- van Roosbroeck, K.; Ferreiro, J.F.; Tousseyn, T.; van der Krogt, J.A.; Michaux, L.; Pienkowska-Grela, B.; Theate, I.; De Paepe, P.; Dierickx, D.; Doyen, C.; et al. Genomic alterations of the JAK2 and PDL loci occur in a broad spectrum of lymphoid malignancies. Genes Chromosomes Cancer 2016, 55, 428–441. [Google Scholar] [CrossRef]
- Roemer, M.G.M.; Redd, R.A.; Cader, F.Z.; Pak, C.J.; Abdelrahman, S.; Ouyang, J.; Sasse, S.; Younes, A.; Fanale, M.; Santoro, A.; et al. Major Histocompatibility Complex Class II and Programmed Death Ligand 1 Expression Predict Outcome After Programmed Death 1 Blockade in Classic Hodgkin Lymphoma. J. Clin. Oncol. 2018, 36, 942–950. [Google Scholar] [CrossRef] [PubMed]
- Ansell, S.M.; Lesokhin, A.M.; Borrello, I.; Halwani, A.; Scott, E.C.; Gutierrez, M.; Schuster, S.J.; Millenson, M.M.; Cattry, D.; Freeman, G.J.; et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N. Engl. J. Med. 2015, 372, 311–319. [Google Scholar] [CrossRef]
- Voorhees, T.J.; Beaven, A.W. Therapeutic updates for relapsed and refractory classical Hodgkin lymphoma. Cancers 2020, 12, 2887. [Google Scholar] [CrossRef] [PubMed]
- Cader, F.Z.; Hu, X.; Goh, W.L.; Wienand, K.; Ouyang, J.; Mandato, E.; Redd, R.; Lawton, L.N.; Chen, P.H.; Weirather, J.L.; et al. A peripheral immune signature of responsiveness to PD-1 blockade in patients with classical Hodgkin lymphoma. Nat. Med. 2020, 26, 1468–1479. [Google Scholar] [CrossRef]
- Roemer, M.G.M.; Advani, R.H.; Redd, R.A.; Pinkus, G.S.; Natkunam, Y.; Ligon, A.H.; Connelly, C.F.; Pak, C.J.; Carey, C.D.; Daadi, S.E.; et al. Classical Hodgkin Lymphoma with Reduced β2M/MHC Class I Expression Is Associated with Inferior Outcome Independent of 9p24.1 Status. Cancer Immunol. Res. 2016, 4, 910–916. [Google Scholar] [CrossRef]
- Nagpal, P.; Descalzi-Montoya, D.B.; Lodhi, N. The circuitry of the tumor microenvironment in adult and pediatric Hodgkin lymphoma: Cellular composition, cytokine profile, EBV, and exosomes. Cancer Rep. (Hoboken) 2020. ahead of print. [Google Scholar] [CrossRef]
- Stelling, A.; Wu, C.T.; Bertram, K.; Hashwah, H.; Theocharides, A.; Manz, M.G.; Tzankov, A.; Müller, A. Pharmacological DNA demethylation restores SMAD1 expression and tumor suppressive signaling in diffuse large B-cell lymphoma. Blood Adv. 2019, 3, 3020–3032. [Google Scholar] [CrossRef]
- Meier, C.; Hoeller, S.; Bourgau, C.; Hirschmann, P.; Schwaller, J.; Went, P.; Pileri, S.A.; Reiter, A.; Dirnhofer, S.; Tzankov, A. Recurrent numerical aberrations of JAK2 and deregulation of the JAK2-STAT cascade in lymphomas. Mod. Pathol. 2009, 22, 476–487. [Google Scholar] [CrossRef]
- Rui, L.; Emre, N.C.; Kruhlak, M.J.; Chung, H.J.; Steidl, C.; Slack, G.; Wright, G.W.; Lenz, G.; Ngo, V.N.; Shaffer, A.L.; et al. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell 2010, 18, 590–605. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Calhoun, H.C.; Xia, F.; Li, J.; Le, L.; Li, W.X. JAK signaling globally counteracts heterochromatic gene silencing. Nat. Genet. 2006, 38, 1071–1076. [Google Scholar] [CrossRef]
- Mottok, A.; Renné, C.; Willenbrock, K.; Hansmann, M.L.; Bräuninger, A. Somatic hypermutation of SOCS1 in lymphocyte-predominant Hodgkin lymphoma is accompanied by high JAK2 expression and activation of STAT6. Blood 2007, 110, 3387–3390. [Google Scholar] [CrossRef] [PubMed]
- Weniger, M.A.; Melzner, I.; Menz, C.K.; Wegener, S.; Bucur, A.J.; Dorsch, K.; Mattfeldt, T.; Barth, T.F.; Möller, P. Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho-STAT5 accumulation. Oncogene 2006, 25, 2679–2684. [Google Scholar] [CrossRef] [PubMed]
- Camus, V.; Stamatoullas, A.; Mareschal, S.; Viailly, P.J.; Sarafan-Vasseur, N.; Bohers, E.; Dubois, S.; Picquenot, J.M.; Ruminy, P.; Maingonnat, C.; et al. Detection and prognostic value of recurrent exportin 1 mutations in tumor and cell-free circulating DNA of patients with classical Hodgkin lymphoma. Haematologica 2016, 101, 1094–1101. [Google Scholar] [CrossRef] [PubMed]
- Stade, K.; Ford, C.S.; Guthrie, C.; Weis, K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 1997, 90, 1041–1050. [Google Scholar] [CrossRef]
- Azizian, N.G.; Li, Y. XPO1-dependent nuclear export as a target for cancer therapy. J. Hematol. Oncol. 2020, 13, 61. [Google Scholar] [CrossRef]
- Deng, M.; Zhang, M.; Xu-Monette, Z.Y.; Pham, L.V.; Tzankov, A.; Visco, C.; Fang, X.; Bhagat, G.; Zhu, F.; Dybkaer, K.; et al. XPO1 expression worsens the prognosis of unfavorable DLBCL that can be effectively targeted by selinexor in the absence of mutant p53. J. Hematol. Oncol. 2020, 13, 148. [Google Scholar] [CrossRef]
- Gunawardana, J.; Chan, F.C.; Telenius, A.; Woolcock, B.; Kridel, R.; Tan, K.L.; Ben-Neriah, S.; Mottok, A.; Lim, R.S.; Boyle, M.; et al. Recurrent somatic mutations of PTPN1 in primary mediastinal B cell lymphoma and Hodgkin lymphoma. Nat. Genet. 2014, 46, 329–335. [Google Scholar] [CrossRef]
- Dhiab, M.B.; Ziadi, S.; Mestiri, S.; Gacem, R.B.; Ksiaa, F.; Trimeche, M. DNA methylation patterns in EBV-positive and EBV-negative Hodgkin lymphomas. Cell Oncol. (Dordr.) 2015, 38, 453–462. [Google Scholar] [CrossRef]
- van Roosbroeck, K.; Cox, L.; Tousseyn, T.; Lahortiga, I.; Gielen, O.; Cauwelier, B.; De Paepe, P.; Verhoef, G.; Marynen, P.; Vandenberghe, P.; et al. JAK2 rearrangements, including the novel SEC31A-JAK2 fusion, are recurrent in classical Hodgkin lymphoma. Blood 2011, 117, 4056–4064. [Google Scholar] [CrossRef]
- Berry, W.L.; Janknecht, R. KDM4/JMJD2 histone demethylases: Epigenetic regulators in cancer cells. Cancer Res. 2013, 73, 2936–2942. [Google Scholar] [CrossRef]
- Schmitz, R.; Hansmann, M.L.; Bohle, V.; Martin-Subero, J.I.; Hartmann, S.; Mechtersheimer, G.; Klapper, W.; Vater, I.; Giefing, M.; Gesk, S.; et al. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J. Exp. Med. 2009, 206, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Emmerich, F.; Theurich, S.; Hummel, M.; Haeffker, A.; Vry, M.S.; Döhner, K.; Bommert, K.; Stein, H.; Dörken, B. Inactivating I kappa B epsilon mutations in Hodgkin/Reed-Sternberg cells. J. Pathol. 2003, 201, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Cabannes, E.; Khan, G.; Aillet, F.; Jarrett, R.F.; Hay, R.T. Mutations in the IkBa gene in Hodgkin’s disease suggest a tumour suppressor role for IkappaBalpha. Oncogene 1999, 18, 3063–3070. [Google Scholar] [CrossRef] [PubMed]
- Emmerich, F.; Meiser, M.; Hummel, M.; Demel, G.; Foss, H.D.; Jundt, F.; Mathas, S.; Krappmann, D.; Scheidereit, C.; Stein, H.; et al. Overexpression of I kappa B alpha without inhibition of NF-kappaB activity and mutations in the I kappa B alpha gene in Reed-Sternberg cells. Blood 1999, 94, 3129–3134. [Google Scholar] [CrossRef]
- Jungnickel, B.; Staratschek-Jox, A.; Bräuninger, A.; Spieker, T.; Wolf, J.; Diehl, V.; Hansmann, M.L.; Rajewsky, K.; Küppers, R. Clonal deleterious mutations in the IkappaBalpha gene in the malignant cells in Hodgkin’s lymphoma. J. Exp. Med. 2000, 191, 395–402. [Google Scholar] [CrossRef]
- Joos, S.; Menz, C.K.; Wrobel, G.; Siebert, R.; Gesk, S.; Ohl, S.; Mechtersheimer, G.; Trümper, L.; Möller, P.; Lichter, P.; et al. Classical Hodgkin lymphoma is characterized by recurrent copy number gains of the short arm of chromosome 2. Blood 2002, 99, 1381–1387. [Google Scholar] [CrossRef]
- Martín-Subero, J.I.; Gesk, S.; Harder, L.; Sonoki, T.; Tucker, P.W.; Schlegelberger, B.; Grote, W.; Novo, F.J.; Calasanz, M.J.; Hansmann, M.L.; et al. Recurrent involvement of the REL and BCL11A loci in classical Hodgkin lymphoma. Blood 2002, 99, 1474–1477. [Google Scholar] [CrossRef]
- Meadows, S.A.; Vega, F.; Kashishian, A.; Johnson, D.; Diehl, V.; Miller, L.L.; Younes, A.; Lannutti, B.J. PI3Kδ inhibitor, GS-1101 (CAL-101), attenuates pathway signaling, induces apoptosis, and overcomes signals from the microenvironment in cellular models of Hodgkin lymphoma. Blood 2012, 119, 1897–1900. [Google Scholar] [CrossRef]
- Healy, J.A.; Nugent, A.; Rempel, R.E.; Moffitt, A.B.; Davis, N.S.; Jiang, X.; Shingleton, J.R.; Zhang, J.; Love, C.; Datta, J.; et al. GNA13 loss in germinal center B cells leads to impaired apoptosis and promotes lymphoma in vivo. Blood 2016, 127, 2723–2731. [Google Scholar] [CrossRef] [PubMed]
- Westernberg, L.; Conche, C.; Huang, Y.H.; Rigaud, S.; Deng, Y.; Siegemund, S.; Mukherjee, S.; Nosaka, L.; Das, J.; Sauer, K. Non-canonical antagonism of PI3K by the kinase Itpkb delays thymocyte β-selection and renders it Notch-dependent. Elife 2016, 5, e10786. [Google Scholar] [CrossRef]
- Rengstl, B.; Newrzela, S.; Heinrich, T.; Weiser, C.; Thalheimer, F.B.; Schmid, F.; Warner, K.; Hartmann, S.; Schroeder, T.; Küppers, R.; et al. Incomplete cytokinesis and re-fusion of small mononucleated Hodgkin cells lead to giant multinucleated Reed-Sternberg cells. Proc. Natl. Acad. Sci. USA 2013, 110, 20729–20734. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, M.; Ishii, Y.; Watanabe, M.; Togano, T.; Umezawa, K.; Higashihara, M.; Watanabe, T.; Horie, R. The side population, as a precursor of Hodgkin and Reed-Sternberg cells and a target for nuclear factor-κB inhibitors in Hodgkin’s lymphoma. Cancer Sci. 2010, 101, 2490–2496. [Google Scholar] [CrossRef]
- Neves, S.R.; Ram, P.T.; Iyengar, R. G protein pathways. Science 2002, 296, 1636–1639. [Google Scholar] [CrossRef]
- Weber-Matthiesen, K.; Deerberg, J.; Poetsch, M.; Grote, W.; Schlegelberger, B. Numerical chromosome aberrations are presentwithin the CD30C Hodgkin and ReedeSternberg cells in 100% ofanalyzed cases of Hodgkin’s disease. Blood 1995, 86, 1464–1468. [Google Scholar] [CrossRef] [PubMed]
- Stokes, D.G.; Perry, R.P. DNA-binding and chromatin localization properties of CHD1. Mol. Cell. Biol. 1995, 15, 2745–2753. [Google Scholar] [CrossRef]
- Liang, W.S.; Vergilio, J.A.; Salhia, B.; Huang, H.J.; Oki, Y.; Garrido-Laguna, I.; Park, H.; Westin, J.R.; Meric-Bernstam, F.; Fabrizio, D.; et al. Comprehensive genomic profiling of Hodgkin lymphoma reveals recurrently mutated genes and increased mutation burden. Oncologist 2019, 24, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xu, E.; Ren, C.; Yang, H.J.; Zhang, Y.; Sun, W.; Kong, X.; Zhang, W.; Chen, M.; Huang, E.; et al. Genetic ablation of Rbm38 promotes lymphomagenesis in the context of mutant p53 by downregulating PTEN. Cancer Res. 2018, 78, 1511–1521. [Google Scholar] [CrossRef]
- Wang, Q.; Dong, S.; Yao, H.; Wen, L.; Qiu, H.; Qin, L.; Ma, L.; Chen, S. ETV6 mutation in a cohort of 970 patients with hematologic malignancies. Haematologica 2014, 99, e176–e178. [Google Scholar] [CrossRef] [PubMed]
- Hannigan, A.; Wilson, J.B. Evaluation of LMP1 of Epstein-Barr virus as a therapeutic target by its inhibition. Mol. Cancer 2010, 9, 184. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.G.; Young, L.S. An etiological role for the Epstein-Barr virus in the pathogenesis of classical Hodgkin lymphoma. Blood 2019, 134, 591–596. [Google Scholar] [CrossRef]
- Mosialos, G.; Birkenbach, M.; Yalamanchili, R.; VanArsdale, T.; Ware, C.; Kieff, E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell 1995, 80, 389–399. [Google Scholar] [CrossRef]
- Mathas, S.; Hartmann, S.; Küppers, R. Hodgkin lymphoma: Pathology and biology. Semin. Hematol. 2016, 53, 139–147. [Google Scholar] [CrossRef]
- Green, M.R.; Rodig, S.; Juszczynski, P.; Ouyang, J.; Sinha, P.; O’Donnell, E.; Neuberg, D.; Shipp, M.A. Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: Implications for targeted therapy. Clin. Cancer Res. 2012, 18, 1611–1618. [Google Scholar] [CrossRef] [PubMed]
- Menter, T.; Tzankov, A. Mechanisms of immune evasion and immune modulation by lymphoma cells. Front. Oncol. 2018, 8, 54. [Google Scholar] [CrossRef]
- Ok, C.Y.; Li, L.; Young, K.H. EBV-driven B-cell lymphoproliferative disorders: From biology, classification and differential diagnosis to clinical management. Exp. Mol. Med. 2015, 47, e132. [Google Scholar] [CrossRef] [PubMed]
- Ushmorov, A.; Leithäuser, F.; Sakk, O.; Weinhaüsel, A.; Popov, S.W.; Möller, P.; Wirth, T. Epigenetic processes play a major role in B-cell-specific gene silencing in classical Hodgkin lymphoma. Blood 2006, 107, 2493–2500. [Google Scholar] [CrossRef]
- Tsai, C.L.; Li, H.P.; Lu, Y.J.; Hsueh, C.; Liang, Y.; Chen, C.L.; Tsao, S.W.; Tse, K.P.; Yu, J.S.; Chang, Y.S. Activation of DNA methyltransferase 1 by EBV LMP1 Involves c-Jun NH(2)-terminal kinase signaling. Cancer Res. 2006, 66, 11668–11676. [Google Scholar] [CrossRef]
- Brune, V.; Tiacci, E.; Pfeil, I.; Döring, C.; Eckerle, S.; van Noesel, C.J.; Klapper, W.; Falini, B.; von Heydebreck, A.; Metzler, D.; et al. Origin and pathogenesis of nodular lymphocyte-predominant Hodgkin lymphoma as revealed by global gene expression analysis. J. Exp. Med. 2008, 205, 2251–2268. [Google Scholar] [CrossRef]
- Hartmann, S.; Eichenauer, D.A. Nodular lymphocyte predominant Hodgkin lymphoma: Pathology, clinical course and relation to T-cell/histiocyte rich large B-cell lymphoma. Pathology 2020, 52, 142–153. [Google Scholar] [CrossRef]
- Liso, A.; Capello, D.; Marafioti, T.; Tiacci, E.; Cerri, M.; Distler, V.; Paulli, M.; Carbone, A.; Delsol, G.; Campo, E.; et al. Aberrant somatic hypermutation in tumor cells of nodular-lymphocyte-predominant and classic Hodgkin lymphoma. Blood 2006, 108, 1013–1020. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schumacher, M.A.; Schmitz, R.; Brune, V.; Tiacci, E.; Döring, C.; Hansmann, M.L.; Siebert, R.; Küppers, R. Mutations in the genes coding for the NF-κB regulating factors IκBα and A20 are uncommon in nodular lymphocyte-predominant Hodgkin’s lymphoma. Haematologica 2010, 95, 153–157. [Google Scholar] [CrossRef]
- Hartmann, S.; Schuhmacher, B.; Rausch, T.; Fuller, L.; Döring, C.; Weniger, M.; Lollies, A.; Weiser, C.; Thurner, L.; Rengstl, B.; et al. Highly recurrent mutations of SGK1, DUSP2 and JUNB in nodular lymphocyte predominant Hodgkin lymphoma. Leukemia 2016, 30, 844–853. [Google Scholar] [CrossRef] [PubMed]
- Wlodarska, I.; Nooyen, P.; Maes, B.; Martin-Subero, J.I.; Siebert, R.; Pauwels, P.; De Wolf-Peeters, C.; Hagemeijer, A. Frequent occurrence of BCL6 rearrangements in nodular lymphocyte predominance Hodgkin lymphoma but not in classical Hodgkin lymphoma. Blood 2003, 101, 706–710. [Google Scholar] [CrossRef] [PubMed]
- Renné, C.; Martín-Subero, J.I.; Hansmann, M.L.; Siebert, R. Molecular cytogenetic analyses of immunoglobulin loci in nodular lymphocyte predominant Hodgkin’s lymphoma reveal a recurrent IGH-BCL6 juxtaposition. J. Mol. Diagn. 2005, 7, 352–356. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brune, M.M.; Juskevicius, D.; Haslbauer, J.; Dirnhofer, S.; Tzankov, A. Genomic Landscape of Hodgkin Lymphoma. Cancers 2021, 13, 682. https://doi.org/10.3390/cancers13040682
Brune MM, Juskevicius D, Haslbauer J, Dirnhofer S, Tzankov A. Genomic Landscape of Hodgkin Lymphoma. Cancers. 2021; 13(4):682. https://doi.org/10.3390/cancers13040682
Chicago/Turabian StyleBrune, Magdalena M., Darius Juskevicius, Jasmin Haslbauer, Stefan Dirnhofer, and Alexandar Tzankov. 2021. "Genomic Landscape of Hodgkin Lymphoma" Cancers 13, no. 4: 682. https://doi.org/10.3390/cancers13040682
APA StyleBrune, M. M., Juskevicius, D., Haslbauer, J., Dirnhofer, S., & Tzankov, A. (2021). Genomic Landscape of Hodgkin Lymphoma. Cancers, 13(4), 682. https://doi.org/10.3390/cancers13040682






