NRF2 Activation Confers Resistance to eIF4A Inhibitors in Cancer Therapy
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
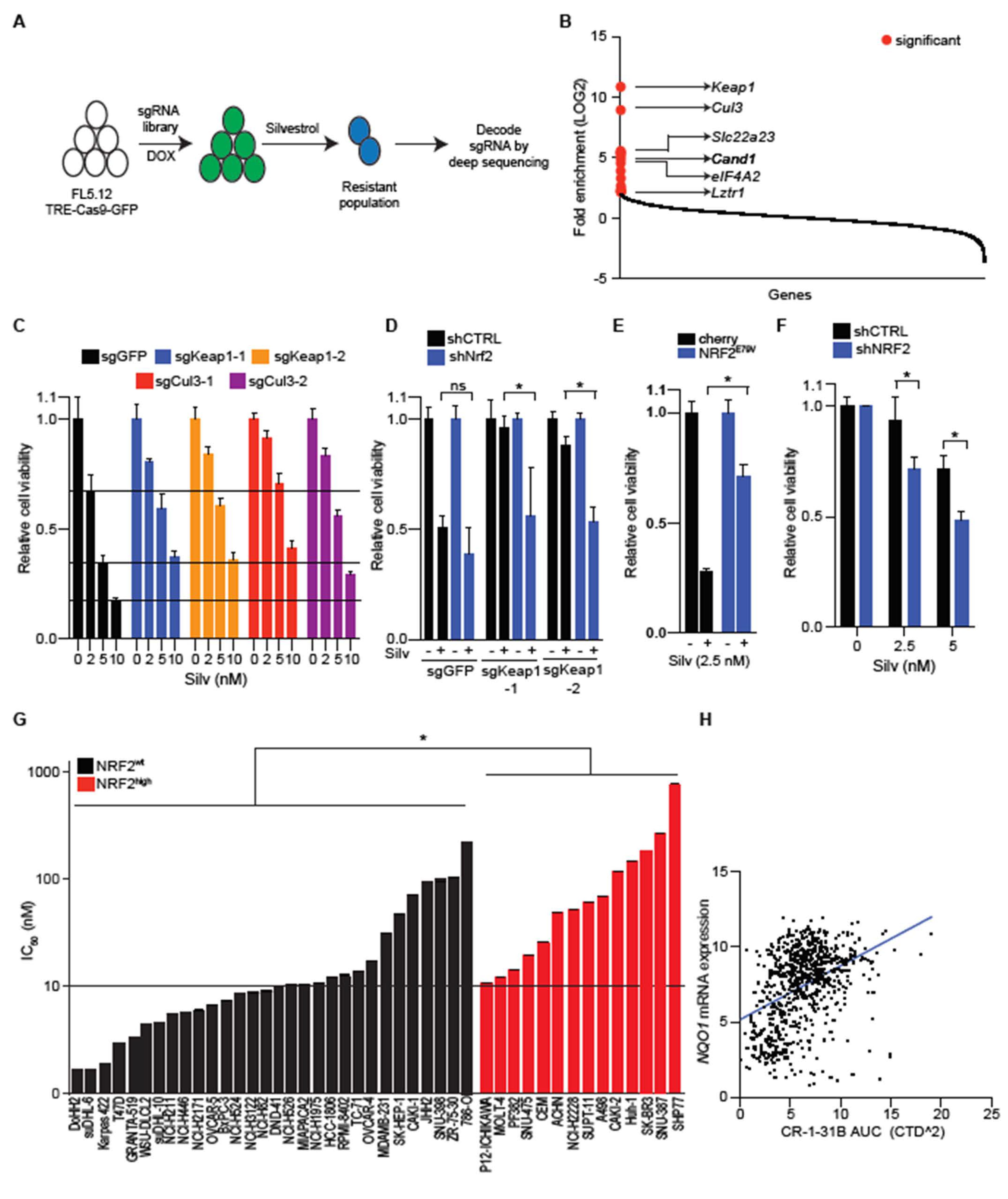
2.1. CR-1-31 B Kills Aggressive Lymphoma Cells at Nanomolar Concentrations
2.2. NRF2 Activation Confers Resistance to EIF4A Inhibition
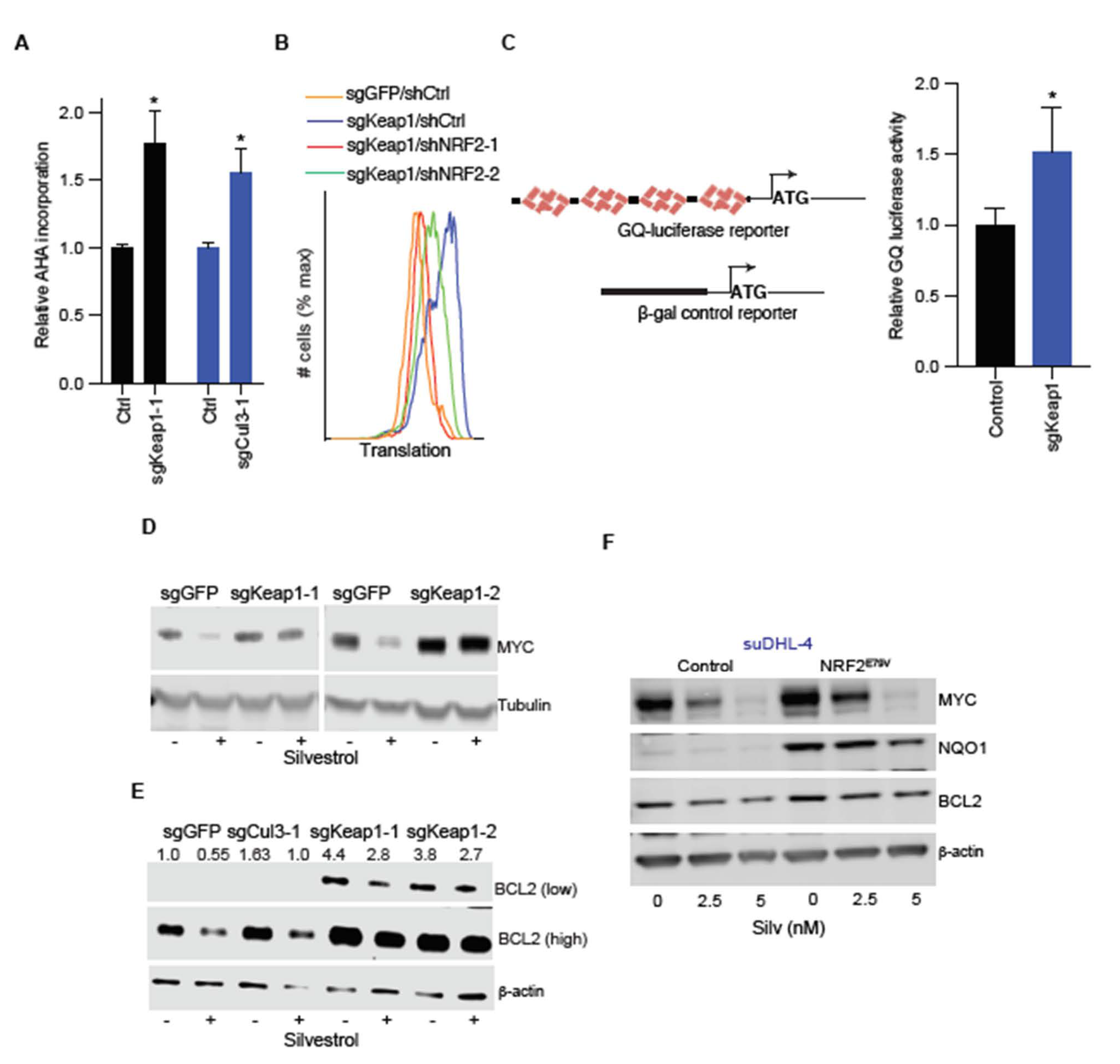
2.3. NRF2 Increases Translation of the eIF4A-Dependent mRNAs MYC and BCL2
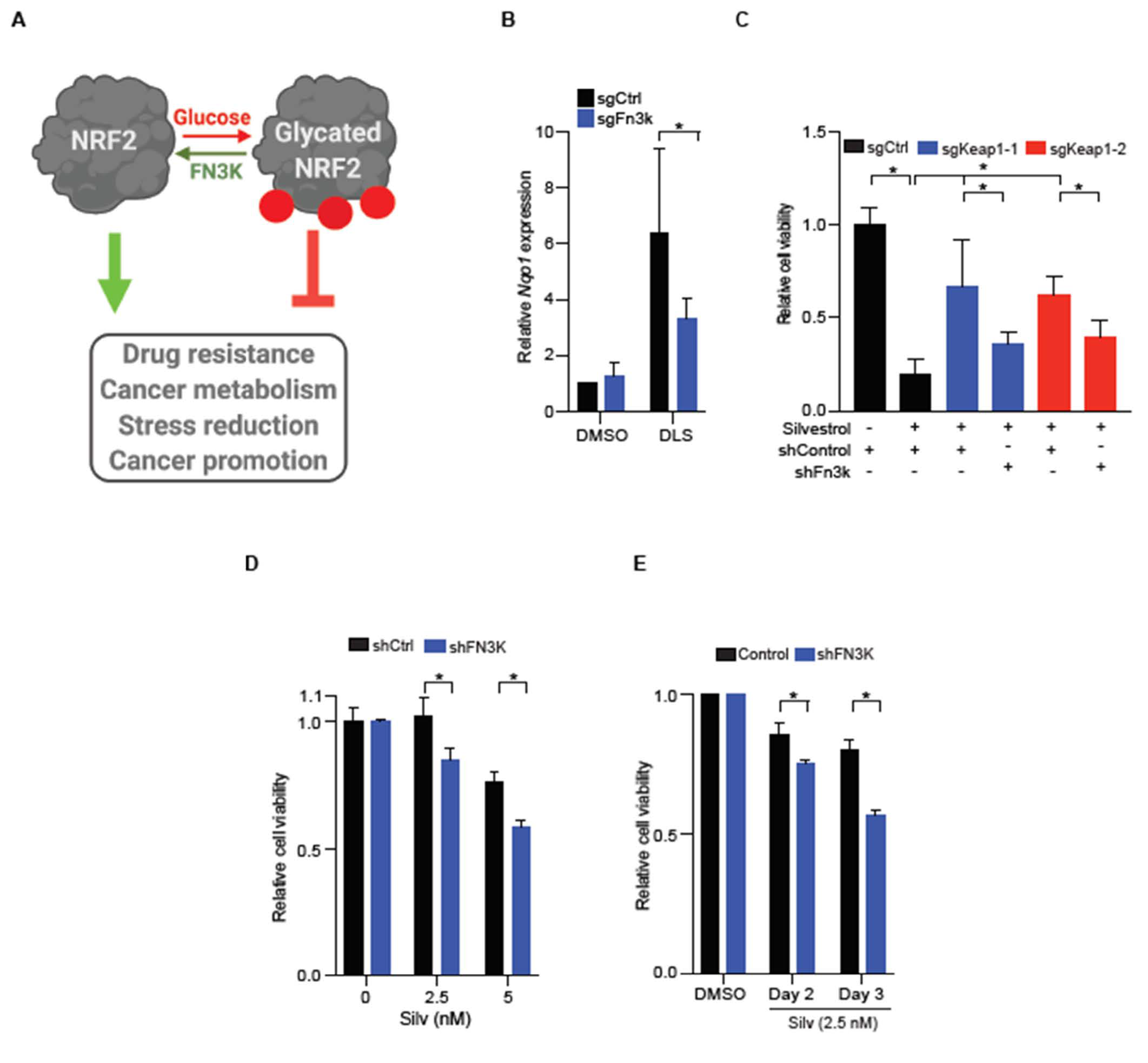
2.4. FN3K Inactivation Reverses NRF2 Driven Drug Resistance
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Animal Modeling
4.2. IC50 Determination
4.3. CRISPR Screen and Analysis
4.4. Gene or Protein Expression and Reporter Assays
4.5. Flow Cytometry and AHA Labeling
4.6. Immunohistochemistry
4.7. Bioinformatics and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jaramillo, M.C.; Zhang, D.D. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013, 27, 2179–2191. [Google Scholar] [CrossRef]
- De La Vega, M.R.; Chapman, E.; Zhang, D.D. NRF2 and the Hallmarks of Cancer. Cancer Cell 2018, 34, 21–43. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.D. Mechanistic Studies of the Nrf2-Keap1 Signaling Pathway. Drug Metab. Rev. 2006, 38, 769–789. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-Y.; Cantley, L.C.; DeNicola, G.M. NRF2 Rewires Cellular Metabolism to Support the Antioxidant Response. Master Regul. Oxidative Stress Transcr. Factor Nrf2 2016. [Google Scholar] [CrossRef]
- Sporn, M.B.; Liby, K.T. NRF2 and cancer: The good, the bad and the importance of context. Nat. Rev. Cancer 2012, 12, 564–571. [Google Scholar] [CrossRef]
- Taguchi, K.; Yamamoto, M. The KEAP1–NRF2 System in Cancer. Front. Oncol. 2017, 7, 85. [Google Scholar] [CrossRef]
- Shibata, T.; Ohta, T.; Tong, K.I.; Kokubu, A.; Odogawa, R.; Tsuta, K.; Asamura, H.; Yamamoto, M.; Hirohashi, S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc. Natl. Acad. Sci. USA 2008, 105, 13568–13573. [Google Scholar] [CrossRef]
- Menegon, S.; Columbano, A.; Giordano, S. The Dual Roles of NRF2 in Cancer. Trends Mol. Med. 2016, 22, 578–593. [Google Scholar] [CrossRef] [PubMed]
- Panieri, E.; Telkoparan-Akillilar, P.; Suzen, S.; Saso, L. The NRF2/KEAP1 Axis in the Regulation of Tumor Metabolism: Mechanisms and Therapeutic Perspectives. Biomolecules 2020, 10, 791. [Google Scholar] [CrossRef]
- Tao, S.; Wang, S.; Moghaddam, S.J.; Ooi, A.; Chapman, E.; Wong, P.K.; Zhang, D.D. Oncogenic KRAS Confers Chemoresistance by Upregulating NRF2. Cancer Res. 2014, 74, 7430–7441. [Google Scholar] [CrossRef]
- Wangy, X.-J.; Suny, Z.; Villeneuve, N.F.; Zhang, S.; Zhao, F.; Li, Y.; Chen, W.; Yi, X.; Zheng, W.; Wondrak, G.T.; et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis 2008, 29, 1235–1243. [Google Scholar] [CrossRef]
- Zanotto-Filho, A.; Masamsetti, V.P.; Loranc, E.; Tonapi, S.S.; Gorthi, A.; Bernard, X.; Gonçalves, R.M.; Moreira, J.C.F.; Chen, Y.; Bishop, A.J.R. Alkylating Agent-Induced NRF2 Blocks Endoplasmic Reticulum Stress-Mediated Apoptosis via Control of Glutathione Pools and Protein Thiol Homeostasis. Mol. Cancer Ther. 2016, 15, 3000–3014. [Google Scholar] [CrossRef]
- Krall, E.B.; Wang, B.; Munoz, D.M.; Ilic, N.; Raghavan, S.; Niederst, M.J.; Yu, K.; A Ruddy, D.; Aguirre, A.J.; Kim, J.W.; et al. KEAP1 loss modulates sensitivity to kinase targeted therapy in lung cancer. eLife 2017, 6, 914. [Google Scholar] [CrossRef]
- Sanghvi, V.R.; Leibold, J.; Mina, M.; Mohan, P.; Berishaj, M.; Li, Z.; Miele, M.M.; Lailler, N.; Zhao, C.; De Stanchina, E.; et al. The Oncogenic Action of NRF2 Depends on De-glycation by Fructosamine-3-Kinase. Cell 2019, 178, 807–819.e21. [Google Scholar] [CrossRef] [PubMed]
- Chio, I.I.C.; Jafarnejad, S.M.; Ponz-Sarvise, M.; Park, Y.; Rivera, K.; Palm, W.; Wilson, J.; Sangar, V.; Hao, Y.; Öhlund, D.; et al. NRF2 Promotes Tumor Maintenance by Modulating mRNA Translation in Pancreatic Cancer. Cell 2016, 166, 963–976. [Google Scholar] [CrossRef]
- Mitsuishi, Y.; Taguchi, K.; Kawatani, Y.; Shibata, T.; Nukiwa, T.; Aburatani, H.; Yamamoto, M.; Motohashi, H. Nrf2 Redirects Glucose and Glutamine into Anabolic Pathways in Metabolic Reprogramming. Cancer Cell 2012, 22, 66–79. [Google Scholar] [CrossRef]
- Chu, J.; Cargnello, M.; Topisirovic, I.; Pelletier, J. Translation Initiation Factors: Reprogramming Protein Synthesis in Cancer. Trends Cell Biol. 2016, 26, 918–933. [Google Scholar] [CrossRef] [PubMed]
- Silvera, D.; Formenti, S.C.; Schneider, R.J. Translational control in cancer. Nat. Rev. Cancer 2010, 10, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Robichaud, N.; Sonenberg, N.; Ruggero, D.; Schneider, R.J. Translational Control in Cancer. Cold Spring Harb. Perspect. Biol. 2018, 11, a032896. [Google Scholar] [CrossRef]
- Bhat, M.; Robichaud, N.; Hulea, L.; Sonenberg, N.; Pelletier, J.; Topisirovic, I. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015, 14, 261–278. [Google Scholar] [CrossRef]
- Wolfe, A.L.; Singh, K.; Zhong, Y.; Drewe, P.; Rajasekhar, V.K.; Sanghvi, V.R.; Mavrakis, K.J.; Jiang, M.; Roderick, J.E.; Van Der Meulen, J.; et al. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nat. Cell Biol. 2014, 513, 65–70. [Google Scholar] [CrossRef]
- Robert, F.; Roman, W.; Bramoullé, A.; Fellmann, C.; Roulston, A.; Shustik, C.; Porco, J.A.; Shore, G.C.; Sebag, M.; Pelletier, J. Translation initiation factor eIF4F modifies the dexamethasone response in multiple myeloma. Proc. Natl. Acad. Sci. USA 2014, 111, 13421–13426. [Google Scholar] [CrossRef] [PubMed]
- A Rubio, C.; Weisburd, B.; Holderfield, M.; Arias, C.; Fang, E.; DeRisi, J.L.; Fanidi, A. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 2014, 15, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.; Robert, F.; Oertlin, C.; Kapeller-Libermann, D.; Avizonis, D.; Gutierrez, J.; Handly-Santana, A.; Doubrovin, M.; Park, J.; Schoepfer, C.; et al. eIF4A supports an oncogenic translation program in pancreatic ductal adenocarcinoma. Nat. Commun. 2019, 10, 5151. [Google Scholar] [CrossRef] [PubMed]
- Ingolia, N.T.; Ghaemmaghami, S.; Newman, J.R.S.; Weissman, J.S. Genome-Wide Analysis in Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling. Science 2009, 324, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Shimada, K.; Muhlich, J.L.; Mitchison, T.J. A tool for browsing the Cancer Dependency Map reveals functional connections between genes and helps predict the efficacy and selectivity of candidate cancer drugs. bioRxiv 2019. [Google Scholar] [CrossRef]
- Rodrigo, C.M.; Cencic, R.; Roche, S.P.; Pelletier, J.; Porco, J.J.A. Synthesis of Rocaglamide Hydroxamates and Related Compounds as Eukaryotic Translation Inhibitors: Synthetic and Biological Studies. J. Med. Chem. 2011, 55, 558–562. [Google Scholar] [CrossRef]
- Koike-Yusa, H.; Li, Y.; Tan, E.-P.; Velasco-Herrera, M.D.C.; Yusa, K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol. 2014, 32, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Stogios, P.J.; Downs, G.S.; Jauhal, J.J.S.; Nandra, S.K.; Privé, G.G. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005, 6, R82. [Google Scholar] [CrossRef] [PubMed]
- Hediger, M.A.; Romero, M.F.; Peng, J.-B.; Rolfs, A.; Takanaga, H.; Bruford, E.A. The ABCs of solute carriers: Physiological, pathological and therapeutic implications of human membrane transport proteins. Pflügers Arch. Eur. J. Physiol. 2003, 447, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Wilczynska, A.; Gillen, S.L.; Schmidt, T.; Meijer, H.A.; Jukes-Jones, R.; Langlais, C.; Kopra, K.; Lu, W.-T.; Godfrey, J.D.; Hawley, B.R.; et al. eIF4A2 drives repression of translation at initiation by Ccr4-Not through purine-rich motifs in the 5′UTR. Genome Biol. 2019, 20, 1–21. [Google Scholar] [CrossRef]
- Villeneuve, N.F.; Lau, A.; Zhang, D.D. Regulation of the Nrf2–Keap1 Antioxidant Response by the Ubiquitin Proteasome System: An Insight into Cullin-Ring Ubiquitin Ligases. Antioxid. Redox Signal. 2010, 13, 1699–1712. [Google Scholar] [CrossRef]
- Gupta, S.V.; Sass, E.J.; Davis, M.E.; Edwards, R.B.; Lozanski, G.; Heerema, N.A.; Lehman, A.; Zhang, X.; Jarjoura, D.; Byrd, J.C.; et al. Resistance to the Translation Initiation Inhibitor Silvestrol is Mediated by ABCB1/P-Glycoprotein Overexpression in Acute Lymphoblastic Leukemia Cells. AAPS J. 2011, 13, 357–364. [Google Scholar] [CrossRef]
- Schatz, J.H.; Oricchio, E.; Wolfe, A.L.; Jiang, M.; Linkov, I.; Maragulia, J.; Shi, W.; Zhang, Z.; Rajasekhar, V.K.; Pagano, N.C.; et al. Targeting cap-dependent translation blocks converging survival signals by AKT and PIM kinases in lymphoma. J. Exp. Med. 2011, 208, 1799–1807. [Google Scholar] [CrossRef] [PubMed]
- Bordeleau, M.-E.; Robert, F.; Gerard, B.; Lindqvist, L.; Chen, S.M.; Wendel, H.-G.; Brem, B.; Greger, H.; Lowe, S.W.; Porco, J.A.; et al. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J. Clin. Investig. 2008, 118, 2651–2660. [Google Scholar] [CrossRef] [PubMed]
- Ernst, J.T.; Thompson, P.A.; Nilewski, C.; Sprengeler, P.A.; Sperry, S.; Packard, G.; Michels, T.; Xiang, A.; Tran, C.; Wegerski, C.J.; et al. Design of Development Candidate eFT226, a First in Class Inhibitor of Eukaryotic Initiation Factor 4A RNA Helicase. J. Med. Chem. 2020, 63, 5879–5955. [Google Scholar] [CrossRef]
- Intlekofer, A.M.; Joffe, E.; Batlevi, C.L.; Hilden, P.; He, J.; Seshan, V.E.; Zelenetz, A.D.; Palomba, M.L.; Moskowitz, C.H.; Portlock, C.; et al. Integrated DNA/RNA targeted genomic profiling of diffuse large B-cell lymphoma using a clinical assay. Blood Cancer J. 2018, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kari, E.J.M.; Teppo, H.-R.; Haapasaari, K.-M.; Kuusisto, M.E.L.; Lemma, A.; Karihtala, P.; Pirinen, R.; Soini, Y.; Jantunen, E.; Turpeenniemi-Hujanen, T.; et al. Nuclear factor erythroid 2-related factors 1 and 2 are able to define the worst prognosis group among high-risk diffuse large B cell lymphomas treated with R-CHOEP. J. Clin. Pathol. 2019, 72, 316–321. [Google Scholar] [CrossRef]
- Rushworth, S.A.; Zaitseva, L.; Murray, M.Y.; Shah, N.M.; Bowles, K.M.; MacEwan, D.J. The high Nrf2 expression in human acute myeloid leukemia is driven by NF-κB and underlies its chemo-resistance. Blood 2012, 120, 5188–5198. [Google Scholar] [CrossRef]
- Helena, A.Y.; Suzawa, K.; Jordan, E.J.; Zehir, A.; Ni, A.; Kim, H.R.; Kris, M.G.; Hellmann, M.D.; Li, B.T.; Somwar, R.; et al. Concurrent Alterations in EGFR-Mutant Lung Cancers Associated with Resistance to EGFR Kinase Inhibitors and Characterization of MTOR as a Mediator of Resistance. Clin. Cancer Res. 2018, 24, 3108–3118. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanghvi, V.R.; Mohan, P.; Singh, K.; Cao, L.; Berishaj, M.; Wolfe, A.L.; Schatz, J.H.; Lailler, N.; de Stanchina, E.; Viale, A.; et al. NRF2 Activation Confers Resistance to eIF4A Inhibitors in Cancer Therapy. Cancers 2021, 13, 639. https://doi.org/10.3390/cancers13040639
Sanghvi VR, Mohan P, Singh K, Cao L, Berishaj M, Wolfe AL, Schatz JH, Lailler N, de Stanchina E, Viale A, et al. NRF2 Activation Confers Resistance to eIF4A Inhibitors in Cancer Therapy. Cancers. 2021; 13(4):639. https://doi.org/10.3390/cancers13040639
Chicago/Turabian StyleSanghvi, Viraj R., Prathibha Mohan, Kamini Singh, Linlin Cao, Marjan Berishaj, Andrew L. Wolfe, Jonathan H. Schatz, Nathalie Lailler, Elisa de Stanchina, Agnes Viale, and et al. 2021. "NRF2 Activation Confers Resistance to eIF4A Inhibitors in Cancer Therapy" Cancers 13, no. 4: 639. https://doi.org/10.3390/cancers13040639
APA StyleSanghvi, V. R., Mohan, P., Singh, K., Cao, L., Berishaj, M., Wolfe, A. L., Schatz, J. H., Lailler, N., de Stanchina, E., Viale, A., & Wendel, H.-G. (2021). NRF2 Activation Confers Resistance to eIF4A Inhibitors in Cancer Therapy. Cancers, 13(4), 639. https://doi.org/10.3390/cancers13040639





