MicroRNAs: Tiny Regulators of Gene Expression with Pivotal Roles in Normal B-Cell Development and B-Cell Chronic Lymphocytic Leukemia
Simple Summary
Abstract
1. Introduction
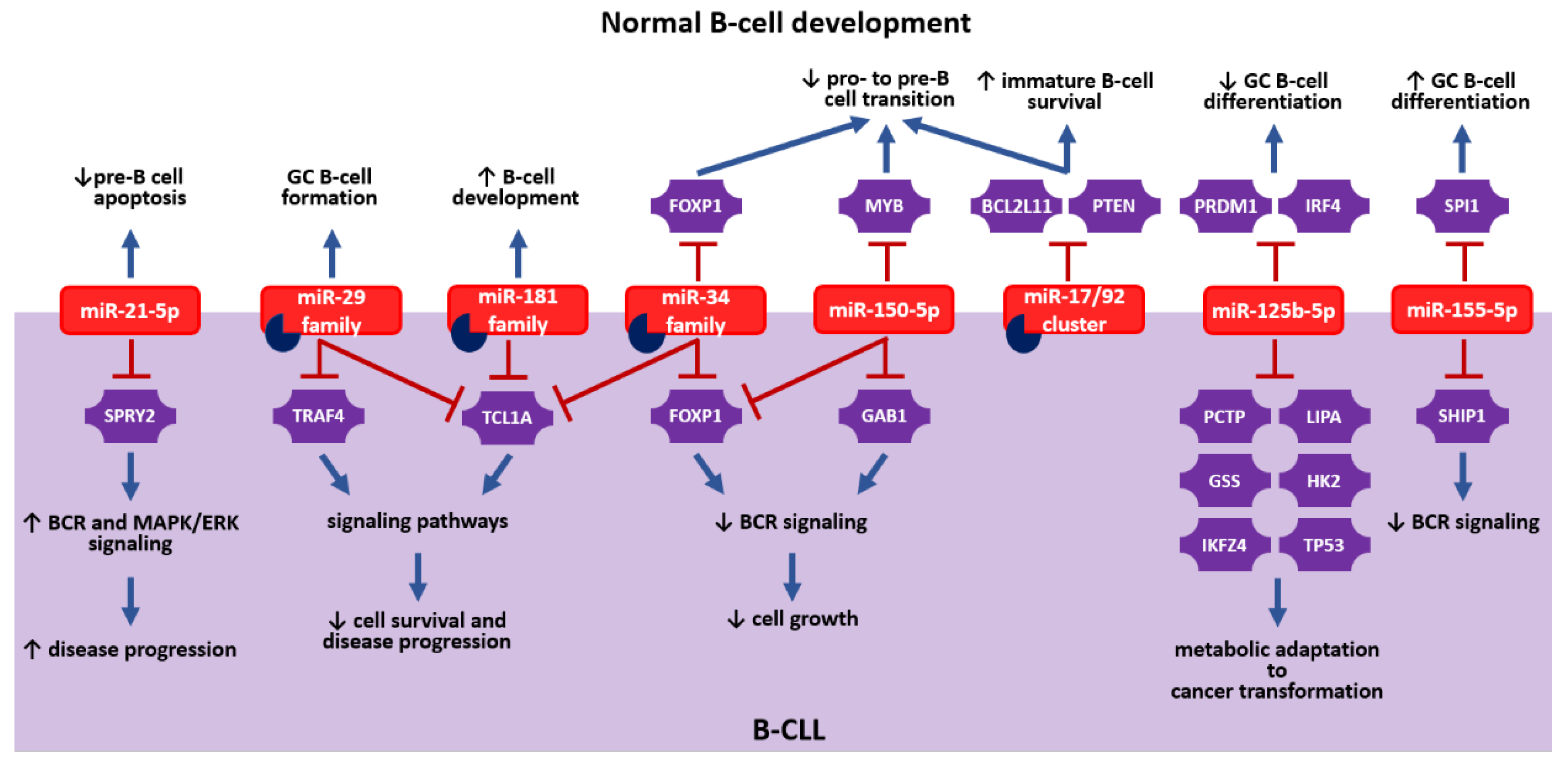
2. miRNAs: How Are They Involved in Normal B-Cell Development?
2.1. miRNAs in Bone Marrow B-Cell Development
2.2. miRNAs in Peripheral B-Cell Development
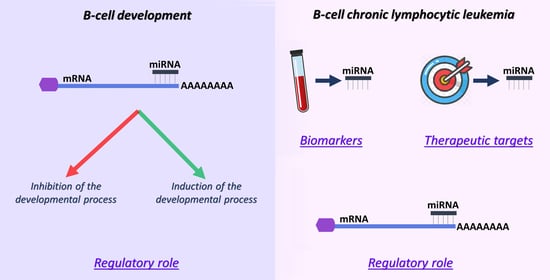
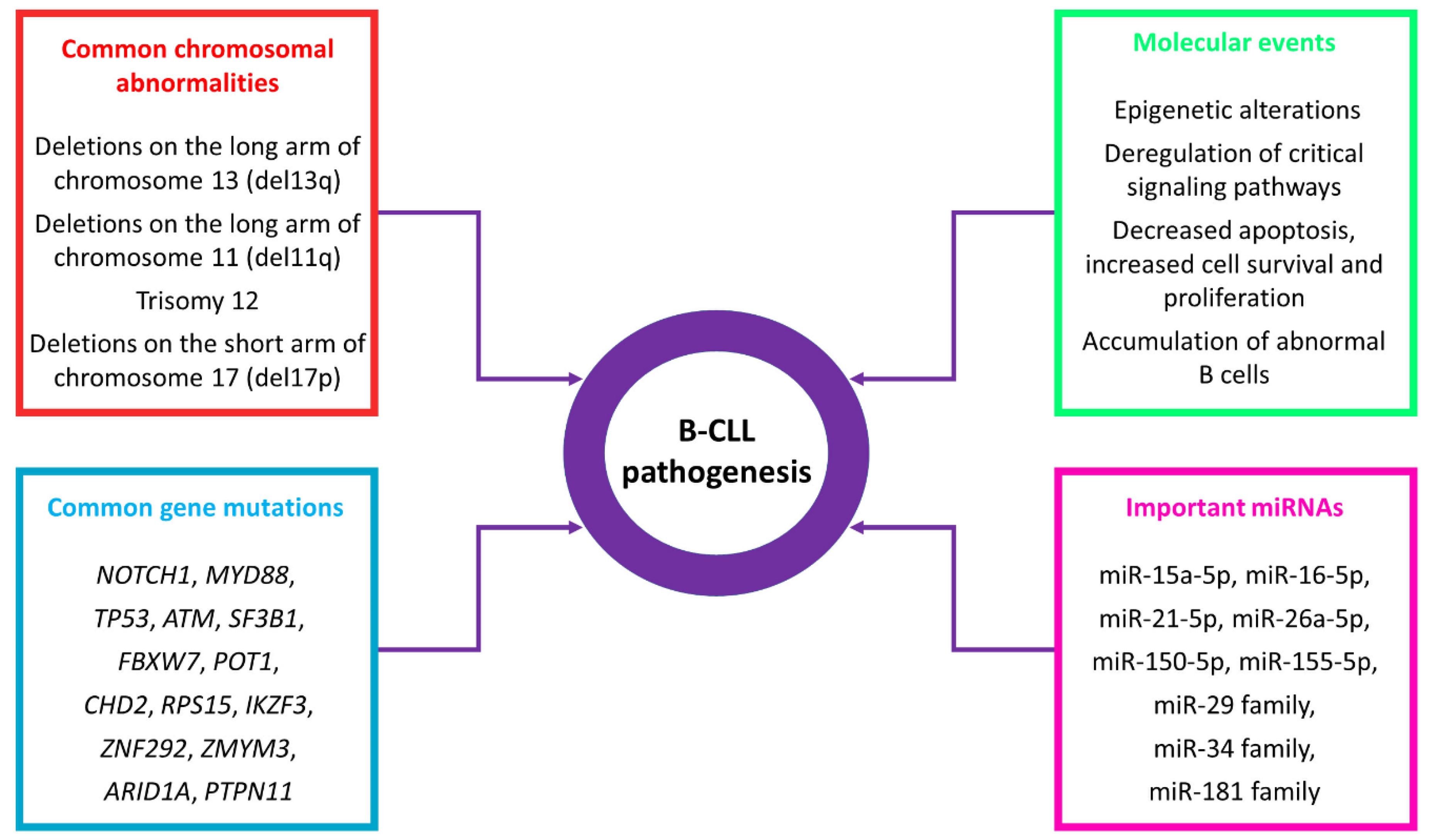
3. miRNAs: Regulators, Biomarkers and Potential Therapeutic Entities in B-CLL
3.1. miRNAs as Regulators in B-CLL
3.2. miRNAs as Diagnostic, Prognostic and Predictive Biomarkers in B-CLL
3.3. miRNAs in B-CLL Therapy
3.4. Viral miRNAs in B-CLL
4. Future Perspectives
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dohner, H.; Stilgenbauer, S.; Benner, A.; Leupolt, E.; Krober, A.; Bullinger, L.; Dohner, K.; Bentz, M.; Lichter, P. Genomic aberrations and survival in chronic lymphocytic leukemia. N. Engl. J. Med. 2000, 343, 1910–1916. [Google Scholar] [CrossRef]
- International CLL-IPI working group. An international prognostic index for patients with chronic lymphocytic leukaemia (CLL-IPI): A meta-analysis of individual patient data. Lancet Oncol. 2016, 17, 779–790. [Google Scholar] [CrossRef]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Vasilatou, D.; Papageorgiou, S.; Pappa, V.; Papageorgiou, E.; Dervenoulas, J. The role of microRNAs in normal and malignant hematopoiesis. Eur. J. Haematol. 2010, 84, 1–16. [Google Scholar] [CrossRef]
- Melchers, F. Checkpoints that control B cell development. J. Clin. Investig. 2015, 125, 2203–2210. [Google Scholar] [CrossRef] [PubMed]
- Koralov, S.B.; Muljo, S.A.; Galler, G.R.; Krek, A.; Chakraborty, T.; Kanellopoulou, C.; Jensen, K.; Cobb, B.S.; Merkenschlager, M.; Rajewsky, N.; et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell 2008, 132, 860–874. [Google Scholar] [CrossRef] [PubMed]
- Brandl, A.; Daum, P.; Brenner, S.; Schulz, S.R.; Yap, D.Y.; Bosl, M.R.; Wittmann, J.; Schuh, W.; Jack, H.M. The microprocessor component, DGCR8, is essential for early B-cell development in mice. Eur. J. Immunol. 2016, 46, 2710–2718. [Google Scholar] [CrossRef] [PubMed]
- Spierings, D.C.; McGoldrick, D.; Hamilton-Easton, A.M.; Neale, G.; Murchison, E.P.; Hannon, G.J.; Green, D.R.; Withoff, S. Ordered progression of stage-specific miRNA profiles in the mouse B2 B-cell lineage. Blood 2011, 117, 5340–5349. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Calado, D.P.; Galler, G.; Thai, T.H.; Patterson, H.C.; Wang, J.; Rajewsky, N.; Bender, T.P.; Rajewsky, K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 2007, 131, 146–159. [Google Scholar] [CrossRef] [PubMed]
- Okuyama, K.; Ikawa, T.; Gentner, B.; Hozumi, K.; Harnprasopwat, R.; Lu, J.; Yamashita, R.; Ha, D.; Toyoshima, T.; Chanda, B.; et al. MicroRNA-126-mediated control of cell fate in B-cell myeloid progenitors as a potential alternative to transcriptional factors. Proc. Natl. Acad. Sci. USA 2013, 110, 13410–13415. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Kurkewich, J.L.; Bikorimana, E.; Nguyen, T.; Klopfenstein, N.; Zhang, H.; Hallas, W.M.; Stayback, G.; McDowell, M.A.; Dahl, R. The mirn23a microRNA cluster antagonizes B cell development. J. Leukoc. Biol. 2016, 100, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Hagman, J.; Lukin, K. Transcription factors drive B cell development. Curr. Opin. Immunol. 2006, 18, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Medvedovic, J.; Ebert, A.; Tagoh, H.; Busslinger, M. Pax5: A master regulator of B cell development and leukemogenesis. Adv. Immunol. 2011, 111, 179–206. [Google Scholar] [CrossRef] [PubMed]
- Kwon, K.; Hutter, C.; Sun, Q.; Bilic, I.; Cobaleda, C.; Malin, S.; Busslinger, M. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity 2008, 28, 751–762. [Google Scholar] [CrossRef]
- Hagman, J.; Ramirez, J.; Lukin, K. B lymphocyte lineage specification, commitment and epigenetic control of transcription by early B cell factor 1. Curr. Top. Microbiol. Immunol. 2012, 356, 17–38. [Google Scholar] [CrossRef]
- Nechanitzky, R.; Akbas, D.; Scherer, S.; Gyory, I.; Hoyler, T.; Ramamoorthy, S.; Diefenbach, A.; Grosschedl, R. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat. Immunol. 2013, 14, 867–875. [Google Scholar] [CrossRef]
- Hu, H.; Wang, B.; Borde, M.; Nardone, J.; Maika, S.; Allred, L.; Tucker, P.W.; Rao, A. Foxp1 is an essential transcriptional regulator of B cell development. Nat. Immunol. 2006, 7, 819–826. [Google Scholar] [CrossRef]
- Contreras, J.R.; Palanichamy, J.K.; Tran, T.M.; Fernando, T.R.; Rodriguez-Malave, N.I.; Goswami, N.; Arboleda, V.A.; Casero, D.; Rao, D.S. MicroRNA-146a modulates B-cell oncogenesis by regulating Egr1. Oncotarget 2015, 6, 11023–11037. [Google Scholar] [CrossRef][Green Version]
- Blume, J.; Zietara, N.; Witzlau, K.; Liu, Y.; Sanchez, O.O.; Puchalka, J.; Winter, S.J.; Kunze-Schumacher, H.; Saran, N.; Duber, S.; et al. miR-191 modulates B-cell development and targets transcription factors E2A, Foxp1, and Egr1. Eur. J. Immunol. 2019, 49, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Wang, S.; Mayr, C.; Bartel, D.P.; Lodish, H.F. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc. Natl. Acad. Sci. USA 2007, 104, 7080–7085. [Google Scholar] [CrossRef] [PubMed]
- Greig, K.T.; de Graaf, C.A.; Murphy, J.M.; Carpinelli, M.R.; Pang, S.H.; Frampton, J.; Kile, B.T.; Hilton, D.J.; Nutt, S.L. Critical roles for c-Myb in lymphoid priming and early B-cell development. Blood 2010, 115, 2796–2805. [Google Scholar] [CrossRef]
- Mehta, A.; Mann, M.; Zhao, J.L.; Marinov, G.K.; Majumdar, D.; Garcia-Flores, Y.; Du, X.; Erikci, E.; Chowdhury, K.; Baltimore, D. The microRNA-212/132 cluster regulates B cell development by targeting Sox4. J. Exp. Med. 2015, 212, 1679–1692. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.Y.; Owens, K.S.; Rogers, J.H.; Mullenix, J.; Velu, C.S.; Grimes, H.L.; Dahl, R. MIR-23A microRNA cluster inhibits B-cell development. Exp. Hematol. 2010, 38, 629–640.e1. [Google Scholar] [CrossRef] [PubMed]
- Lai, M.; Gonzalez-Martin, A.; Cooper, A.B.; Oda, H.; Jin, H.Y.; Shepherd, J.; He, L.; Zhu, J.; Nemazee, D.; Xiao, C. Regulation of B-cell development and tolerance by different members of the miR-17 approximately 92 family microRNAs. Nat. Commun. 2016, 7, 12207. [Google Scholar] [CrossRef] [PubMed]
- Benhamou, D.; Labi, V.; Getahun, A.; Benchetrit, E.; Dowery, R.; Rajewsky, K.; Cambier, J.C.; Melamed, D. The c-Myc/miR17-92/PTEN Axis Tunes PI3K Activity to Control Expression of Recombination Activating Genes in Early B Cell Development. Front. Immunol. 2018, 9, 2715. [Google Scholar] [CrossRef]
- Gonzalez-Martin, A.; Adams, B.D.; Lai, M.; Shepherd, J.; Salvador-Bernaldez, M.; Salvador, J.M.; Lu, J.; Nemazee, D.; Xiao, C. The microRNA miR-148a functions as a critical regulator of B cell tolerance and autoimmunity. Nat. Immunol. 2016, 17, 433–440. [Google Scholar] [CrossRef]
- Xiao, C.; Srinivasan, L.; Calado, D.P.; Patterson, H.C.; Zhang, B.; Wang, J.; Henderson, J.M.; Kutok, J.L.; Rajewsky, K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 2008, 9, 405–414. [Google Scholar] [CrossRef]
- Benhamou, D.; Labi, V.; Novak, R.; Dai, I.; Shafir-Alon, S.; Weiss, A.; Gaujoux, R.; Arnold, R.; Shen-Orr, S.S.; Rajewsky, K.; et al. A c-Myc/miR17-92/Pten Axis Controls PI3K-Mediated Positive and Negative Selection in B Cell Development and Reconstitutes CD19 Deficiency. Cell Rep. 2016, 16, 419–431. [Google Scholar] [CrossRef]
- Lu, D.; Nakagawa, R.; Lazzaro, S.; Staudacher, P.; Abreu-Goodger, C.; Henley, T.; Boiani, S.; Leyland, R.; Galloway, A.; Andrews, S.; et al. The miR-155-PU.1 axis acts on Pax5 to enable efficient terminal B cell differentiation. J. Exp. Med. 2014, 211, 2183–2198. [Google Scholar] [CrossRef] [PubMed]
- Porstner, M.; Winkelmann, R.; Daum, P.; Schmid, J.; Pracht, K.; Corte-Real, J.; Schreiber, S.; Haftmann, C.; Brandl, A.; Mashreghi, M.F.; et al. miR-148a promotes plasma cell differentiation and targets the germinal center transcription factors Mitf and Bach2. Eur. J. Immunol. 2015, 45, 1206–1215. [Google Scholar] [CrossRef] [PubMed]
- Gururajan, M.; Haga, C.L.; Das, S.; Leu, C.M.; Hodson, D.; Josson, S.; Turner, M.; Cooper, M.D. MicroRNA 125b inhibition of B cell differentiation in germinal centers. Int. Immunol. 2010, 22, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; So, A.Y.; Sookram, R.; Wong, S.; Wang, J.K.; Ouyang, Y.; He, P.; Su, Y.; Casellas, R.; Baltimore, D. Epigenetic silencing of miR-125b is required for normal B-cell development. Blood 2018, 131, 1920–1930. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, Y.; Kitaura, J.; Hatakeyama, K.; Watanuki, J.; Akasaka, T.; Kato, N.; Shimanuki, M.; Nishimura, K.; Takahashi, M.; Taniwaki, M.; et al. Emu/miR-125b transgenic mice develop lethal B-cell malignancies. Leukemia 2011, 25, 1849–1856. [Google Scholar] [CrossRef]
- Zhang, J.; Jima, D.D.; Jacobs, C.; Fischer, R.; Gottwein, E.; Huang, G.; Lugar, P.L.; Lagoo, A.S.; Rizzieri, D.A.; Friedman, D.R.; et al. Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood 2009, 113, 4586–4594. [Google Scholar] [CrossRef]
- He, Y.; Pear, W.S. Notch signalling in B cells. Semin. Cell Dev. Biol. 2003, 14, 135–142. [Google Scholar] [CrossRef]
- King, J.K.; Ung, N.M.; Paing, M.H.; Contreras, J.R.; Alberti, M.O.; Fernando, T.R.; Zhang, K.; Pellegrini, M.; Rao, D.S. Regulation of Marginal Zone B-Cell Differentiation by MicroRNA-146a. Front. Immunol. 2016, 7, 670. [Google Scholar] [CrossRef]
- Luo, Z.; Mu, L.; Zheng, Y.; Shen, W.; Li, J.; Xu, L.; Zhong, B.; Liu, Y.; Zhou, Y. NUMB enhances Notch signaling by repressing ubiquitination of NOTCH1 intracellular domain. J. Mol. Cell Biol. 2020, 12, 345–358. [Google Scholar] [CrossRef]
- Kramer, N.J.; Wang, W.L.; Reyes, E.Y.; Kumar, B.; Chen, C.C.; Ramakrishna, C.; Cantin, E.M.; Vonderfecht, S.L.; Taganov, K.D.; Chau, N.; et al. Altered lymphopoiesis and immunodeficiency in miR-142 null mice. Blood 2015, 125, 3720–3730. [Google Scholar] [CrossRef]
- Rao, D.S.; O’Connell, R.M.; Chaudhuri, A.A.; Garcia-Flores, Y.; Geiger, T.L.; Baltimore, D. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity 2010, 33, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Medina, P.P.; Nolde, M.; Slack, F.J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 2010, 467, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Mok, Y.; Schwierzeck, V.; Thomas, D.C.; Vigorito, E.; Rayner, T.F.; Jarvis, L.B.; Prosser, H.M.; Bradley, A.; Withers, D.R.; Martensson, I.L.; et al. MiR-210 is induced by Oct-2, regulates B cells, and inhibits autoantibody production. J. Immunol. 2013, 191, 3037–3048. [Google Scholar] [CrossRef]
- van Nieuwenhuijze, A.; Dooley, J.; Humblet-Baron, S.; Sreenivasan, J.; Koenders, M.; Schlenner, S.M.; Linterman, M.; Liston, A. Defective germinal center B-cell response and reduced arthritic pathology in microRNA-29a-deficient mice. Cell. Mol. Life Sci. CMLS 2017, 74, 2095–2106. [Google Scholar] [CrossRef] [PubMed]
- Underbayev, C.; Kasar, S.; Ruezinsky, W.; Degheidy, H.; Schneider, J.S.; Marti, G.; Bauer, S.R.; Fraidenraich, D.; Lightfoote, M.M.; Parashar, V.; et al. Role of mir-15a/16-1 in early B cell development in a mouse model of chronic lymphocytic leukemia. Oncotarget 2016, 7, 60986–60999. [Google Scholar] [CrossRef]
- Kovaleva, V.; Mora, R.; Park, Y.J.; Plass, C.; Chiramel, A.I.; Bartenschlager, R.; Dohner, H.; Stilgenbauer, S.; Pscherer, A.; Lichter, P.; et al. miRNA-130a targets ATG2B and DICER1 to inhibit autophagy and trigger killing of chronic lymphocytic leukemia cells. Cancer Res. 2012, 72, 1763–1772. [Google Scholar] [CrossRef]
- Tili, E.; Michaille, J.J.; Luo, Z.; Volinia, S.; Rassenti, L.Z.; Kipps, T.J.; Croce, C.M. The down-regulation of miR-125b in chronic lymphocytic leukemias leads to metabolic adaptation of cells to a transformed state. Blood 2012, 120, 2631–2638. [Google Scholar] [CrossRef]
- Baer, C.; Oakes, C.C.; Ruppert, A.S.; Claus, R.; Kim-Wanner, S.Z.; Mertens, D.; Zenz, T.; Stilgenbauer, S.; Byrd, J.C.; Plass, C. Epigenetic silencing of miR-708 enhances NF-kappaB signaling in chronic lymphocytic leukemia. Int. J. Cancer 2015, 137, 1352–1361. [Google Scholar] [CrossRef]
- Farahani, M.; Rubbi, C.; Liu, L.; Slupsky, J.R.; Kalakonda, N. CLL Exosomes Modulate the Transcriptome and Behaviour of Recipient Stromal Cells and Are Selectively Enriched in miR-202-3p. PLoS ONE 2015, 10, e0141429. [Google Scholar] [CrossRef]
- Bottoni, A.; Rizzotto, L.; Lai, T.H.; Liu, C.; Smith, L.L.; Mantel, R.; Reiff, S.; El-Gamal, D.; Larkin, K.; Johnson, A.J.; et al. Targeting BTK through microRNA in chronic lymphocytic leukemia. Blood 2016, 128, 3101–3112. [Google Scholar] [CrossRef]
- Pekarsky, Y.; Santanam, U.; Cimmino, A.; Palamarchuk, A.; Efanov, A.; Maximov, V.; Volinia, S.; Alder, H.; Liu, C.G.; Rassenti, L.; et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006, 66, 11590–11593. [Google Scholar] [CrossRef] [PubMed]
- Sivina, M.; Hartmann, E.; Vasyutina, E.; Boucas, J.M.; Breuer, A.; Keating, M.J.; Wierda, W.G.; Rosenwald, A.; Herling, M.; Burger, J.A. Stromal cells modulate TCL1 expression, interacting AP-1 components and TCL1-targeting micro-RNAs in chronic lymphocytic leukemia. Leukemia 2012, 26, 1812–1820. [Google Scholar] [CrossRef] [PubMed]
- Vasyutina, E.; Boucas, J.M.; Bloehdorn, J.; Aszyk, C.; Crispatzu, G.; Stiefelhagen, M.; Breuer, A.; Mayer, P.; Lengerke, C.; Dohner, H.; et al. The regulatory interaction of EVI1 with the TCL1A oncogene impacts cell survival and clinical outcome in CLL. Leukemia 2015, 29, 2003–2014. [Google Scholar] [CrossRef] [PubMed]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef] [PubMed]
- Rampazzo, E.; Bojnik, E.; Trentin, L.; Bonaldi, L.; Del Bianco, P.; Frezzato, F.; Visentin, A.; Facco, M.; Semenzato, G.; De Rossi, A. Role of miR-15a/miR-16-1 and the TP53 axis in regulating telomerase expression in chronic lymphocytic leukemia. Haematologica 2017, 102, e253–e256. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Vicente, A.E.; Quwaider, D.; Benito, R.; Misiewicz-Krzeminska, I.; Hernandez-Sanchez, M.; de Coca, A.G.; Fisac, R.; Alonso, J.M.; Zato, C.; de Paz, J.F.; et al. MicroRNA-223 is a novel negative regulator of HSP90B1 in CLL. BMC Cancer 2015, 15, 238. [Google Scholar] [CrossRef] [PubMed]
- Rozovski, U.; Calin, G.A.; Setoyama, T.; D’Abundo, L.; Harris, D.M.; Li, P.; Liu, Z.; Grgurevic, S.; Ferrajoli, A.; Faderl, S.; et al. Signal transducer and activator of transcription (STAT)-3 regulates microRNA gene expression in chronic lymphocytic leukemia cells. Mol. Cancer 2013, 12, 50. [Google Scholar] [CrossRef] [PubMed]
- Kluiver, J.L.; Chen, C.Z. MicroRNAs regulate B-cell receptor signaling-induced apoptosis. Genes Immun. 2012, 13, 239–244. [Google Scholar] [CrossRef]
- Cui, B.; Chen, L.; Zhang, S.; Mraz, M.; Fecteau, J.F.; Yu, J.; Ghia, E.M.; Zhang, L.; Bao, L.; Rassenti, L.Z.; et al. MicroRNA-155 influences B-cell receptor signaling and associates with aggressive disease in chronic lymphocytic leukemia. Blood 2014, 124, 546–554. [Google Scholar] [CrossRef]
- Mraz, M.; Chen, L.; Rassenti, L.Z.; Ghia, E.M.; Li, H.; Jepsen, K.; Smith, E.N.; Messer, K.; Frazer, K.A.; Kipps, T.J. miR-150 influences B-cell receptor signaling in chronic lymphocytic leukemia by regulating expression of GAB1 and FOXP1. Blood 2014, 124, 84–95. [Google Scholar] [CrossRef]
- Cerna, K.; Oppelt, J.; Chochola, V.; Musilova, K.; Seda, V.; Pavlasova, G.; Radova, L.; Arigoni, M.; Calogero, R.A.; Benes, V.; et al. MicroRNA miR-34a downregulates FOXP1 during DNA damage response to limit BCR signalling in chronic lymphocytic leukaemia B cells. Leukemia 2019, 33, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Pavlasova, G.M.; Seda, V.; Cerna, K.A.; Vojackova, E.; Filip, D.; Ondrisova, L.; Sandova, V.; Kostalova, L.; Zeni, P.F.; et al. miR-29 Modulates CD40 Signaling in Chronic Lymphocytic Leukemia by Targeting TRAF4: An Axis Affected by BCR inhibitors. Blood 2020. [Google Scholar] [CrossRef]
- Shukla, A.; Rai, K.; Shukla, V.; Chaturvedi, N.K.; Bociek, R.G.; Pirruccello, S.J.; Band, H.; Lu, R.; Joshi, S.S. Sprouty 2: A novel attenuator of B-cell receptor and MAPK-Erk signaling in CLL. Blood 2016, 127, 2310–2321. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.Q.; Wong, K.Y.; Rosen, A.; Chim, C.S. Epigenetic silencing of tumor suppressor miR-3151 contributes to Chinese chronic lymphocytic leukemia by constitutive activation of MADD/ERK and PIK3R2/AKT signaling pathways. Oncotarget 2015, 6, 44422–44436. [Google Scholar] [CrossRef]
- Guinn, D.; Lehman, A.; Fabian, C.; Yu, L.; Maddocks, K.; Andritsos, L.A.; Jones, J.A.; Flynn, J.M.; Jaglowski, S.M.; Woyach, J.A.; et al. The regulation of tumor-suppressive microRNA, miR-126, in chronic lymphocytic leukemia. Cancer Med. 2017, 6, 778–787. [Google Scholar] [CrossRef]
- Palacios, F.; Abreu, C.; Prieto, D.; Morande, P.; Ruiz, S.; Fernandez-Calero, T.; Naya, H.; Libisch, G.; Robello, C.; Landoni, A.I.; et al. Activation of the PI3K/AKT pathway by microRNA-22 results in CLL B-cell proliferation. Leukemia 2015, 29, 115–125. [Google Scholar] [CrossRef]
- Zou, Z.J.; Fan, L.; Wang, L.; Xu, J.; Zhang, R.; Tian, T.; Li, J.Y.; Xu, W. miR-26a and miR-214 down-regulate expression of the PTEN gene in chronic lymphocytic leukemia, but not PTEN mutation or promoter methylation. Oncotarget 2015, 6, 1276–1285. [Google Scholar] [CrossRef]
- Fabbri, M.; Bottoni, A.; Shimizu, M.; Spizzo, R.; Nicoloso, M.S.; Rossi, S.; Barbarotto, E.; Cimmino, A.; Adair, B.; Wojcik, S.E.; et al. Association of a microRNA/TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemia. JAMA 2011, 305, 59–67. [Google Scholar] [CrossRef]
- Baer, C.; Claus, R.; Frenzel, L.P.; Zucknick, M.; Park, Y.J.; Gu, L.; Weichenhan, D.; Fischer, M.; Pallasch, C.P.; Herpel, E.; et al. Extensive promoter DNA hypermethylation and hypomethylation is associated with aberrant microRNA expression in chronic lymphocytic leukemia. Cancer Res. 2012, 72, 3775–3785. [Google Scholar] [CrossRef]
- Deneberg, S.; Kanduri, M.; Ali, D.; Bengtzen, S.; Karimi, M.; Qu, Y.; Kimby, E.; Mansouri, L.; Rosenquist, R.; Lennartsson, A.; et al. microRNA-34b/c on chromosome 11q23 is aberrantly methylated in chronic lymphocytic leukemia. Epigenetics 2014, 9, 910–917. [Google Scholar] [CrossRef]
- Wang, L.Q.; Kwong, Y.L.; Wong, K.F.; Kho, C.S.; Jin, D.Y.; Tse, E.; Rosen, A.; Chim, C.S. Epigenetic inactivation of mir-34b/c in addition to mir-34a and DAPK1 in chronic lymphocytic leukemia. J. Transl. Med. 2014, 12, 52. [Google Scholar] [CrossRef]
- Sampath, D.; Liu, C.; Vasan, K.; Sulda, M.; Puduvalli, V.K.; Wierda, W.G.; Keating, M.J. Histone deacetylases mediate the silencing of miR-15a, miR-16, and miR-29b in chronic lymphocytic leukemia. Blood 2012, 119, 1162–1172. [Google Scholar] [CrossRef] [PubMed]
- Gassner, F.J.; Zaborsky, N.; Feldbacher, D.; Greil, R.; Geisberger, R. RNA Editing Alters miRNA Function in Chronic Lymphocytic Leukemia. Cancers 2020, 12, 1159. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Wu, L.; Bao, J.; Li, Q.; Chen, X.; Xia, H.; Xia, R. Circular RNA circ-CBFB promotes proliferation and inhibits apoptosis in chronic lymphocytic leukemia through regulating miR-607/FZD3/Wnt/beta-catenin pathway. Biochem. Biophys. Res. Commun. 2018, 503, 385–390. [Google Scholar] [CrossRef]
- Wu, W.; Wu, Z.; Xia, Y.; Qin, S.; Li, Y.; Wu, J.; Liang, J.; Wang, L.; Zhu, H.; Fan, L.; et al. Downregulation of circ_0132266 in chronic lymphocytic leukemia promoted cell viability through miR-337-3p/PML axis. Aging 2019, 11, 3561–3573. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Feng, L.; Lu, K.; Li, P.; Lv, X.; Wang, X. STAT6 phosphorylation upregulates microRNA-155 expression and subsequently enhances the pathogenesis of chronic lymphocytic leukemia. Oncology Lett. 2019, 18, 95–100. [Google Scholar] [CrossRef]
- Frenquelli, M.; Muzio, M.; Scielzo, C.; Fazi, C.; Scarfo, L.; Rossi, C.; Ferrari, G.; Ghia, P.; Caligaris-Cappio, F. MicroRNA and proliferation control in chronic lymphocytic leukemia: Functional relationship between miR-221/222 cluster and p27. Blood 2010, 115, 3949–3959. [Google Scholar] [CrossRef]
- Rassenti, L.Z.; Balatti, V.; Ghia, E.M.; Palamarchuk, A.; Tomasello, L.; Fadda, P.; Pekarsky, Y.; Widhopf, G.F., 2nd; Kipps, T.J.; Croce, C.M. MicroRNA dysregulation to identify therapeutic target combinations for chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2017, 114, 10731–10736. [Google Scholar] [CrossRef]
- Kopparapu, P.K.; Bhoi, S.; Mansouri, L.; Arabanian, L.S.; Plevova, K.; Pospisilova, S.; Wasik, A.M.; Croci, G.A.; Sander, B.; Paulli, M.; et al. Epigenetic silencing of miR-26A1 in chronic lymphocytic leukemia and mantle cell lymphoma: Impact on EZH2 expression. Epigenetics 2016, 11, 335–343. [Google Scholar] [CrossRef]
- Sampath, D.; Calin, G.A.; Puduvalli, V.K.; Gopisetty, G.; Taccioli, C.; Liu, C.G.; Ewald, B.; Liu, C.; Keating, M.J.; Plunkett, W. Specific activation of microRNA106b enables the p73 apoptotic response in chronic lymphocytic leukemia by targeting the ubiquitin ligase Itch for degradation. Blood 2009, 113, 3744–3753. [Google Scholar] [CrossRef]
- Yang, Y.Q.; Tian, T.; Zhu, H.Y.; Liang, J.H.; Wu, W.; Wu, J.Z.; Xia, Y.; Wang, L.; Fan, L.; Li, J.Y.; et al. NDRG2 mRNA levels and miR-28-5p and miR-650 activity in chronic lymphocytic leukemia. BMC Cancer 2018, 18, 1009. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Liu, C.G.; Sevignani, C.; Ferracin, M.; Felli, N.; Dumitru, C.D.; Shimizu, M.; Cimmino, A.; Zupo, S.; Dono, M.; et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl. Acad. Sci. USA 2004, 101, 11755–11760. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Ferracin, M.; Cimmino, A.; Di Leva, G.; Shimizu, M.; Wojcik, S.E.; Iorio, M.V.; Visone, R.; Sever, N.I.; Fabbri, M.; et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005, 353, 1793–1801. [Google Scholar] [CrossRef] [PubMed]
- Visone, R.; Veronese, A.; Rassenti, L.Z.; Balatti, V.; Pearl, D.K.; Acunzo, M.; Volinia, S.; Taccioli, C.; Kipps, T.J.; Croce, C.M. miR-181b is a biomarker of disease progression in chronic lymphocytic leukemia. Blood 2011, 118, 3072–3079. [Google Scholar] [CrossRef]
- Ferrajoli, A.; Shanafelt, T.D.; Ivan, C.; Shimizu, M.; Rabe, K.G.; Nouraee, N.; Ikuo, M.; Ghosh, A.K.; Lerner, S.; Rassenti, L.Z.; et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood 2013, 122, 1891–1899. [Google Scholar] [CrossRef]
- Papageorgiou, S.G.; Kontos, C.K.; Diamantopoulos, M.A.; Bouchla, A.; Glezou, E.; Bazani, E.; Pappa, V.; Scorilas, A. MicroRNA-155-5p Overexpression in Peripheral Blood Mononuclear Cells of Chronic Lymphocytic Leukemia Patients Is a Novel, Independent Molecular Biomarker of Poor Prognosis. Dis. Markers 2017, 2017, 2046545. [Google Scholar] [CrossRef]
- Mraz, M.; Malinova, K.; Kotaskova, J.; Pavlova, S.; Tichy, B.; Malcikova, J.; Stano Kozubik, K.; Smardova, J.; Brychtova, Y.; Doubek, M.; et al. miR-34a, miR-29c and miR-17-5p are downregulated in CLL patients with TP53 abnormalities. Leukemia 2009, 23, 1159–1163. [Google Scholar] [CrossRef]
- Mertens, D.; Philippen, A.; Ruppel, M.; Allegra, D.; Bhattacharya, N.; Tschuch, C.; Wolf, S.; Idler, I.; Zenz, T.; Stilgenbauer, S. Chronic lymphocytic leukemia and 13q14: miRs and more. Leuk. Lymphoma 2009, 50, 502–505. [Google Scholar] [CrossRef]
- Visone, R.; Rassenti, L.Z.; Veronese, A.; Taccioli, C.; Costinean, S.; Aguda, B.D.; Volinia, S.; Ferracin, M.; Palatini, J.; Balatti, V.; et al. Karyotype-specific microRNA signature in chronic lymphocytic leukemia. Blood 2009, 114, 3872–3879. [Google Scholar] [CrossRef]
- Stamatopoulos, B.; Meuleman, N.; De Bruyn, C.; Pieters, K.; Anthoine, G.; Mineur, P.; Bron, D.; Lagneaux, L. A molecular score by quantitative PCR as a new prognostic tool at diagnosis for chronic lymphocytic leukemia patients. PLoS ONE 2010, 5. [Google Scholar] [CrossRef]
- Rossi, S.; Shimizu, M.; Barbarotto, E.; Nicoloso, M.S.; Dimitri, F.; Sampath, D.; Fabbri, M.; Lerner, S.; Barron, L.L.; Rassenti, L.Z.; et al. microRNA fingerprinting of CLL patients with chromosome 17p deletion identify a miR-21 score that stratifies early survival. Blood 2010, 116, 945–952. [Google Scholar] [CrossRef] [PubMed]
- Moussay, E.; Palissot, V.; Vallar, L.; Poirel, H.A.; Wenner, T.; El Khoury, V.; Aouali, N.; Van Moer, K.; Leners, B.; Bernardin, F.; et al. Determination of genes and microRNAs involved in the resistance to fludarabine in vivo in chronic lymphocytic leukemia. Mol. Cancer 2010, 9, 115. [Google Scholar] [CrossRef] [PubMed]
- Gagez, A.L.; Duroux-Richard, I.; Lepretre, S.; Orsini-Piocelle, F.; Letestu, R.; De Guibert, S.; Tuaillon, E.; Leblond, V.; Khalifa, O.; Gouilleux-Gruart, V.; et al. miR-125b and miR-532-3p predict the efficiency of rituximab-mediated lymphodepletion in chronic lymphocytic leukemia patients. A French Innovative Leukemia Organization study. Haematologica 2017, 102, 746–754. [Google Scholar] [CrossRef]
- Zenz, T.; Mohr, J.; Eldering, E.; Kater, A.P.; Buhler, A.; Kienle, D.; Winkler, D.; Durig, J.; van Oers, M.H.; Mertens, D.; et al. miR-34a as part of the resistance network in chronic lymphocytic leukemia. Blood 2009, 113, 3801–3808. [Google Scholar] [CrossRef] [PubMed]
- Ferracin, M.; Zagatti, B.; Rizzotto, L.; Cavazzini, F.; Veronese, A.; Ciccone, M.; Saccenti, E.; Lupini, L.; Grilli, A.; De Angeli, C.; et al. MicroRNAs involvement in fludarabine refractory chronic lymphocytic leukemia. Mol. Cancer 2010, 9, 123. [Google Scholar] [CrossRef]
- Papageorgiou, S.G.; Kontos, C.K.; Tsiakanikas, P.; Stavroulaki, G.; Bouchla, A.; Vasilatou, D.; Bazani, E.; Lazarakou, A.; Scorilas, A.; Pappa, V. Elevated miR-20b-5p expression in peripheral blood mononuclear cells: A novel, independent molecular biomarker of favorable prognosis in chronic lymphocytic leukemia. Leuk. Res. 2018, 70, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Ruhela, V.; Rani, L.; Gupta, A.; Sriram, K.; Gogia, A.; Sharma, A.; Kumar, L.; Gupta, R. RNA-Seq profiling of deregulated miRs in CLL and their impact on clinical outcome. Blood Cancer J. 2020, 10, 6. [Google Scholar] [CrossRef]
- Papageorgiou, S.G.; Diamantopoulos, M.A.; Kontos, C.K.; Bouchla, A.; Vasilatou, D.; Bazani, E.; Scorilas, A.; Pappa, V. MicroRNA-92a-3p overexpression in peripheral blood mononuclear cells is an independent predictor of prolonged overall survival of patients with chronic lymphocytic leukemia. Leuk. Lymphoma 2019, 60, 658–667. [Google Scholar] [CrossRef]
- Casabonne, D.; Benavente, Y.; Seifert, J.; Costas, L.; Armesto, M.; Arestin, M.; Besson, C.; Hosnijeh, F.S.; Duell, E.J.; Weiderpass, E.; et al. Serum levels of hsa-miR-16-5p, hsa-miR-29a-3p, hsa-miR-150-5p, hsa-miR-155-5p and hsa-miR-223-3p and subsequent risk of chronic lymphocytic leukemia in the EPIC study. Int. J. Cancer 2020, 147, 1315–1324. [Google Scholar] [CrossRef]
- Moussay, E.; Wang, K.; Cho, J.H.; van Moer, K.; Pierson, S.; Paggetti, J.; Nazarov, P.V.; Palissot, V.; Hood, L.E.; Berchem, G.; et al. MicroRNA as biomarkers and regulators in B-cell chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2011, 108, 6573–6578. [Google Scholar] [CrossRef]
- Negrini, M.; Cutrona, G.; Bassi, C.; Fabris, S.; Zagatti, B.; Colombo, M.; Ferracin, M.; D’Abundo, L.; Saccenti, E.; Matis, S.; et al. microRNAome expression in chronic lymphocytic leukemia: Comparison with normal B-cell subsets and correlations with prognostic and clinical parameters. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 4141–4153. [Google Scholar] [CrossRef] [PubMed]
- Salerno, E.; Scaglione, B.J.; Coffman, F.D.; Brown, B.D.; Baccarini, A.; Fernandes, H.; Marti, G.; Raveche, E.S. Correcting miR-15a/16 genetic defect in New Zealand Black mouse model of CLL enhances drug sensitivity. Mol. Cancer Ther. 2009, 8, 2684–2692. [Google Scholar] [CrossRef] [PubMed]
- Kasar, S.; Salerno, E.; Yuan, Y.; Underbayev, C.; Vollenweider, D.; Laurindo, M.F.; Fernandes, H.; Bonci, D.; Addario, A.; Mazzella, F.; et al. Systemic in vivo lentiviral delivery of miR-15a/16 reduces malignancy in the NZB de novo mouse model of chronic lymphocytic leukemia. Genes Immun. 2012, 13, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Fonte, E.; Apollonio, B.; Scarfo, L.; Ranghetti, P.; Fazi, C.; Ghia, P.; Caligaris-Cappio, F.; Muzio, M. In vitro sensitivity of CLL cells to fludarabine may be modulated by the stimulation of Toll-like receptors. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Dehkordi, K.A.; Chaleshtori, M.H.; Sharifi, M.; Jalili, A.; Fathi, F.; Roshani, D.; Nikkhoo, B.; Hakhamaneshi, M.S.; Sani, M.R.M.; Ganji-Arjenaki, M. Inhibition of MicroRNA miR-222 with LNA Inhibitor Can Reduce Cell Proliferation in B Chronic Lymphoblastic Leukemia. Indian J. Hematol. Blood Transfus. Off. J. Indian Soc. Hematol. Blood Transfus. 2017, 33, 327–332. [Google Scholar] [CrossRef]
- Zhu, D.X.; Zhu, W.; Fang, C.; Fan, L.; Zou, Z.J.; Wang, Y.H.; Liu, P.; Hong, M.; Miao, K.R.; Liu, P.; et al. miR-181a/b significantly enhances drug sensitivity in chronic lymphocytic leukemia cells via targeting multiple anti-apoptosis genes. Carcinogenesis 2012, 33, 1294–1301. [Google Scholar] [CrossRef]
- Bresin, A.; Callegari, E.; D’Abundo, L.; Cattani, C.; Bassi, C.; Zagatti, B.; Narducci, M.G.; Caprini, E.; Pekarsky, Y.; Croce, C.M.; et al. miR-181b as a therapeutic agent for chronic lymphocytic leukemia in the Emicro-TCL1 mouse model. Oncotarget 2015, 6, 19807–19818. [Google Scholar] [CrossRef]
- Saleh, L.M.; Wang, W.; Herman, S.E.; Saba, N.S.; Anastas, V.; Barber, E.; Corrigan-Cummins, M.; Farooqui, M.; Sun, C.; Sarasua, S.M.; et al. Ibrutinib downregulates a subset of miRNA leading to upregulation of tumor suppressors and inhibition of cell proliferation in chronic lymphocytic leukemia. Leukemia 2017, 31, 340–349. [Google Scholar] [CrossRef]
- Dereani, S.; Macor, P.; D’Agaro, T.; Mezzaroba, N.; Dal-Bo, M.; Capolla, S.; Zucchetto, A.; Tissino, E.; Del Poeta, G.; Zorzet, S.; et al. Potential therapeutic role of antagomiR17 for the treatment of chronic lymphocytic leukemia. J. Hematol. Oncol. 2014, 7, 79. [Google Scholar] [CrossRef]
- D’Abundo, L.; Callegari, E.; Bresin, A.; Chillemi, A.; Elamin, B.K.; Guerriero, P.; Huang, X.; Saccenti, E.; Hussein, E.; Casciano, F.; et al. Anti-leukemic activity of microRNA-26a in a chronic lymphocytic leukemia mouse model. Oncogene 2017, 36, 6617–6626. [Google Scholar] [CrossRef]
- Chiang, C.L.; Goswami, S.; Frissora, F.W.; Xie, Z.; Yan, P.S.; Bundschuh, R.; Walker, L.A.; Huang, X.; Mani, R.; Mo, X.M.; et al. ROR1-targeted delivery of miR-29b induces cell cycle arrest and therapeutic benefit in vivo in a CLL mouse model. Blood 2019, 134, 432–444. [Google Scholar] [CrossRef] [PubMed]
- Cutrona, G.; Matis, S.; Colombo, M.; Massucco, C.; Baio, G.; Valdora, F.; Emionite, L.; Fabris, S.; Recchia, A.G.; Gentile, M.; et al. Effects of miRNA-15 and miRNA-16 expression replacement in chronic lymphocytic leukemia: Implication for therapy. Leukemia 2017, 31, 1894–1904. [Google Scholar] [CrossRef] [PubMed]
- Dolcetti, R.; Carbone, A. Epstein-Barr virus infection and chronic lymphocytic leukemia: A possible progression factor? Infect. Agents Cancer 2010, 5, 22. [Google Scholar] [CrossRef] [PubMed]
- Visco, C.; Falisi, E.; Young, K.H.; Pascarella, M.; Perbellini, O.; Carli, G.; Novella, E.; Rossi, D.; Giaretta, I.; Cavallini, C.; et al. Epstein-Barr virus DNA load in chronic lymphocytic leukemia is an independent predictor of clinical course and survival. Oncotarget 2015, 6, 18653–18663. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.H.; Gao, R.; Xia, Y.; Gale, R.P.; Chen, R.Z.; Yang, Y.Q.; Wang, L.; Qu, X.Y.; Qiu, H.R.; Cao, L.; et al. Prognostic impact of Epstein-Barr virus (EBV)-DNA copy number at diagnosis in chronic lymphocytic leukemia. Oncotarget 2016, 7, 2135–2142. [Google Scholar] [CrossRef] [PubMed]
- Iizasa, H.; Kim, H.; Kartika, A.V.; Kanehiro, Y.; Yoshiyama, H. Role of Viral and Host microRNAs in Immune Regulation of Epstein-Barr Virus-Associated Diseases. Front. Immunol. 2020, 11, 367. [Google Scholar] [CrossRef]
- Ferrajoli, A.; Ivan, C.; Ciccone, M.; Shimizu, M.; Kita, Y.; Ohtsuka, M.; D’Abundo, L.; Qiang, J.; Lerner, S.; Nouraee, N.; et al. Epstein-Barr Virus MicroRNAs are Expressed in Patients with Chronic Lymphocytic Leukemia and Correlate with Overall Survival. EBioMedicine 2015, 2, 572–582. [Google Scholar] [CrossRef]
- Linnstaedt, S.D.; Gottwein, E.; Skalsky, R.L.; Luftig, M.A.; Cullen, B.R. Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr virus. J. Virol. 2010, 84, 11670–11678. [Google Scholar] [CrossRef]
- Xu, D.M.; Kong, Y.L.; Wang, L.; Zhu, H.Y.; Wu, J.Z.; Xia, Y.; Li, Y.; Qin, S.C.; Fan, L.; Li, J.Y.; et al. EBV-miR-BHRF1-1 Targets p53 Gene: Potential Role in Epstein-Barr Virus Associated Chronic Lymphocytic Leukemia. Cancer Res. Treat. Off. J. Korean Cancer Assoc. 2020, 52, 492–504. [Google Scholar] [CrossRef]
- Stamatopoulos, B.; Van Damme, M.; Crompot, E.; Dessars, B.; Housni, H.E.; Mineur, P.; Meuleman, N.; Bron, D.; Lagneaux, L. Opposite Prognostic Significance of Cellular and Serum Circulating MicroRNA-150 in Patients with Chronic Lymphocytic Leukemia. Mol. Med. 2015, 21, 123–133. [Google Scholar] [CrossRef]
- Balatti, V.; Tomasello, L.; Rassenti, L.Z.; Veneziano, D.; Nigita, G.; Wang, H.Y.; Thorson, J.A.; Kipps, T.J.; Pekarsky, Y.; Croce, C.M. miR-125a and miR-34a expression predicts Richter syndrome in chronic lymphocytic leukemia patients. Blood 2018, 132, 2179–2182. [Google Scholar] [CrossRef] [PubMed]
- Martin-Guerrero, I.; Gutierrez-Camino, A.; Lopez-Lopez, E.; Bilbao-Aldaiturriaga, N.; Pombar-Gomez, M.; Ardanaz, M.; Garcia-Orad, A. Genetic variants in miRNA processing genes and pre-miRNAs are associated with the risk of chronic lymphocytic leukemia. PLoS ONE 2015, 10, e0118905. [Google Scholar] [CrossRef] [PubMed]

| miRNA | Target | Effect | References | |
|---|---|---|---|---|
| Bone marrow | miR-181a-5p | - | Promotes B-cell development | [12] |
| miR-23a-5p | - | Inhibits B-cell development | [13] | |
| miR-191-5p | Tcf3, Foxp1, and Egr1 | Acts as a rheostat for early B-cell development in mice | [21] | |
| miR-126-3p | RAG1 and RAG2 | Rescues B-cell development in absence of EBF1 | [11] | |
| miR-146a-5p | Egr1 | Downregulates Egr1, leading to B-cell malignancies | [20] | |
| miR-150-5p | MYB | Inhibits pro- to pre-B-cell transition | [10,22] | |
| miR-132-3p | Sox4 | [24] | ||
| miR-34a-5p | FOXP1 | [41] | ||
| miR-24-3p | MYC | [13,25] | ||
| miR-17/92 cluster | BCL2L11 and PTEN | Inhibits pro- to pre-B-cell transition; promotes the survival of immature B cells | [26] | |
| miR-21-5p | - | Inhibits pre-B cell apoptosis | [42] | |
| miR-148a-3p | BCL2L11, PTEN, and GADD45A | Promotes the survival of immature B cells | [28] | |
| miR-210-3p | - | Inhibits autoantibody production in mice | [43] | |
| Periphery | miR-29a-3p | - | Is essential for the formation of germinal center B cells in mice | [44] |
| miR-125b-5p | PRDM1 and IRF4 | Inhibits the differentiation of germinal center B cells | [33,34,35] | |
| miR-9-5p | PRDM1 | [36] | ||
| miR-30b-5p/miR-30d-5p | ||||
| miR-223-3p | LMO2 | [36] | ||
| miR-155-5p | SPI1 | Promotes the differentiation of germinal center B cells | [31] | |
| miR-148a-3p | BACH2 and MITF | [32] | ||
| miR-146a-5p | NUMB | Inhibits the formation of marginal zone B cells | [38] | |
| miR-142-5p | BAFFR | Inhibits the formation of marginal zone B cells; regulates gene expression in mature B cells | [40] |
| miRNA | Target | Effect | References |
|---|---|---|---|
| miR-29 family | TRAF4 | Suppresses CD40 signaling | [62] |
| miR-202-3p | SUFU | Regulates Hedgehog (Hh) signaling | [49] |
| miR-607 | FZD3 | Enhances WNT/β-catenin signaling | [74] |
| miR-708-5p | IKBKB | Suppresses NFkB signaling | [48] |
| miR-22-3p | PTEN, CDKN1B, and BIRC5 | Enhances PI3K/AKT signaling | [66] |
| miR-3151-5p | MADD and PIK3R2 | Suppresses MAPK/ERK and PI3K/AKT signaling | [64] |
| miR-126-3p | PIK3R2 | Suppresses MAPK/ERK signaling | [65] |
| miR-21-5p | SPRY2 | Enhances BCR and MAPK/ERK signaling | [63] |
| miR-150-5p | GAB1 and FOXP1 | Suppresses BCR signaling | [58,60] |
| miR-34a-5p | FOXP1 | [61] | |
| miR-155-5p | SHIP1 | Enhances BCR signaling | [59] |
| - | Regulates cell survival | [76] | |
| miR-221-3p; miR-222-3p | CDKN1B | Regulates cell proliferation | [77] |
| miR-15a-5p; miR-16-5p | BCL2 | Regulates cell survival | [54] |
| TP53 | Regulates cell proliferation | [55,68] | |
| ROR1 | Regulates cell survival and proliferation | [78] | |
| miR-26a-5p | EZH2 | Regulates cell survival and proliferation | [79] |
| PTEN | [67] | ||
| miR-214-3p | PTEN | [67] | |
| miR-337-3p | PML | [75] | |
| miR-106b-5p | ITCH | [80] | |
| miR-28-5p | NDRG2 | [81] | |
| miR-650 | |||
| miR-181a-5p/miR-181b-5p | - | Suppresses cell growth | [58] |
| miR-210-3p; miR-425-5p; miR-1253; miR-4269; miR-4667-3p | BTK | Promotes apoptosis | [50] |
| miR-130a-3p | ATG2B and DICER1 | Inhibits autophagy and regulates cell survival | [46] |
| miR-125b-5p | PCTP, LIPA, GSS, HK2, IKZF4, and TP53 | Regulates metabolic adaptation to cancer transformation | [47] |
| miR-29b-3p; miR-34b-5p; miR-181b-5p; miR-484 | TCL1A | Regulates multiple signaling pathways and cell survival | [51,52,53] |
| miRNA | Localization | miRNA Expression | Biomarker Utility | References |
|---|---|---|---|---|
| miR-20b-5p | PBMCs 1 | Lower levels in patients with poor prognosis | Prognosis | [96] |
| miR-21-5p; miR-125b-5p; miR-148a-3p; miR-181a-5p; miR-221-3p; miR-222-3p; miR-532-3p | PBMCs 1 | Lower levels in responsive patients | Prediction of response | [92,93,95] |
| miR-29a-3p; miR-34a-5p | Higher levels in responsive patients | [92,94] | ||
| miR-181b-5p | PBMCs 1 | Higher levels in indolent vs. aggressive disease | Prediction of progression | [84] |
| miR-744-5p | Lower levels in patients with shorter time to first treatment | [97] | ||
| miR-4524a-5p | High levels in patients with shorter time to first treatment | |||
| miR-92a-3p | PBMCs 1 | Lower levels in B-CLL patients vs. non-leukemic controls | Diagnosis | [98] |
| Lower levels in patients with poor prognosis | Prognosis | |||
| miR-155-5p | Plasma | Higher levels in B-CLL patients vs. non-leukemic controls | Diagnosis | [86] |
| PBMCs 1 | Higher levels in patients with poor prognosis | Prognosis | ||
| Purified B cells | Lower levels in responsive patients | Prediction of response | [85] | |
| miRNA signature | Serum | - | Diagnosis | [99] |
| miRNA signature | - | [100] | ||
| miRNA signature | - | Prediction of progression | [89] | |
| miRNA signature | PBMCs 1 | Diagnosis; prognosis | [82] | |
| miRNA signature | Purified B cells | [101] | ||
| miRNA signature | PBMCs 1 | Prognosis; prediction of progression | [83] | |
| Scoring system, including miR-21-5p | PBMCs 1 | Higher levels in patients with poor prognosis | Prognosis | [91] |
| Scoring system, including miR-29c-3p | Lower levels in patients with poor prognosis | [90] |
| miRNA | Experimental Approach | Effect | References |
|---|---|---|---|
| miR-15a-5p; miR-16-5p | Human cells | Restoration of cell cycle control | [102] |
| Mouse model | Drug sensitization and induction of apoptosis in mice upon its upregulation | [102,103,112] | |
| miR-155-3p | Human cells | Regulation of chemoresistance | [104] |
| miR-222-3p | Human cells | Reduced cell viability and proliferation upon its downregulation | [105] |
| miR-181a-5p/miR-181b-5p | Human cells | Increased apoptosis of cells | [106,107] |
| Mouse model | Reduced leukemic cell expansion and increase of survival in mice upon its upregulation | [107] | |
| miR-34a-5p; miR-146b-5p | Human cells | Inhibition of cell proliferation upon its downregulation | [108] |
| miR-17-5p | Human cells | Reduced cell proliferation | [109] |
| Mouse model | Reduced tumor growth and increased survival in mice upon its downregulation | ||
| miR-26a-5p | Human cells | Induction of apoptosis with CD38-targeted delivery | [110] |
| Mouse model | |||
| miR-29b-3p | Mouse model | Induction of cell cycle arrest with ROR1-targeted delivery | [111] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Katsaraki, K.; Karousi, P.; Artemaki, P.I.; Scorilas, A.; Pappa, V.; Kontos, C.K.; Papageorgiou, S.G. MicroRNAs: Tiny Regulators of Gene Expression with Pivotal Roles in Normal B-Cell Development and B-Cell Chronic Lymphocytic Leukemia. Cancers 2021, 13, 593. https://doi.org/10.3390/cancers13040593
Katsaraki K, Karousi P, Artemaki PI, Scorilas A, Pappa V, Kontos CK, Papageorgiou SG. MicroRNAs: Tiny Regulators of Gene Expression with Pivotal Roles in Normal B-Cell Development and B-Cell Chronic Lymphocytic Leukemia. Cancers. 2021; 13(4):593. https://doi.org/10.3390/cancers13040593
Chicago/Turabian StyleKatsaraki, Katerina, Paraskevi Karousi, Pinelopi I. Artemaki, Andreas Scorilas, Vasiliki Pappa, Christos K. Kontos, and Sotirios G. Papageorgiou. 2021. "MicroRNAs: Tiny Regulators of Gene Expression with Pivotal Roles in Normal B-Cell Development and B-Cell Chronic Lymphocytic Leukemia" Cancers 13, no. 4: 593. https://doi.org/10.3390/cancers13040593
APA StyleKatsaraki, K., Karousi, P., Artemaki, P. I., Scorilas, A., Pappa, V., Kontos, C. K., & Papageorgiou, S. G. (2021). MicroRNAs: Tiny Regulators of Gene Expression with Pivotal Roles in Normal B-Cell Development and B-Cell Chronic Lymphocytic Leukemia. Cancers, 13(4), 593. https://doi.org/10.3390/cancers13040593