Extracellular miRNAs as Predictive Biomarkers for Glypican-3-Derived Peptide Vaccine Therapy Response in Ovarian Clear Cell Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
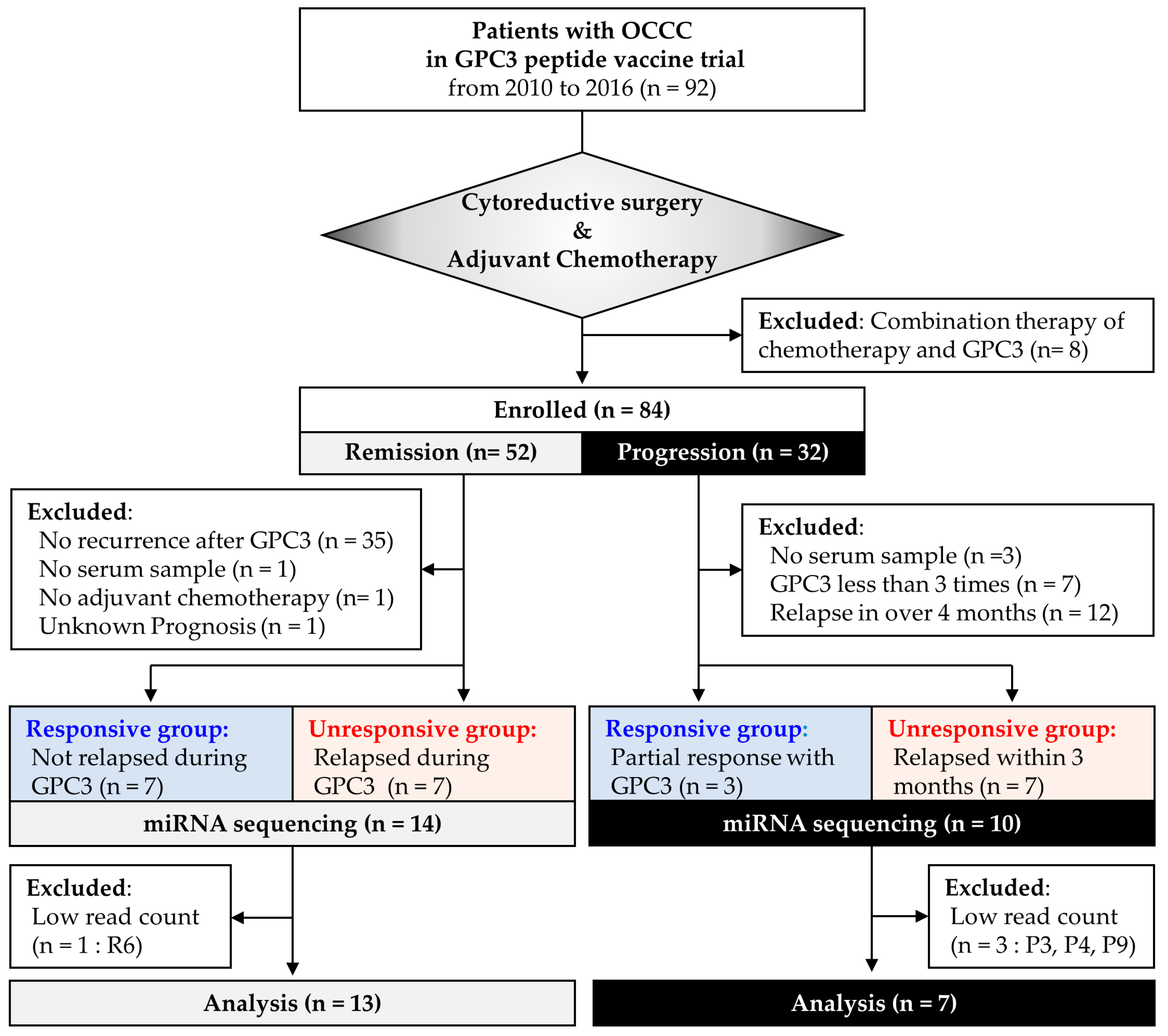
2.1. Study Design
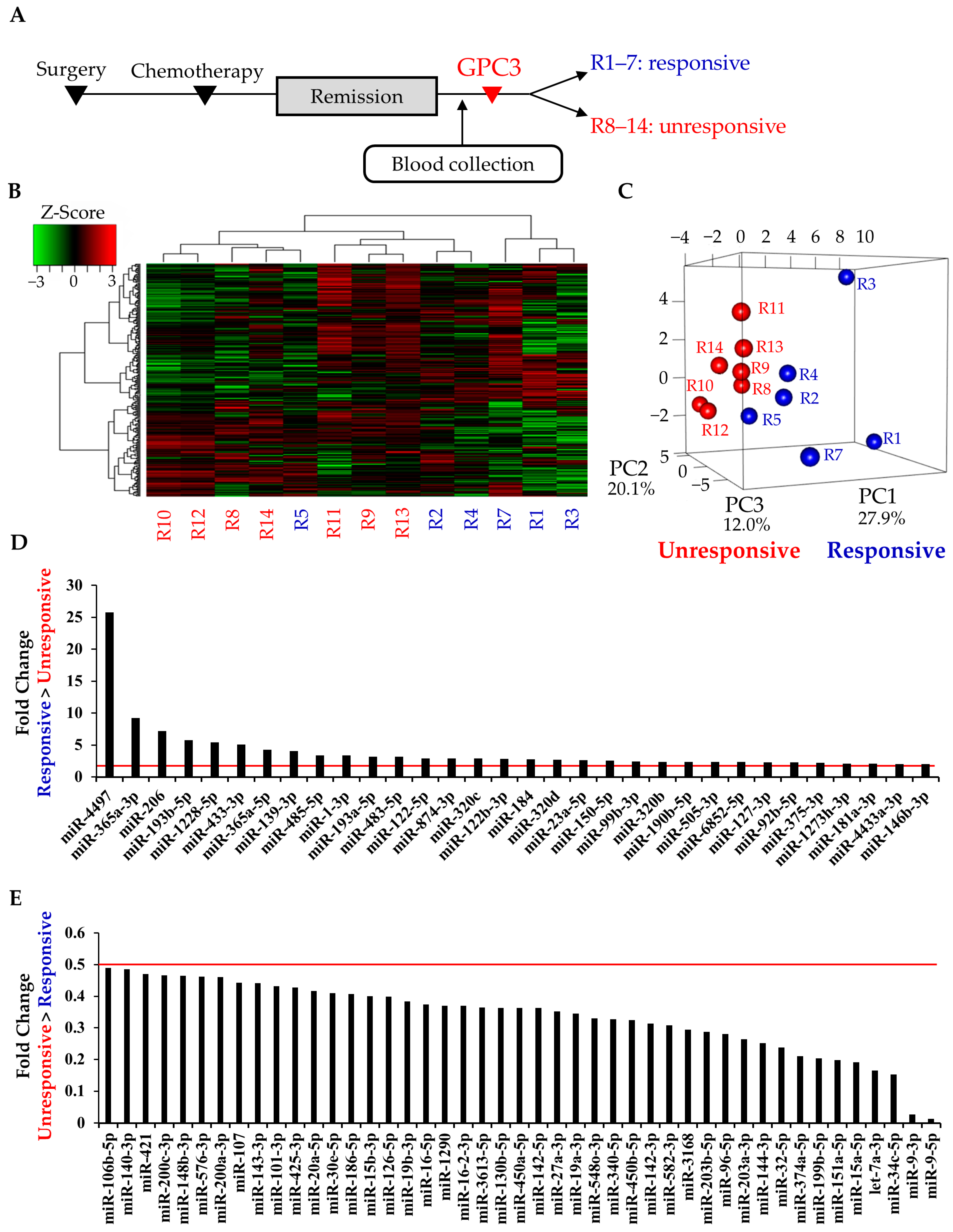
2.2. Study 1: Identification of Circulating miRNAs that Predict GPC3 Peptide Vaccine Efficacy
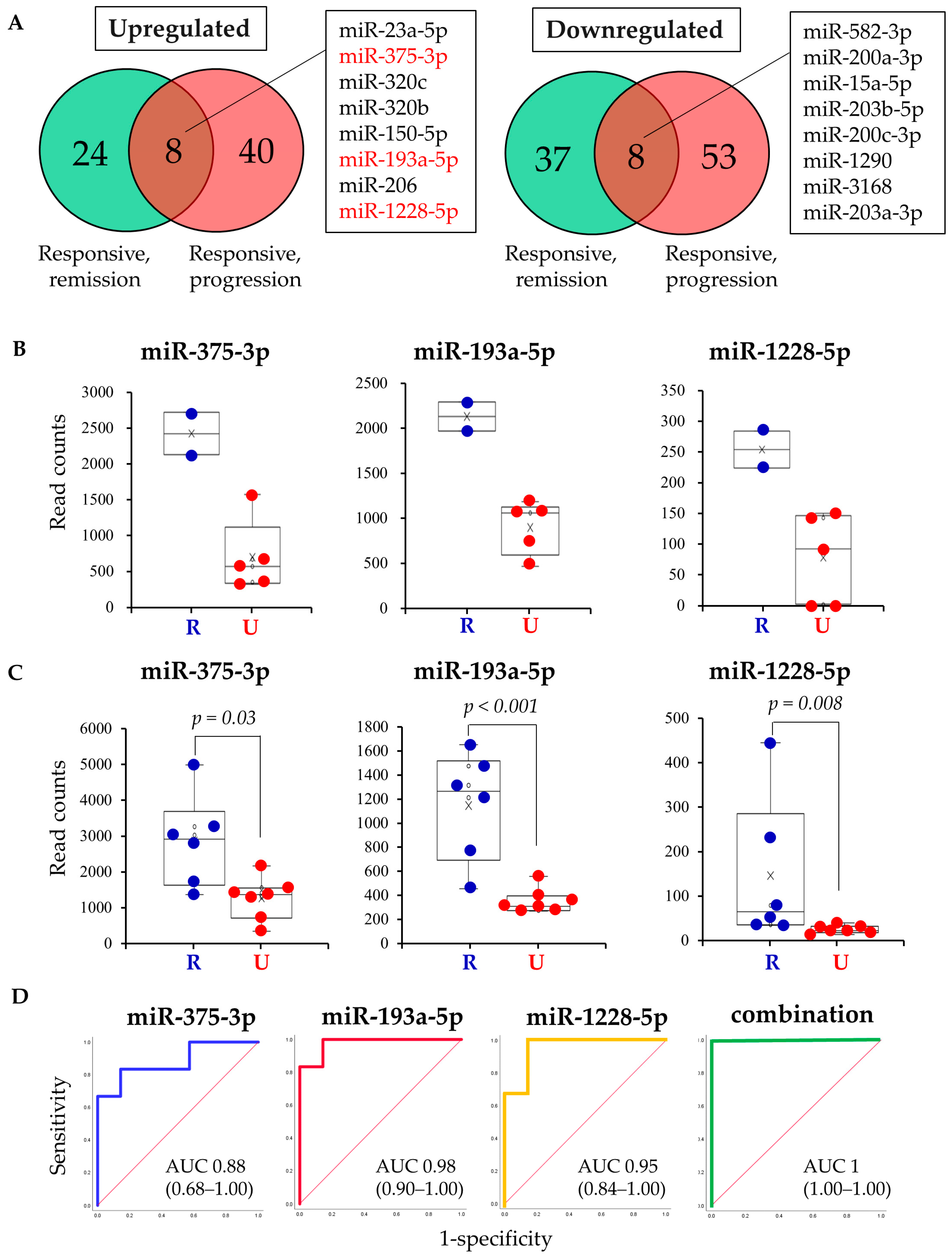
2.3. Study 2: Validation of Candidate miRNAs in the Patients with Progressive Disease
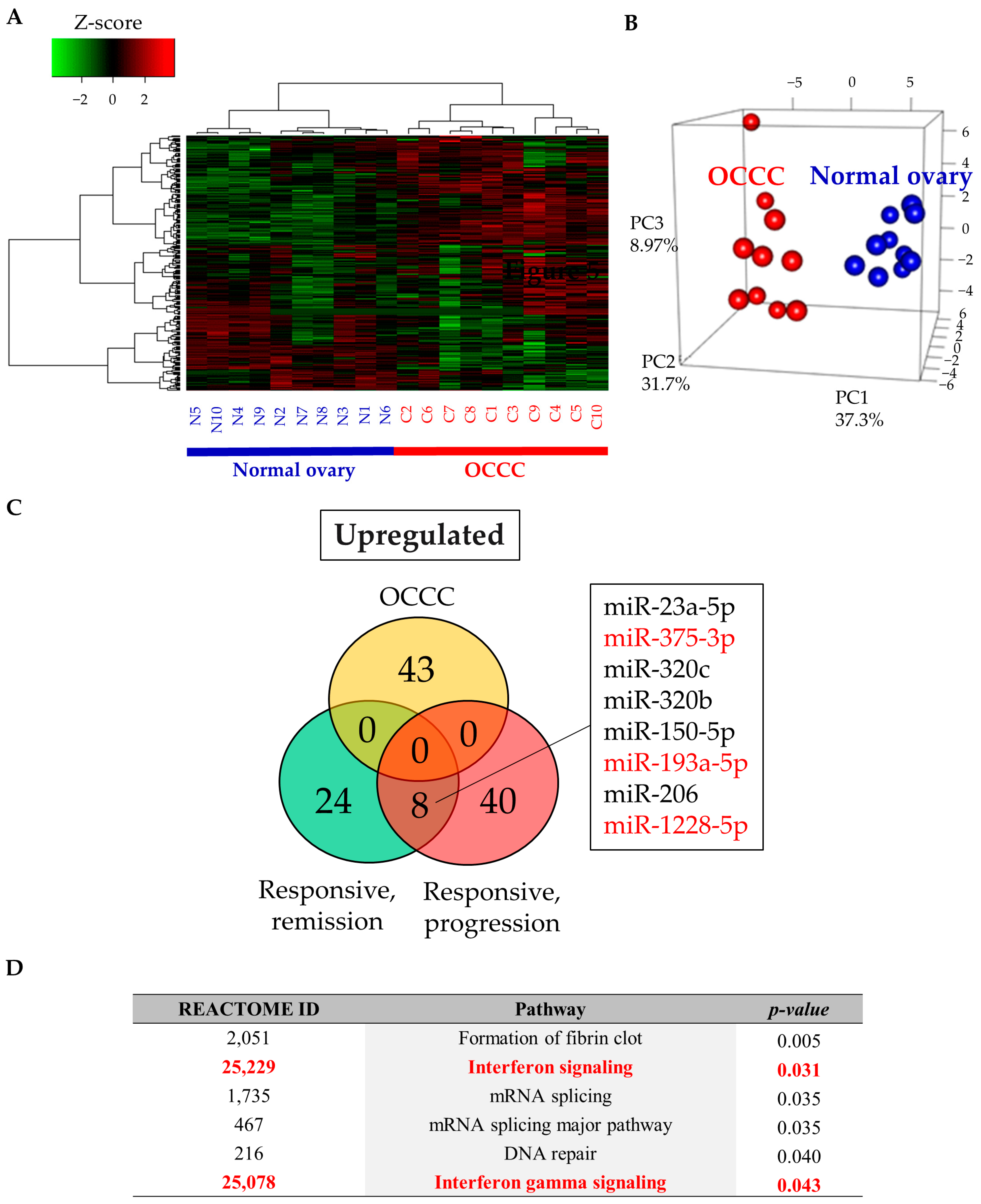
2.4. Study 3: Assessing the Function of Candidate miRNAs
3. Discussion
4. Materials and Methods
4.1. Patients and Sample
4.2. RNA Extraction and miRNA Sequencing
4.3. Data Analysis
4.4. In Silico Analysis
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Odunsi, K. Immunotherapy in ovarian cancer. Ann. Oncol. 2017, 28, viii1–viii7. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, K.; Shintani, D.; Nishikawa, T. Clear-cell carcinoma of the ovary. Ann. Oncol. 2016, 27, i50–i52. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, T.; Kamura, T.; Kigawa, J.; Terakawa, N.; Kikuchi, Y.; Kita, T.; Suzuki, M.; Sato, I.; Taguchi, K. Clinical characteristics of clear cell carcinoma of the ovary: A distinct histologic type with poor prognosis and resistance to platinum-based chemotherapy. Cancer 2000, 88, 2584–2589. [Google Scholar] [CrossRef]
- Pujade-Lauraine, E.; Ledermann, J.A.; Selle, F.; Gebski, V.; Penson, R.T.; Oza, A.M.; Korach, J.; Huzarski, T.; Poveda, A.; Pignata, S.; et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet. Oncol. 2017, 18, 1274–1284. [Google Scholar] [CrossRef]
- Ledermann, J.; Harter, P.; Gourley, C.; Friedlander, M.; Vergote, I.; Rustin, G.; Scott, C.; Meier, W.; Shapira-Frommer, R.; Safra, T.; et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N. Engl. J. Med. 2012, 366, 1382–1392. [Google Scholar] [CrossRef]
- Perren, T.J.; Swart, A.M.; Pfisterer, J.; Ledermann, J.A.; Pujade-Lauraine, E.; Kristensen, G.; Carey, M.S.; Beale, P.; Cervantes, A.; Kurzeder, C.; et al. A phase 3 trial of bevacizumab in ovarian cancer. N. Engl. J. Med. 2011, 365, 2484–2496. [Google Scholar] [CrossRef]
- Burger, R.A.; Brady, M.F.; Bookman, M.A.; Fleming, G.F.; Monk, B.J.; Huang, H.; Mannel, R.S.; Homesley, H.D.; Fowler, J.; Greer, B.E.; et al. Incorporation of bevacizumab in the primary treatment of ovarian cancer. N. Engl. J. Med. 2011, 365, 2473–2483. [Google Scholar] [CrossRef]
- Suzuki, S.; Yoshikawa, T.; Hirosawa, T.; Shibata, K.; Kikkawa, F.; Akatsuka, Y.; Nakatsura, T. Glypican-3 could be an effective target for immunotherapy combined with chemotherapy against ovarian clear cell carcinoma. Cancer Sci. 2011, 102, 1622–1629. [Google Scholar] [CrossRef]
- Suzuki, S.; Sakata, J.; Utsumi, F.; Sekiya, R.; Kajiyama, H.; Shibata, K.; Kikkawa, F.; Nakatsura, T. Efficacy of glypican-3-derived peptide vaccine therapy on the survival of patients with refractory ovarian clear cell carcinoma. Oncoimmunology 2016, 5, e1238542. [Google Scholar] [CrossRef]
- Kim, V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005, 6, 376–385. [Google Scholar] [CrossRef]
- Matsuyama, H.; Suzuki, H.I.; Nishimori, H.; Noguchi, M.; Yao, T.; Komatsu, N.; Mano, H.; Sugimoto, K.; Miyazono, K. miR-135b mediates NPM-ALK-driven oncogenicity and renders IL-17-producing immunophenotype to anaplastic large cell lymphoma. Blood 2011, 118, 6881–6892. [Google Scholar] [CrossRef] [PubMed]
- Png, K.J.; Halberg, N.; Yoshida, M.; Tavazoie, S.F. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature 2011, 481, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.I.; Katsura, A.; Matsuyama, H.; Miyazono, K. MicroRNA regulons in tumor microenvironment. Oncogene 2015, 34, 3085–3094. [Google Scholar] [CrossRef] [PubMed]
- Au Yeung, C.L.; Co, N.N.; Tsuruga, T.; Yeung, T.L.; Kwan, S.Y.; Leung, C.S.; Li, Y.; Lu, E.S.; Kwan, K.; Wong, K.K.; et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat. Commun. 2016, 7, 11150. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Ochiya, T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010, 101, 2087–2092. [Google Scholar] [CrossRef]
- Yoshida, K.; Yokoi, A.; Kato, T.; Ochiya, T.; Yamamoto, Y. The clinical impact of intra- and extracellular miRNAs in ovarian cancer. Cancer Sci. 2020, 111, 3435–3444. [Google Scholar] [CrossRef]
- Rapado-González, Ó.; Álvarez-Castro, A.; López-López, R.; Iglesias-Canle, J.; Suárez-Cunqueiro, M.M.; Muinelo-Romay, L. Circulating microRNAs as Promising Biomarkers in Colorectal Cancer. Cancers 2019, 11. [Google Scholar] [CrossRef]
- Yokoi, A.; Matsuzaki, J.; Yamamoto, Y.; Yoneoka, Y.; Takahashi, K.; Shimizu, H.; Uehara, T.; Ishikawa, M.; Ikeda, S.I.; Sonoda, T.; et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat. Commun. 2018, 9, 4319. [Google Scholar] [CrossRef]
- Goff, B.A. Advanced ovarian cancer: What should be the standard of care? J. Gynecol. Oncol. 2013, 24, 83–91. [Google Scholar] [CrossRef]
- Lim, H.J.; Ledger, W. Targeted therapy in ovarian cancer. Womens Health 2016, 12, 363–378. [Google Scholar] [CrossRef]
- Hwang, W.T.; Adams, S.F.; Tahirovic, E.; Hagemann, I.S.; Coukos, G. Prognostic significance of tumor-infiltrating T cells in ovarian cancer: A meta-analysis. Gynecol. Oncol. 2012, 124, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Gadducci, A.; Guerrieri, M.E. Immune Checkpoint Inhibitors in Gynecological Cancers: Update of Literature and Perspectives of Clinical Research. Anticancer Res. 2017, 37, 5955–5965. [Google Scholar] [CrossRef] [PubMed]
- Palaia, I.; Tomao, F.; Sassu, C.M.; Musacchio, L.; Benedetti Panici, P. Immunotherapy For Ovarian Cancer: Recent Advances And Combination Therapeutic Approaches. OncoTargets Ther. 2020, 13, 6109–6129. [Google Scholar] [CrossRef]
- Romano, G.; Kwong, L.N. Diagnostic and therapeutic applications of miRNA-based strategies to cancer immunotherapy. Cancer Metastasis Rev. 2018, 37, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, M.; Westergaard, M.C.W.; Milne, K.; Nielsen, M.; Borch, T.H.; Poulsen, L.G.; Hendel, H.W.; Kennedy, M.; Briggs, G.; Ledoux, S.; et al. Adoptive cell therapy with tumor-infiltrating lymphocytes in patients with metastatic ovarian cancer: A pilot study. Oncoimmunology 2018, 7, e1502905. [Google Scholar] [CrossRef]
- Aoki, Y.; Takakuwa, K.; Kodama, S.; Tanaka, K.; Takahashi, M.; Tokunaga, A.; Takahashi, T. Use of adoptive transfer of tumor-infiltrating lymphocytes alone or in combination with cisplatin-containing chemotherapy in patients with epithelial ovarian cancer. Cancer Res. 1991, 51, 1934–1939. [Google Scholar]
- Garofalo, M.; Croce, C.M. MicroRNAs as therapeutic targets in chemoresistance. Drug Resist. Updat. 2013, 16, 47–59. [Google Scholar] [CrossRef]
- Rodríguez-Galán, A.; Fernández-Messina, L.; Sánchez-Madrid, F. Control of Immunoregulatory Molecules by miRNAs in T Cell Activation. Front. Immunol. 2018, 9, 2148. [Google Scholar] [CrossRef]
- Wu, Q.; Zhao, Y.; Sun, Y.; Yan, X.; Wang, P. miR-375 inhibits IFN-γ-induced programmed death 1 ligand 1 surface expression in head and neck squamous cell carcinoma cells by blocking JAK2/STAT1 signaling. Oncol. Rep. 2018, 39, 1461–1468. [Google Scholar] [CrossRef]
- Yu, H.; Jiang, L.; Sun, C.; Guo, L.; Lin, M.; Huang, J.; Zhu, L. Decreased circulating miR-375: A potential biomarker for patients with non-small-cell lung cancer. Gene 2014, 534, 60–65. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, J.S.; Wen, J.K.; Gao, H.T.; Zheng, B.; Qu, C.B.; Liu, K.L.; Zhang, M.L.; Gu, J.F.; Li, J.D.; et al. Silencing of miR-193a-5p increases the chemosensitivity of prostate cancer cells to docetaxel. J. Exp. Clin. Cancer Res. CR 2017, 36, 178. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.P.; Liao, J.Z.; Lu, Y.Q.; Tian, D.A.; Ye, F.; Zhao, P.X.; Xiang, G.Y.; Tang, W.X.; He, X.X. MiR-375 and Doxorubicin Co-delivered by Liposomes for Combination Therapy of Hepatocellular Carcinoma. Mol. Ther. Nucleic Acids 2017, 7, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.; Balwierz, A.; Zhang, J.D.; Küblbeck, M.; Pawitan, Y.; Hielscher, T.; Wiemann, S.; Sahin, Ö. Re-expression of microRNA-375 reverses both tamoxifen resistance and accompanying EMT-like properties in breast cancer. Oncogene 2013, 32, 1173–1182. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Ye, M.L.; Zhang, Y.P.; Li, W.J.; Li, M.T.; Wang, H.Z.; Qiu, X.; Xu, Y.; Yin, J.W.; Hu, Q.; et al. MicroRNA-375-3p enhances chemosensitivity to 5-fluorouracil by targeting thymidylate synthase in colorectal cancer. Cancer Sci. 2020, 111, 1528–1541. [Google Scholar] [CrossRef]
- Xu, X.; Chen, X.; Xu, M.; Liu, X.; Pan, B.; Qin, J.; Xu, T.; Zeng, K.; Pan, Y.; He, B.; et al. miR-375-3p suppresses tumorigenesis and partially reverses chemoresistance by targeting YAP1 and SP1 in colorectal cancer cells. Aging 2019, 11, 7357–7385. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lieberman, R.; Pan, J.; Zhang, Q.; Du, M.; Zhang, P.; Nevalainen, M.; Kohli, M.; Shenoy, N.K.; Meng, H.; et al. miR-375 induces docetaxel resistance in prostate cancer by targeting SEC23A and YAP1. Mol. Cancer 2016, 15, 70. [Google Scholar] [CrossRef]
- Lindberg, R.L.; Hoffmann, F.; Mehling, M.; Kuhle, J.; Kappos, L. Altered expression of miR-17-5p in CD4+ lymphocytes of relapsing-remitting multiple sclerosis patients. Eur. J. Immunol. 2010, 40, 888–898. [Google Scholar] [CrossRef] [PubMed]
- Graff, J.W.; Dickson, A.M.; Clay, G.; McCaffrey, A.P.; Wilson, M.E. Identifying functional microRNAs in macrophages with polarized phenotypes. J. Biol. Chem. 2012, 287, 21816–21825. [Google Scholar] [CrossRef]
- Kao, S.C.; Cheng, Y.Y.; Williams, M.; Kirschner, M.B.; Madore, J.; Lum, T.; Sarun, K.H.; Linton, A.; McCaughan, B.; Klebe, S.; et al. Tumor Suppressor microRNAs Contribute to the Regulation of PD-L1 Expression in Malignant Pleural Mesothelioma. J. Thorac. Oncol. 2017, 12, 1421–1433. [Google Scholar] [CrossRef]
- Deng, H.; Lv, L.; Li, Y.; Zhang, C.; Meng, F.; Pu, Y.; Xiao, J.; Qian, L.; Zhao, W.; Liu, Q.; et al. miR-193a-3p regulates the multi-drug resistance of bladder cancer by targeting the LOXL4 gene and the oxidative stress pathway. Mol. Cancer 2014, 13, 234. [Google Scholar] [CrossRef]
- Bagnoli, M.; Canevari, S.; Califano, D.; Losito, S.; Maio, M.D.; Raspagliesi, F.; Carcangiu, M.L.; Toffoli, G.; Cecchin, E.; Sorio, R.; et al. Development and validation of a microRNA-based signature (MiROvaR) to predict early relapse or progression of epithelial ovarian cancer: A cohort study. Lancet Oncol. 2016, 17, 1137–1146. [Google Scholar] [CrossRef]
- Xue, W.X.; Zhang, M.Y.; Rui, L.; Liu, X.; Yin, Y.H.; Qu, Y.Q. Serum miR-1228-3p and miR-181a-5p as Noninvasive Biomarkers for Non-Small Cell Lung Cancer Diagnosis and Prognosis. BioMed Res. Int. 2020, 2020, 9601876. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Hazama, S.; Iida, M.; Tsunedomi, R.; Takenouchi, H.; Nakajima, M.; Tokumitsu, Y.; Kanekiyo, S.; Shindo, Y.; Tomochika, S.; et al. miR-125b-1 and miR-378a are predictive biomarkers for the efficacy of vaccine treatment against colorectal cancer. Cancer Sci. 2017, 108, 2229–2238. [Google Scholar] [CrossRef] [PubMed]
- Kijima, T.; Hazama, S.; Tsunedomi, R.; Tanaka, H.; Takenouchi, H.; Kanekiyo, S.; Inoue, Y.; Nakashima, M.; Iida, M.; Sakamoto, K.; et al. MicroRNA-6826 and -6875 in plasma are valuable non-invasive biomarkers that predict the efficacy of vaccine treatment against metastatic colorectal cancer. Oncol. Rep. 2017, 37, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Birkhäuser, F.D.; Koya, R.C.; Neufeld, C.; Rampersaud, E.N.; Lu, X.; Micewicz, E.D.; Chodon, T.; Atefi, M.; Kroeger, N.; Chandramouli, G.V.R.; et al. Dendritic cell-based immunotherapy in prevention and treatment of renal cell carcinoma: Efficacy, safety, and activity of Ad-GM·CAIX in immunocompetent mouse models. J. Immunother. 2013, 36, 102–111. [Google Scholar] [CrossRef]
- Suzuki, S.; Shibata, K.; Kikkawa, F.; Nakatsura, T. Significant clinical response of progressive recurrent ovarian clear cell carcinoma to glypican-3-derived peptide vaccine therapy: Two case reports. Hum. Vaccin. Immunother. 2014, 10, 338–343. [Google Scholar] [CrossRef]
- Rustin, G.J.; Vergote, I.; Eisenhauer, E.; Pujade-Lauraine, E.; Quinn, M.; Thigpen, T.; du Bois, A.; Kristensen, G.; Jakobsen, A.; Sagae, S.; et al. Definitions for response and progression in ovarian cancer clinical trials incorporating RECIST 1.1 and CA 125 agreed by the Gynecological Cancer Intergroup (GCIG). Int. J. Gynecol. Cancer 2011, 21, 419–423. [Google Scholar] [CrossRef]

| Case | Age | Stage | Surgery | Chemotherapy | Number of Vaccination |
|---|---|---|---|---|---|
| Responsive subgroup | |||||
| R1 | 61 | IIIc | ATH + BSO + OM + PEN + PAN | TC | 10 |
| R2 | 47 | IIIc | ATH + BSO + OM + PEN + PAN | TC | 10 |
| R3 | 62 | IIc(b) | ATH + BSO + OM + PEN + PAN | TC | 10 |
| R4 | 56 | IIIc | RSO + OM | TC | 10 |
| R5 | 63 | Ic(a) | BSO | TC | 10 |
| R6 | 56 | Ic(b) | RSO + OM | TC | 10 |
| R7 | 59 | IIIc | ATH + BSO + OM + PEN + PAN | TC | 10 |
| Unresponsive subgroup | |||||
| R8 | 49 | IIIa | ATH + BSO + OM + PEN + PAN | TC | 8 |
| R9 | 51 | IIc(1) | ATH + BSO + OM + PEN + PAN | TC | 4 |
| R10 | 40 | IIIa | ATH + BSO + OM + PEN + PAN | TC | 4 |
| R11 | 54 | IIIc | ATH + BSO + OM + PEN + PAN | TC, DC | 3 |
| R12 | 37 | Ic(2) | ATH + BSO + OM + PEN + PAN | TC | 8 |
| R13 | 46 | IIc(1) | ATH + BSO + OM + PEN + PAN | TC | 6 |
| R14 | 57 | Ic(a) | ATH + BSO + OM + PEN + PAN | TC | 7 |
| Case | Age | Stage | Surgery | Chemotherapy | Site of Recurrence | Number of Vaccination |
|---|---|---|---|---|---|---|
| Responsive subgroup | ||||||
| P1 | 43 | IIIc | ATH + BSO + OM + PEN + PAN | CPT-11 + CDDP | liver, retroperitoneal LN, peritoneum | 13 |
| P2 | 67 | IIIc | ATH + BSO + OM + PEN + PAN | TC, CPT-11 + NDP, GEM + DTX | multiple LN | 27 |
| P3 | 65 | IIc | ATH + BSO + OM + PEN + PAN | PLD + CBDCA | peritoneum | 13 |
| Unresponsive subgroup | ||||||
| P4 | 50 | IIIc | ATH + BSO + OM + PEN | TC, CPT-11, GEM | cancerous peritonitis | 4 |
| P5 | 51 | IIIb | ATH + BSO + OM | TC, NGT, ETP | liver, multiple LN, peritoneum | 3 |
| P6 | 53 | IIIb | ATH + BSO + OM | TCB, GEM | multiple LN peritoneum | 3 |
| P7 | 61 | IIIc | ATH + BSO + OM | TCB, GEM | brain | 3 |
| P8 | 48 | Ic | ATH + BSO + OM + PEN + PAN | TC, DC, PLD | liver, lung | 4 |
| P9 | 40 | IVb | ATH + BSO + OM + PEN + PAN | TC, Radiation | bone | 5 |
| P10 | 63 | IV | ATH + BSO + OM + PEN + PAN | TC, CPT-11 + CDDP, PLD, NGT, GEM, Radiation | multiple LN | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ukai, M.; Yokoi, A.; Yoshida, K.; Suzuki, S.; Shibata, K.; Kikkawa, F.; Nakatsura, T.; Kajiyama, H. Extracellular miRNAs as Predictive Biomarkers for Glypican-3-Derived Peptide Vaccine Therapy Response in Ovarian Clear Cell Carcinoma. Cancers 2021, 13, 550. https://doi.org/10.3390/cancers13030550
Ukai M, Yokoi A, Yoshida K, Suzuki S, Shibata K, Kikkawa F, Nakatsura T, Kajiyama H. Extracellular miRNAs as Predictive Biomarkers for Glypican-3-Derived Peptide Vaccine Therapy Response in Ovarian Clear Cell Carcinoma. Cancers. 2021; 13(3):550. https://doi.org/10.3390/cancers13030550
Chicago/Turabian StyleUkai, Mayu, Akira Yokoi, Kosuke Yoshida, Shiro Suzuki, Kiyosumi Shibata, Fumitaka Kikkawa, Tetsuya Nakatsura, and Hiroaki Kajiyama. 2021. "Extracellular miRNAs as Predictive Biomarkers for Glypican-3-Derived Peptide Vaccine Therapy Response in Ovarian Clear Cell Carcinoma" Cancers 13, no. 3: 550. https://doi.org/10.3390/cancers13030550
APA StyleUkai, M., Yokoi, A., Yoshida, K., Suzuki, S., Shibata, K., Kikkawa, F., Nakatsura, T., & Kajiyama, H. (2021). Extracellular miRNAs as Predictive Biomarkers for Glypican-3-Derived Peptide Vaccine Therapy Response in Ovarian Clear Cell Carcinoma. Cancers, 13(3), 550. https://doi.org/10.3390/cancers13030550






