Bile Acid Dysregulation Is Intrinsically Related to Cachexia in Tumor-Bearing Mice
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Mouse Experiments
2.3. TGR5 Activation in a Cell Reporter Assay
2.4. Bile Acid Quantification
2.5. Tissue mRNA Analysis
2.6. Statistical Analyses
3. Results
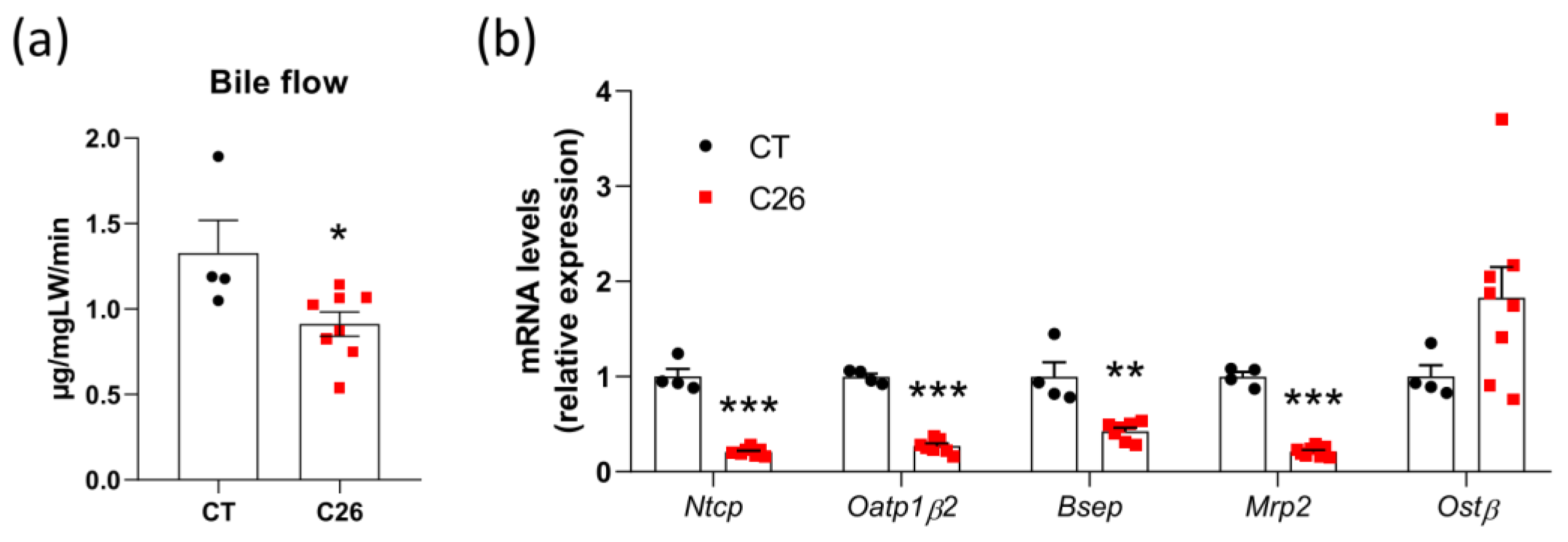
3.1. Decreased Bile Flow and Alterations in the Hepatobiliary Transport System in C26 Cachectic Mice
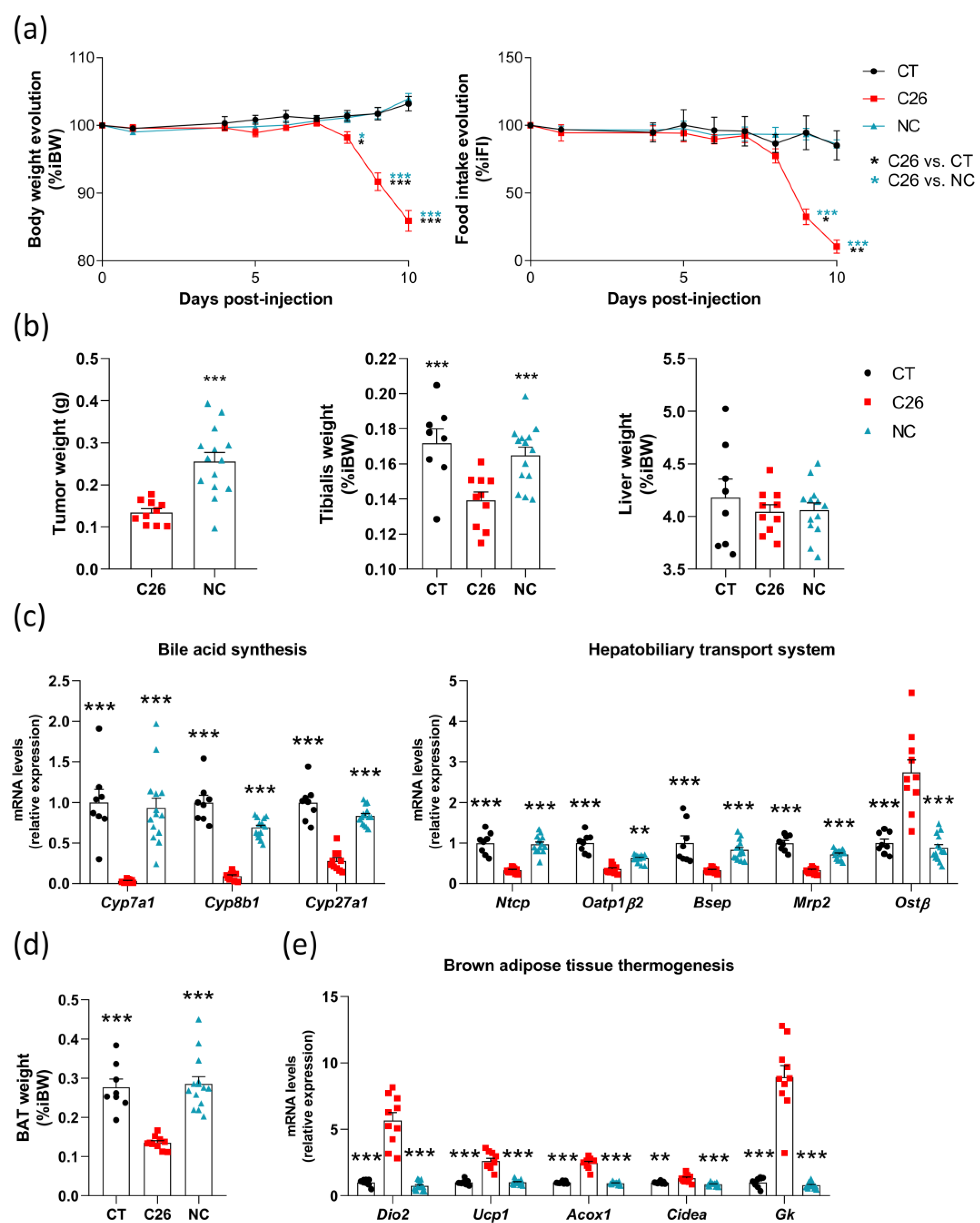
3.2. Many Alterations in the Liver, Brown Adipose Tissue and Muscle Are Intrinsically Related to Cachexia in the C26 Model
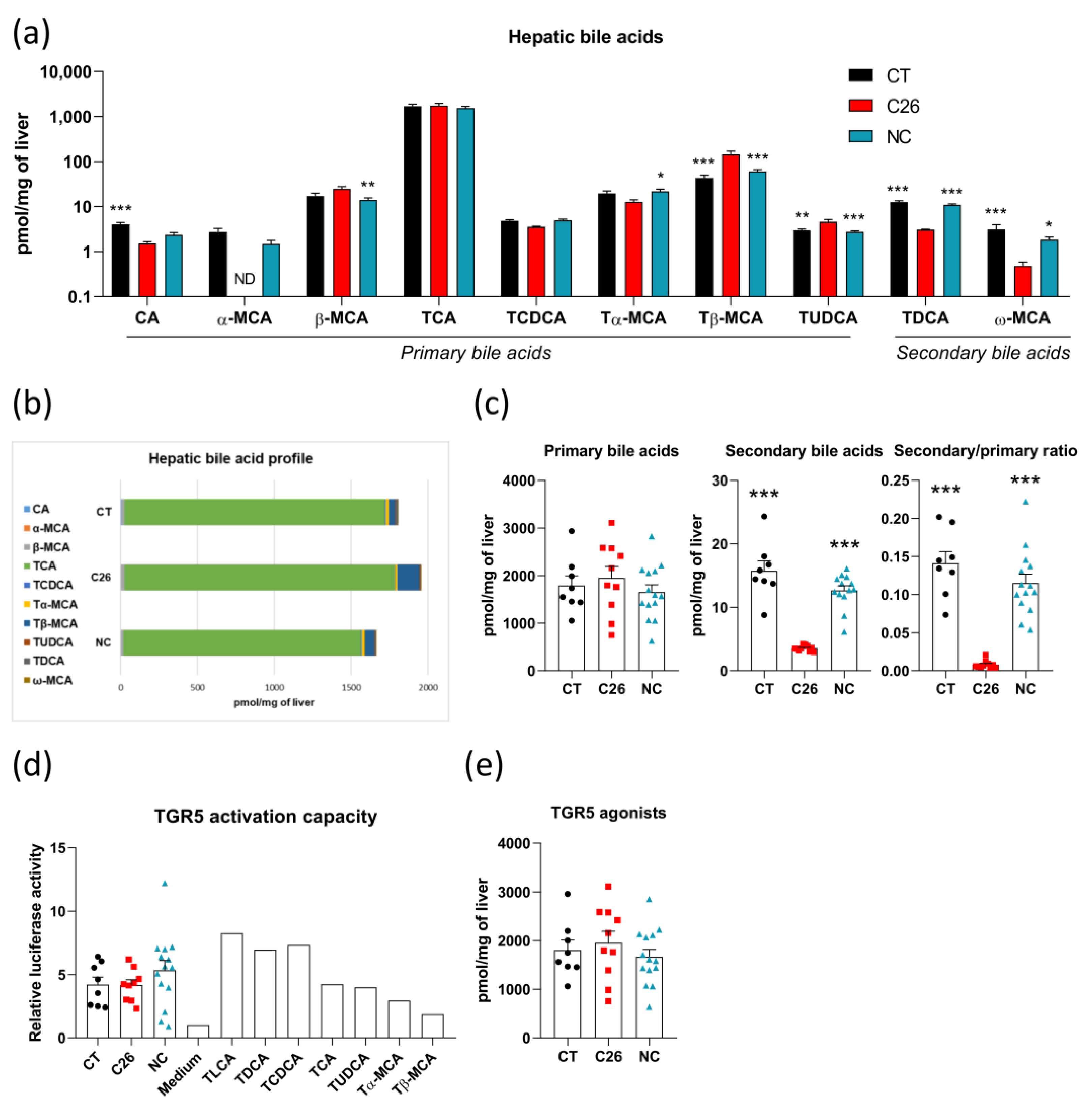
3.3. Alterations in Bile Acid Profile Are Intrinsically Related to Cachexia without Any Modification of TGR5 Activation Capacity in C26 Cachectic Mice
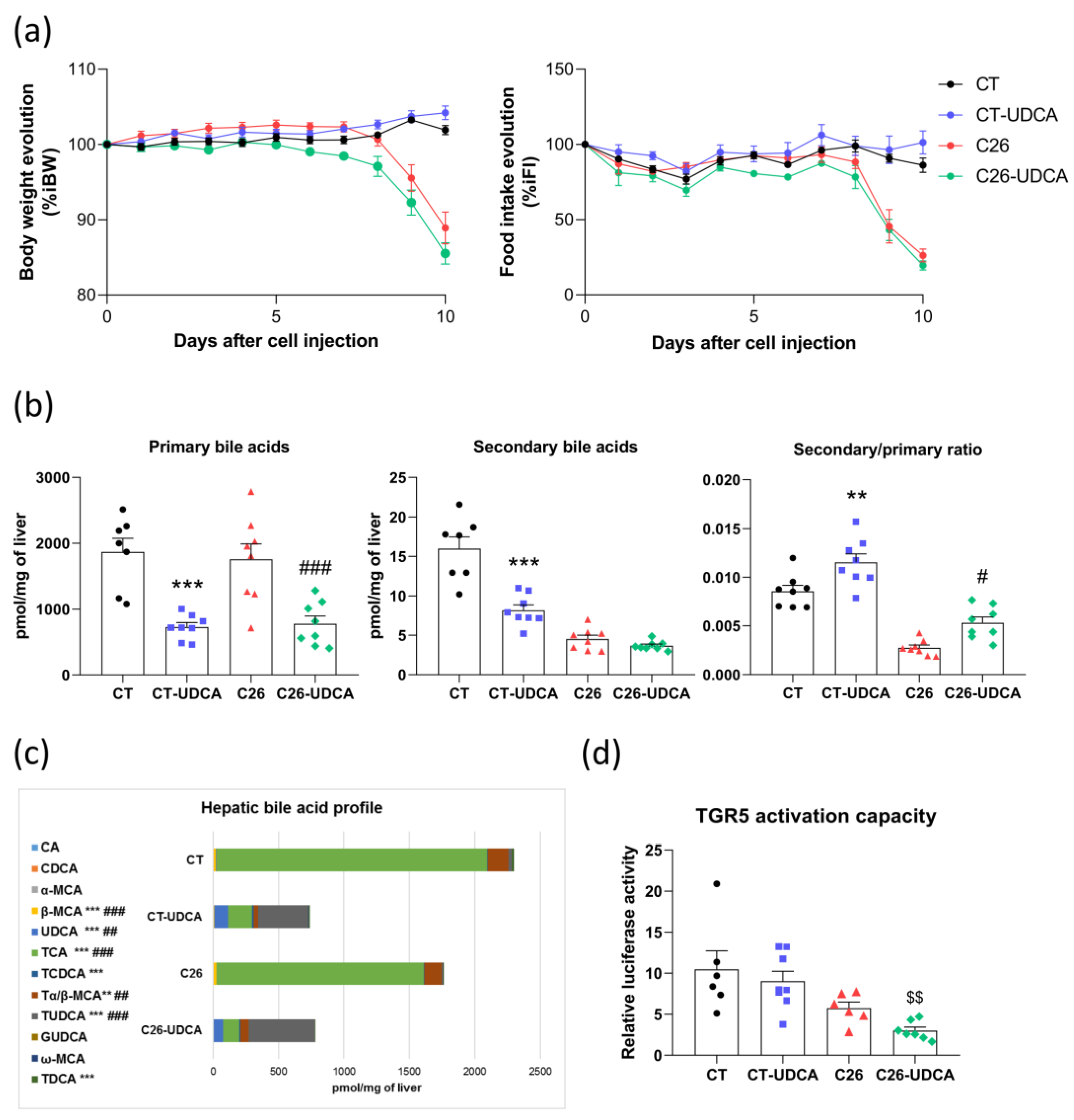
3.4. UDCA Treatment Changes the Bile Acid Profile and Decreases TGR5 Activation Capacity in C26 Cachectic Mice
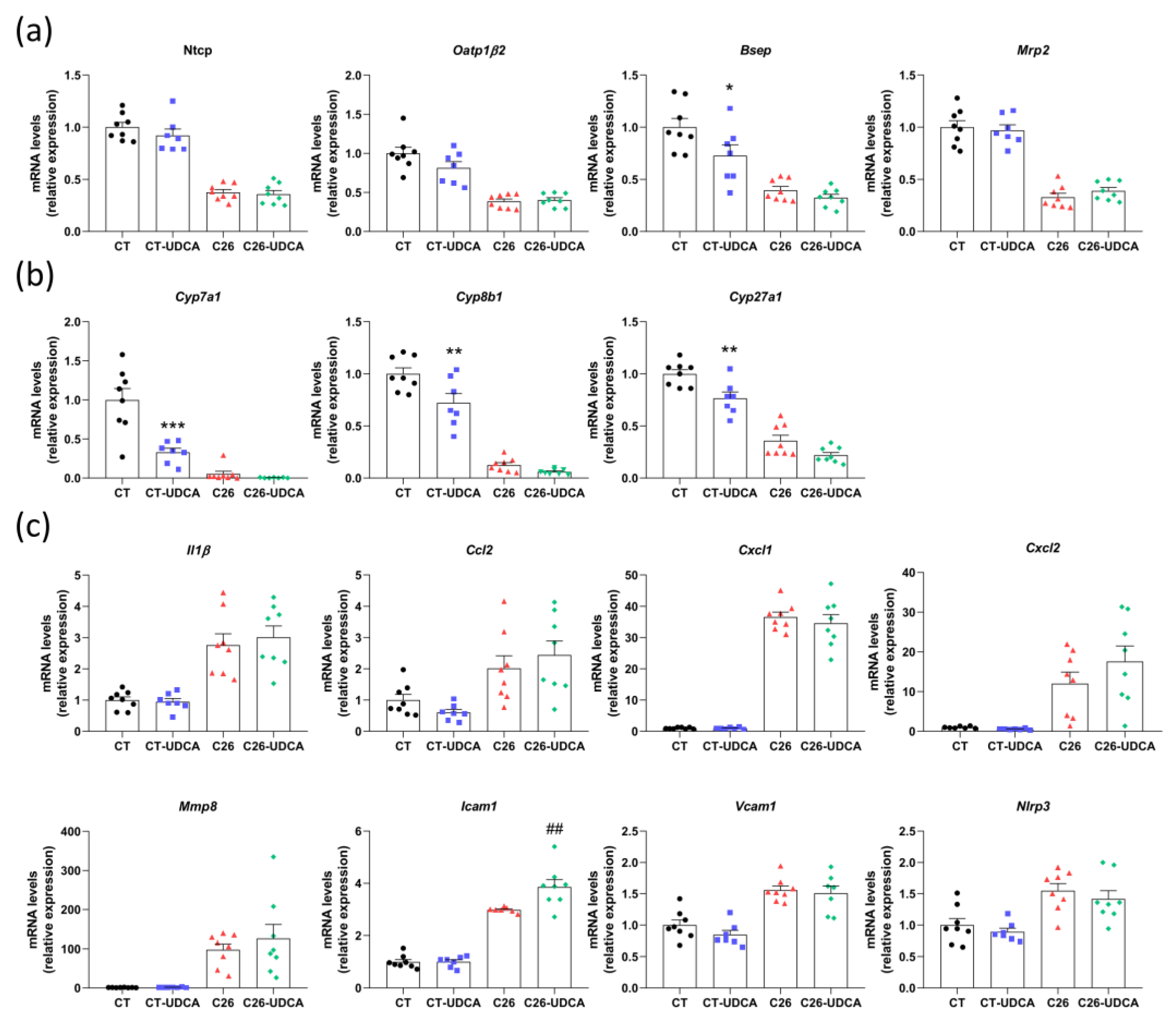
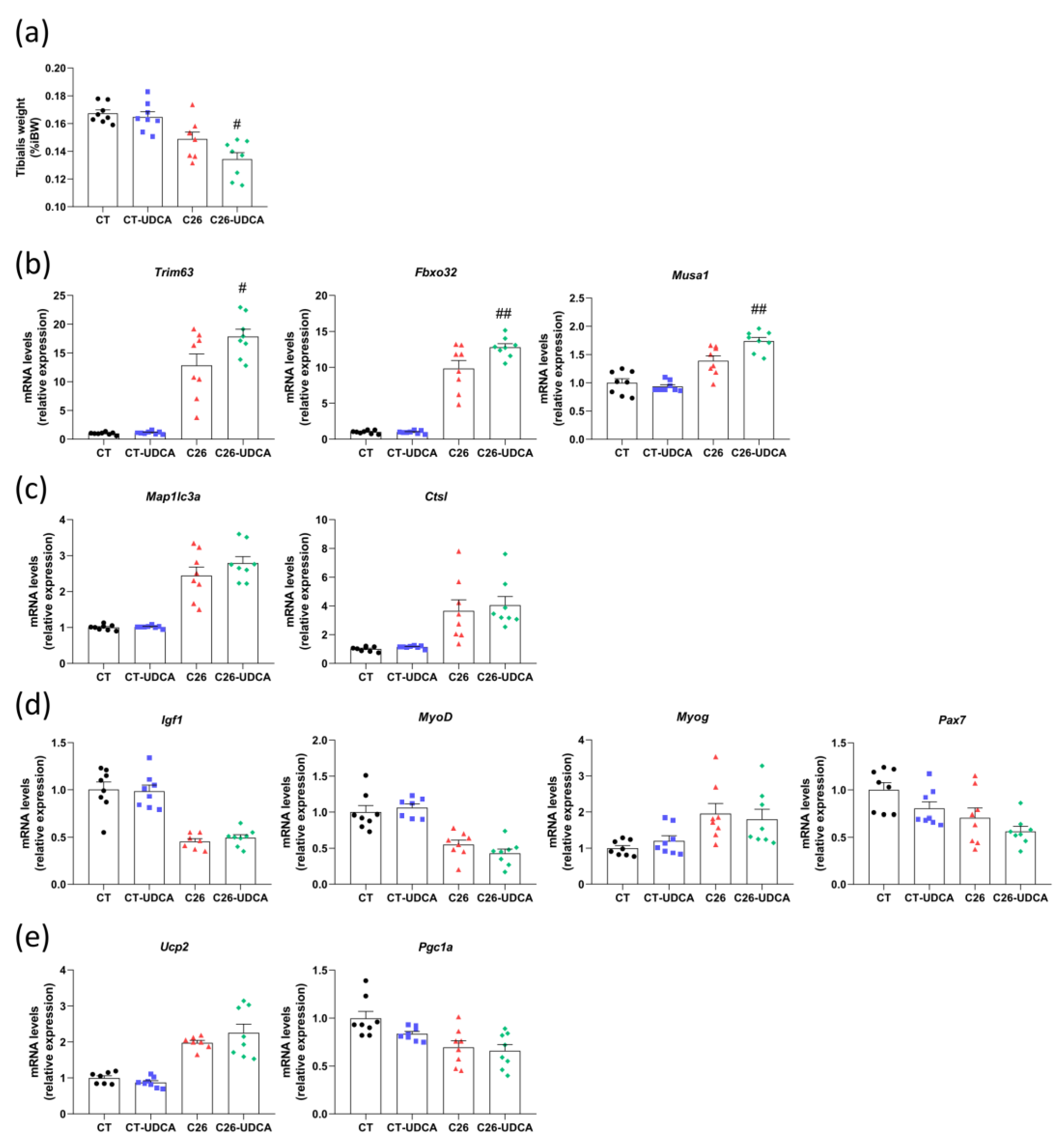
3.5. UDCA Treatment Does Not Improve Hepatic Inflammation and Exacerbates Muscle Atrophy in C26 Cachectic Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baracos, V.E.; Martin, L.; Korc, M.; Guttridge, D.C.; Fearon, K.C.H. Cancer-associated cachexia. Nat. Rev. Dis. Prim. 2018, 4, 17105. [Google Scholar] [CrossRef]
- Fearon, K.; Strasser, F.; Anker, S.D.; Bosaeus, I.; Bruera, E.; Fainsinger, R.L.; Jatoi, A.; Loprinzi, C.; MacDonald, N.; Mantovani, G.; et al. Definition and classification of cancer cachexia: An international consensus. Lancet Oncol. 2011, 12, 489–495. [Google Scholar] [CrossRef]
- Dolly, A.; Dumas, J.; Servais, S. Cancer cachexia and skeletal muscle atrophy in clinical studies: What do we really know? J. Cachexia Sarcopenia Muscle 2020, 11, 1413–1428. [Google Scholar] [CrossRef]
- Petruzzelli, M.; Schweiger, M.; Schreiber, R.; Campos-Olivas, R.; Tsoli, M.; Allen, J.; Swarbrick, M.; Rose-John, S.; Rincon, M.; Robertson, G.; et al. A switch from white to brown fat increases energy expenditure in cancer-associated cachexia. Cell Metab. 2014, 20, 433–447. [Google Scholar] [CrossRef]
- Rohm, M.; Zeigerer, A.; Machado, J.; Herzig, S. Energy metabolism in cachexia. EMBO Rep. 2019, 20, e47258. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.F.; Rohm, M.; Herzig, S.; Diaz, M.B. Cancer Cachexia: More than skeletal muscle wasting. Trends Cancer 2018, 4, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Argiles, J.M.; Stemmler, B.; López-Soriano, F.J.; Busquets, S. Inter-tissue communication in cancer cachexia. Nat. Rev. Endocrinol. 2019, 15, 9–20. [Google Scholar] [CrossRef]
- Farkas, J.; von Haehling, S.; Kalantar-Zadeh, K.; Morley, J.E.; Anker, S.D.; Lainscak, M. Cachexia as a major public health problem: Frequent, costly, and deadly. J. Cachexia Sarcopenia Muscle 2013, 4, 173–178. [Google Scholar] [CrossRef]
- Von Haehling, S.; Anker, M.S.; Anker, S.D. Prevalence and clinical impact of cachexia in chronic illness in Europe, USA, and Japan: Facts and numbers update 2016. J. Cachexia Sarcopenia Muscle 2016, 7, 507–509. [Google Scholar] [CrossRef] [PubMed]
- Guzior, D.V.; Quinn, R.A. Review: Microbial transformations of human bile acids. Microbiome 2021, 9, 1–13. [Google Scholar] [CrossRef]
- Wahlström, A.; Sayin, S.I.; Marschall, H.-U.; Bäckhed, F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016, 24, 41–50. [Google Scholar] [CrossRef]
- Fiorucci, S.; Distrutti, E.; Carino, A.; Zampella, A.; Biagioli, M. Bile acids and their receptors in metabolic disorders. Prog. Lipid Res. 2021, 82, 101094. [Google Scholar] [CrossRef] [PubMed]
- Chávez-Talavera, O.; Tailleux, A.; Lefebvre, P.; Staels, B. Bile acid control of metabolism and inflammation in obesity, type 2 diabetes, dyslipidemia, and nonalcoholic fatty liver disease. Gastroenterology 2017, 152, 1679–1694.e3. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, T.; Miyamoto, Y.; Nakamura, T.; Tamai, Y.; Okada, H.; Sugiyama, E.; Nakamura, T.; Itadani, H.; Tanaka, K. Identification of membrane-type receptor for bile acids (M-BAR). Biochem. Biophys. Res. Commun. 2002, 298, 714–719. [Google Scholar] [CrossRef]
- Fiorucci, S.; Distrutti, E. The pharmacology of bile acids and their receptors. Handb. Exp. Pharmacol. 2019, 256, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Fiorucci, S.; Biagioli, M.; Zampella, A.; Distrutti, E. Bile acids activated receptors regulate innate immunity. Front. Immunol. 2018, 9, 1853. [Google Scholar] [CrossRef]
- Keitel, V.; Stindt, J.; Häussinger, D. Bile acid-activated receptors: GPBAR1 (TGR5) and other G protein-coupled receptors. Transgenic Models Pharmacol. 2019, 256, 19–49. [Google Scholar] [CrossRef]
- Kawamata, Y.; Fujii, R.; Hosoya, M.; Harada, M.; Yoshida, H.; Miwa, M.; Fukusumi, S.; Habata, Y.; Itoh, T.; Shintani, Y.; et al. A G protein-coupled receptor responsive to bile acids. J. Biol. Chem. 2003, 278, 9435–9440. [Google Scholar] [CrossRef]
- Pols, T.W.; Nomura, M.; Harach, T.; Sasso, G.L.; Oosterveer, M.H.; Thomas, C.; Rizzo, G.; Gioiello, A.; Adorini, L.; Pellicciari, R.; et al. TGR5 activation inhibits Atherosclerosis by reducing macrophage inflammation and lipid loading. Cell Metab. 2011, 14, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-D.; Chen, W.-D.; Yu, D.; Forman, B.M.; Huang, W. The G-protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor kappa light-chain enhancer of activated B cells (NF-κB) in mice. Hepatology 2011, 54, 1421–1432. [Google Scholar] [CrossRef]
- Watanabe, M.; Houten, S.; Mataki, C.; Christoffolete, M.; Kim, B.W.; Sato, H.; Messaddeq, N.; Harney, J.W.; Ezaki, O.; Kodama, T.; et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nat. Cell Biol. 2006, 439, 484–489. [Google Scholar] [CrossRef]
- Velazquez-Villegas, L.; Perino, A.; Lemos, V.; Ziętak, M.; Nomura, M.; Pols, T.W.H.; Schoonjans, K. TGR5 signalling promotes mitochondrial fission and beige remodelling of white adipose tissue. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Broeders, E.P.; Nascimento, E.; Havekes, B.; Brans, B.; Roumans, K.H.; Tailleux, A.; Schaart, G.; Kouach, M.; Charton, J.; Deprez, B.; et al. The bile acid chenodeoxycholic acid increases human brown adipose tissue activity. Cell Metab. 2015, 22, 418–426. [Google Scholar] [CrossRef]
- Tsoli, M.; Robertson, G. Cancer cachexia: Malignant inflammation, tumorkines, and metabolic mayhem. Trends Endocrinol. Metab. 2013, 24, 174–183. [Google Scholar] [CrossRef]
- Zimmers, T.A.; Fishel, M.L.; Bonetto, A. STAT3 in the systemic inflammation of cancer cachexia. Semin. Cell Dev. Biol. 2016, 54, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Trauner, M.; Fickert, P.; Stauber, E.R. Inflammation-induced cholestasis. J. Gastroenterol. Hepatol. 1999, 14, 946–959. [Google Scholar] [CrossRef] [PubMed]
- Mulder, J.; Karpen, S.J.; Tietge, U.J.F.; Kuipers, F. Nuclear receptors: Mediators and modifiers of inflammation-induced cholestasis. Front. Biosci. 2009, 14, 2599–2630. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wagner, M.; Zollner, G.; Trauner, M. New molecular insights into the mechanisms of cholestasis. J. Hepatol. 2009, 51, 565–580. [Google Scholar] [CrossRef]
- Thibaut, M.M.; Sboarina, M.; Roumain, M.; Pötgens, S.A.; Neyrinck, A.M.; Destrée, F.; Gillard, J.; Leclercq, I.A.; Dachy, G.; Demoulin, J.; et al. Inflammation-induced cholestasis in cancer cachexia. J. Cachexia Sarcopenia Muscle 2021, 12, 70–90. [Google Scholar] [CrossRef]
- Schwarz, S.; Prokopchuk, O.; Esefeld, K.; Gröschel, S.; Bachmann, J.; Lorenzen, S.; Friess, H.; Halle, M.; Martignoni, M.E. The clinical picture of cachexia: A mosaic of different parameters (experience of 503 patients). BMC Cancer 2017, 17, 130. [Google Scholar] [CrossRef]
- Lazaridis, K.N.; Gores, G.J.; Lindor, K.D. Ursodeoxycholic acid ‘mechanisms of action and clinical use in hepatobiliary disorders’. J. Hepatol. 2001, 35, 134–146. [Google Scholar] [CrossRef]
- Floreani, A.; Mangini, C. Primary biliary cholangitis: Old and novel therapy. Eur. J. Intern. Med. 2018, 47, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Goulis, J.; Leandro, G.; Burroughs, A.K. Randomised controlled trials of ursodeoxycholic-acid therapy for primary biliary cirrhosis: A meta-analysis. Lancet 1999, 354, 1053–1060. [Google Scholar] [CrossRef]
- Vesterhus, M.; Karlsen, T.H. Emerging therapies in primary sclerosing cholangitis: Pathophysiological basis and clinical opportunities. J. Gastroenterol. 2020, 55, 588–614. [Google Scholar] [CrossRef] [PubMed]
- Paumgartner, G. Ursodeoxycholic acid in cholestatic liver disease: Mechanisms of action and therapeutic use revisited. Hepatology 2002, 36, 525–531. [Google Scholar] [CrossRef]
- Bonetto, A.; Rupert, J.E.; Barreto, R.; Zimmers, T.A. The Colon-26 carcinoma tumor-bearing mouse as a model for the study of cancer cachexia. J. Vis. Exp. 2016, e54893. [Google Scholar] [CrossRef]
- Ballarò, R.; Lopalco, P.; Audrito, V.; Beltrà, M.; Pin, F.; Angelini, R.; Costelli, P.; Corcelli, A.; Bonetto, A.; Szeto, H.; et al. Targeting Mitochondria by SS-31 Ameliorates the whole body energy status in cancer- and chemotherapy-induced cachexia. Cancers 2021, 13, 850. [Google Scholar] [CrossRef]
- Massart, I.S.; Paulissen, G.; Loumaye, A.; Lause, P.; Pötgens, S.A.; Thibaut, M.M.; Balan, E.; Deldicque, L.; Atfi, A.; Louis, E.; et al. Marked increased production of acute phase reactants by skeletal muscle during cancer cachexia. Cancers 2020, 12, 3221. [Google Scholar] [CrossRef] [PubMed]
- Santoveña, A.; Sánchez-Negrín, E.; Charola, L.; Llabrés, M.; Fariña, J. Study of quality and stability of ursodeoxycholic acid formulations for oral pediatric administration. Int. J. Pharm. 2014, 477, 32–38. [Google Scholar] [CrossRef]
- Duboc, H.; Taché, Y.; Hofmann, A.F. The bile acid TGR5 membrane receptor: From basic research to clinical application. Dig. Liver Dis. 2014, 46, 302–312. [Google Scholar] [CrossRef]
- Guillemot-Legris, O.; Mutemberezi, V.; Cani, P.; Muccioli, G.G. Obesity is associated with changes in oxysterol metabolism and levels in mice liver, hypothalamus, adipose tissue and plasma. Sci. Rep. 2016, 6, 19694. [Google Scholar] [CrossRef]
- Lazar, C.; Gatto, L.; Ferro, M.; Bruley, C.; Burger, T. Accounting for the multiple natures of missing values in label-free quantitative proteomics data sets to compare imputation strategies. J. Proteome Res. 2016, 15, 1116–1125. [Google Scholar] [CrossRef]
- Woolbright, B.L.; Jaeschke, H. Therapeutic targets for cholestatic liver injury. Expert Opin. Ther. Targets 2016, 20, 463–475. [Google Scholar] [CrossRef]
- Sasaki, T.; Kuboyama, A.; Mita, M.; Murata, S.; Shimizu, M.; Inoue, J.; Mori, K.; Sato, R. The exercise-inducible bile acid receptor Tgr5 improves skeletal muscle function in mice. J. Biol. Chem. 2018, 293, 10322–10332. [Google Scholar] [CrossRef] [PubMed]
- Reddel, C.J.; Allen, J.D.; Ehteda, A.; Taylor, R.; Chen, V.M.Y.; Curnow, J.L.; Kritharides, L.; Robertson, G. Increased thrombin generation in a mouse model of cancer cachexia is partially interleukin-6 dependent. J. Thromb. Haemost. 2017, 15, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Bindels, L.B.; Neyrinck, A.M.; Claus, S.P.; Le Roy, C.I.; Grangette, C.; Pot, B.; Martinez, I.; Walter, J.; Cani, P.D.; Delzenne, N.M. Synbiotic approach restores intestinal homeostasis and prolongs survival in leukaemic mice with cachexia. ISME J. 2015, 10, 1456–1470. [Google Scholar] [CrossRef]
- Bindels, L.B.; Neyrinck, A.M.; Salazar, N.; Taminiau, B.; Druart, C.; Muccioli, G.G.; François, E.; Blecker, C.; Richel, A.; Daube, G.; et al. Non digestible Oligosaccharides modulate the gut microbiota to control the development of leukemia and associated cachexia in mice. PLoS ONE 2015, 10, e0131009. [Google Scholar] [CrossRef]
- Shao, J.-W.; Ge, T.-T.; Chen, S.-Z.; Wang, G.; Yang, Q.; Huang, C.-H.; Xu, L.-C.; Chen, Z. Role of bile acids in liver diseases mediated by the gut microbiome. World J. Gastroenterol. 2021, 27, 3010–3021. [Google Scholar] [CrossRef]
- Zarrabi, K.; Masic, S.; Schaefer, C.; Bartel, M.J.; Kutikov, A.; Zibelman, M. Neoadjuvant checkpoint inhibition in renal cell carcinoma associated Stauffer’s syndrome. Urol. Case Rep. 2019, 29, 101077. [Google Scholar] [CrossRef] [PubMed]
- Koruk, M.; Büyükberber, M.; Savaş, C.; Kadayifçi, A. Paraneoplastic cholestasis associated with prostate carcinoma. Turk. J. Gastroenterol. 2004, 15, 53–55. [Google Scholar] [PubMed]
- Barta, S.K.; Yahalom, J.; Shia, J.; Hamlin, P.A. Idiopathic cholestasis as a paraneoplastic phenomenon in Hodgkin’s Lymphoma. Clin. Lymphoma Myeloma 2006, 7, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Elferink, R.O. Cholestasis. Gut 2003, 52, 42ii–48. [Google Scholar] [CrossRef] [PubMed]
- Esteller, A. Physiology of bile secretion. World J. Gastroenterol. 2008, 14, 5641–5649. [Google Scholar] [CrossRef] [PubMed]
- Boyer, J.L. Bile formation and secretion. Compr. Physiol. 2013, 3, 1035–1078. [Google Scholar] [CrossRef]
- Ghibellini, G.; Leslie, E.; Brouwer, K.L.R. Methods to evaluate biliary excretion of drugs in humans: An updated review. Mol. Pharm. 2006, 3, 198–211. [Google Scholar] [CrossRef]
- Tchambaz, L.; Schlatter, C.; Jakob, M.; Krähenbühl, A.; Wolf, P.; Krähenbühl, S. Dose adaptation of antineoplastic drugs in patients with liver disease. Drug Saf. 2006, 29, 509–522. [Google Scholar] [CrossRef]
- Kurz, A.K.; Graf, D.; Schmitt, M.; Dahl, S.V.; Häussinger, D. Tauroursodesoxycholate-induced choleresis involves p38MAPK activation and translocation of the bile salt export pump in rats. Gastroenterology 2001, 121, 407–419. [Google Scholar] [CrossRef]
- Häussinger, D.; Kurz, A.K.; Wettstein, M.; Graf, D.; Dahl, S.V.; Schliess, F. Involvement of integrins and Src in tauroursodeoxycholate-induced and swelling-induced choleresis. Gastroenterology 2003, 124, 1476–1487. [Google Scholar] [CrossRef]
- Beuers, U.; Bilzer, M.; Chittattu, A.; Kullak-Ublick, G.A.; Keppler, D.; Paumgartner, G.; Dombrowski, F. Tauroursodeoxycholic acid inserts the apical conjugate export pump, Mrp2, into canalicular membranes and stimulates organic anion secretion by protein kinase C–dependent mechanisms in cholestatic rat liver. Hepatology 2001, 33, 1206–1216. [Google Scholar] [CrossRef]
- Wimmer, R.; Hohenester, S.; Pusl, T.; Denk, G.U.; Rust, C.; Beuers, U. Tauroursodeoxycholic acid exerts anticholestatic effects by a cooperative cPKC -/PKA-dependent mechanism in rat liver. Gut 2008, 57, 1448–1454. [Google Scholar] [CrossRef]
- Cruz, L.N.; Guerra, M.T.; Kruglov, E.; Mennone, A.; Garcia, C.R.S.; Chen, J.; Nathanson, M.H. Regulation of multidrug resistance-associated protein 2 by calcium signaling in mouse liver. Hepatology 2010, 52, 327–337. [Google Scholar] [CrossRef]
- Beuers, U.; Trauner, M.; Jansen, P.; Poupon, R. New paradigms in the treatment of hepatic cholestasis: From UDCA to FXR, PXR and beyond. J. Hepatol. 2015, 62, S25–S37. [Google Scholar] [CrossRef] [PubMed]
- Marschall, H.-U.; Wagner, M.; Zollner, G.; Fickert, P.; Diczfalusy, U.; Gumhold, J.; Silbert, D.; Fuchsbichler, A.; Benthin, L.; Grundström, R. Complementary stimulation of hepatobiliary transport and detoxification systems by rifampicin and Ursodeoxycholic Acid in humans. Gastroenterology 2005, 129, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Goossens, J.-F.; Bailly, C. Ursodeoxycholic acid and cancer: From chemoprevention to chemotherapy. Pharmacol. Ther. 2019, 203, 107396. [Google Scholar] [CrossRef] [PubMed]
- Fickert, P.; Zollner, G.; Fuchsbichler, A.; Stumptner, C.; Weiglein, A.H.; Lammert, F.; Marschall, H.-U.; Tsybrovskyy, O.; Zatloukal, K.; Denk, H.; et al. Ursodeoxycholic acid aggravates bile infarcts in bile duct–ligated and Mdr2 knockout mice via disruption of cholangioles. Gastroenterology 2002, 123, 1238–1251. [Google Scholar] [CrossRef]
- Fickert, P.; Wagner, M.; Marschall, H.; Fuchsbichler, A.; Zollner, G.; Tsybrovskyy, O.; Zatloukal, K.; Liu, J.; Waalkes, M.P.; Cover, C.; et al. 24-norUrsodeoxycholic acid is superior to Ursodeoxycholic acid in the treatment of Sclerosing Cholangitis in Mdr2 (Abcb4) knockout mice. Gastroenterology 2006, 130, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Fickert, P.; Pollheimer, M.J.; Silbert, D.; Moustafa, T.; Halilbasic, E.; Krones, E.; Durchschein, F.; Thüringer, A.; Zollner, G.; Denk, H.; et al. Differential effects of norUDCA and UDCA in obstructive cholestasis in mice. J. Hepatol. 2013, 58, 1201–1208. [Google Scholar] [CrossRef] [PubMed]
- Baluyut, A.R.; Sherman, S.; Lehman, G.A.; Hoen, H.; Chalasani, N. Impact of endoscopic therapy on the survival of patients with primary sclerosing cholangitis. Gastrointest. Endosc. 2001, 53, 308–312. [Google Scholar] [CrossRef]
- Rupp, C.; Hippchen, T.; Bruckner, T.; Klöters-Plachky, P.; Schaible, A.; Koschny, R.; Stiehl, A.; Gotthardt, D.N.; Sauer, P. Effect of scheduled endoscopic dilatation of dominant strictures on outcome in patients with primary sclerosing cholangitis. Gut 2019, 68, 2170–2178. [Google Scholar] [CrossRef]
- Keitel, V.; Donner, M.; Winandy, S.; Kubitz, R.; Häussinger, D. Expression and function of the bile acid receptor TGR5 in Kupffer cells. Biochem. Biophys. Res. Commun. 2008, 372, 78–84. [Google Scholar] [CrossRef]
- Guo, C.; Xie, S.; Chi, Z.; Zhang, J.; Liu, Y.; Zhang, L.; Zheng, M.; Zhang, X.; Xia, D.; Ke, Y.; et al. Bile acids control inflammation and metabolic disorder through Inhibition of NLRP3 inflammasome. Immunity 2016, 45, 802–816. [Google Scholar] [CrossRef] [PubMed]
- Perino, A.; Pols, T.W.H.; Nomura, M.; Stein, S.; Pellicciari, R.; Schoonjans, K. TGR5 reduces macrophage migration through mTOR-induced C/EBPβ differential translation. J. Clin. Investig. 2014, 124, 5424–5436. [Google Scholar] [CrossRef] [PubMed]
- McMahan, R.H.; Wang, X.X.; Cheng, L.L.; Krisko, T.; Smith, M.; El Kasmi, K.; Pruzanski, M.; Adorini, L.; Golden-Mason, L.; Levi, M.; et al. Bile acid receptor activation modulates hepatic monocyte activity and improves nonalcoholic fatty liver disease. J. Biol. Chem. 2013, 288, 11761–11770. [Google Scholar] [CrossRef] [PubMed]
- Reich, M.; Deutschmann, K.; Sommerfeld, A.; Klindt, C.; Kluge, S.; Kubitz, R.; Ullmer, C.; Knoefel, W.T.; Herebian, D.; Mayatepek, E.; et al. TGR5 is essential for bile acid-dependent cholangiocyte proliferation in vivo and in vitro. Gut 2016, 65, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Reich, M.; Klindt, C.; Deutschmann, K.; Spomer, L.; Häussinger, D.; Keitel, V. Role of the G protein-coupled bile acid receptor TGR5 in liver damage. Dig. Dis. 2017, 35, 235–240. [Google Scholar] [CrossRef]
- Tschirner, A.; von Haehling, S.; Palus, S.; Doehner, W.; Anker, S.D.; Springer, J. Ursodeoxycholic acid treatment in a rat model of cancer cachexia. J. Cachexia Sarcopenia Muscle 2012, 3, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Abrigo, J.; Gonzalez, F.; Aguirre, F.; Tacchi, F.; González, A.; Meza, M.P.; Simon, F.; Cabrera, D.; Arrese, M.; Karpen, S.; et al. Cholic acid and deoxycholic acid induce skeletal muscle atrophy through a mechanism dependent on TGR5 receptor. J. Cell. Physiol. 2021, 236, 260–272. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thibaut, M.M.; Gillard, J.; Dolly, A.; Roumain, M.; Leclercq, I.A.; Delzenne, N.M.; Muccioli, G.G.; Bindels, L.B. Bile Acid Dysregulation Is Intrinsically Related to Cachexia in Tumor-Bearing Mice. Cancers 2021, 13, 6389. https://doi.org/10.3390/cancers13246389
Thibaut MM, Gillard J, Dolly A, Roumain M, Leclercq IA, Delzenne NM, Muccioli GG, Bindels LB. Bile Acid Dysregulation Is Intrinsically Related to Cachexia in Tumor-Bearing Mice. Cancers. 2021; 13(24):6389. https://doi.org/10.3390/cancers13246389
Chicago/Turabian StyleThibaut, Morgane M., Justine Gillard, Adeline Dolly, Martin Roumain, Isabelle A. Leclercq, Nathalie M. Delzenne, Giulio G. Muccioli, and Laure B. Bindels. 2021. "Bile Acid Dysregulation Is Intrinsically Related to Cachexia in Tumor-Bearing Mice" Cancers 13, no. 24: 6389. https://doi.org/10.3390/cancers13246389
APA StyleThibaut, M. M., Gillard, J., Dolly, A., Roumain, M., Leclercq, I. A., Delzenne, N. M., Muccioli, G. G., & Bindels, L. B. (2021). Bile Acid Dysregulation Is Intrinsically Related to Cachexia in Tumor-Bearing Mice. Cancers, 13(24), 6389. https://doi.org/10.3390/cancers13246389






