Transcriptome Analysis in Vulvar Squamous Cell Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Tumor Tissue
2.2. Subgroup Definition
2.3. RNA Extraction
2.4. Sequencing and Bioinformatic Analysis
2.5. Statistical Analysis
3. Results
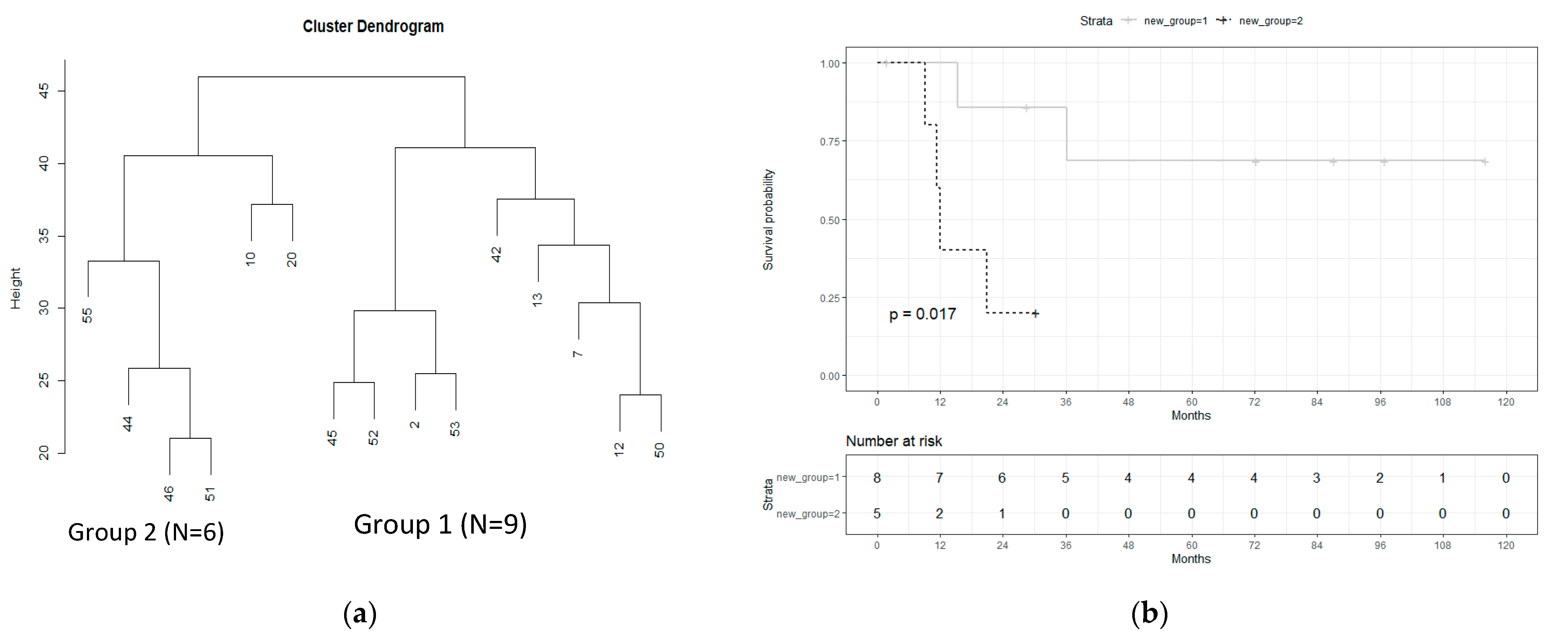
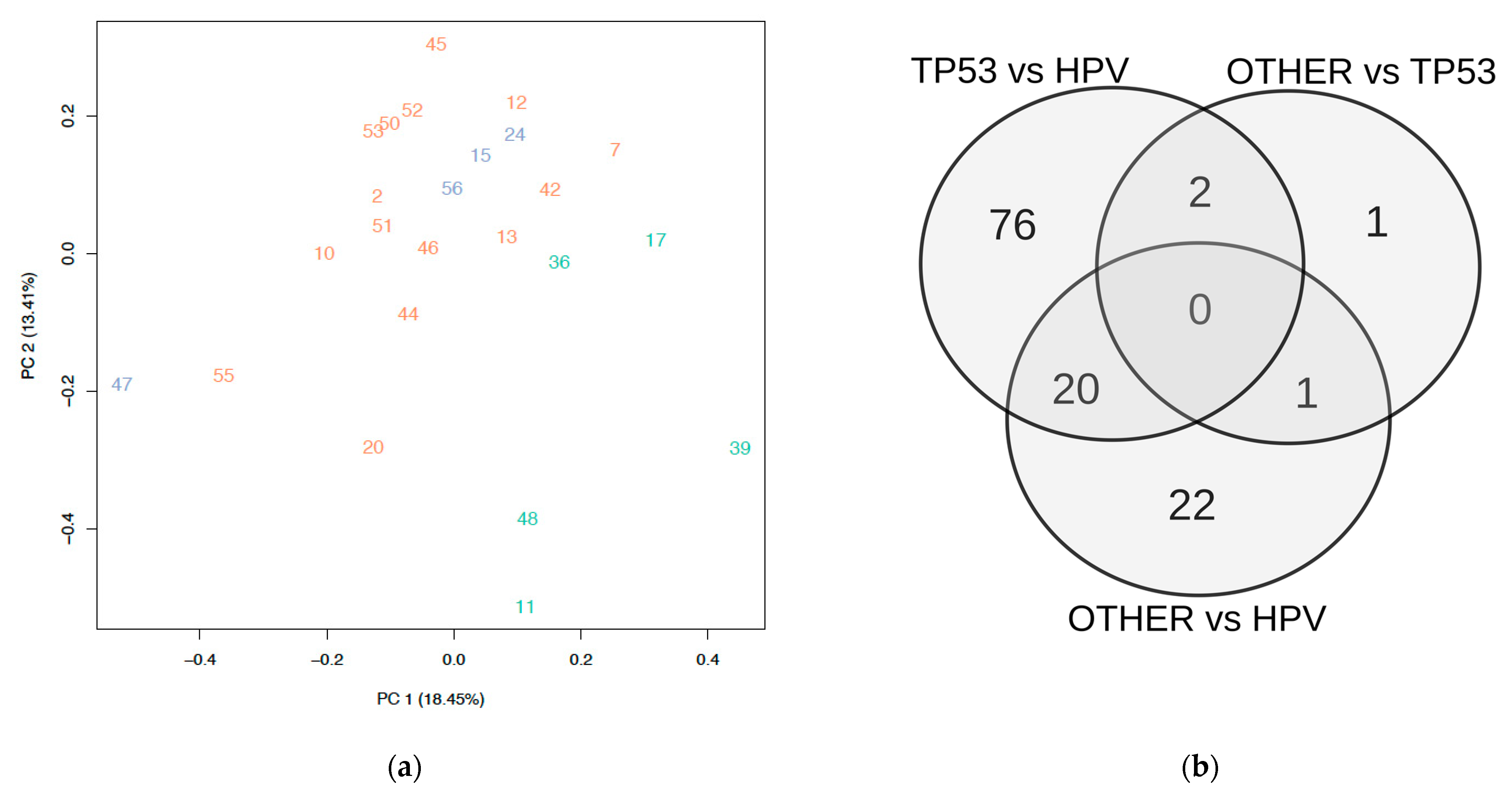
3.1. Subgroup Definition
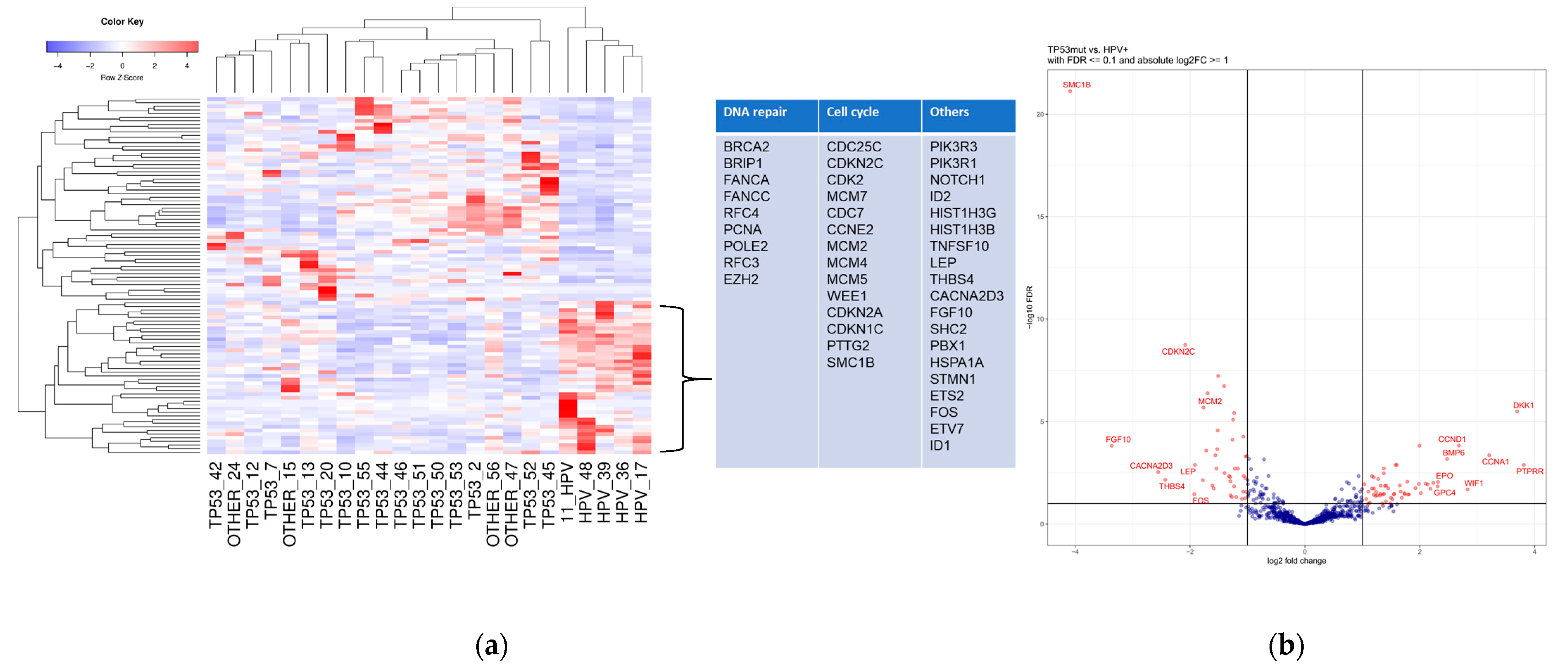
3.2. Differential Expression in Subgroups
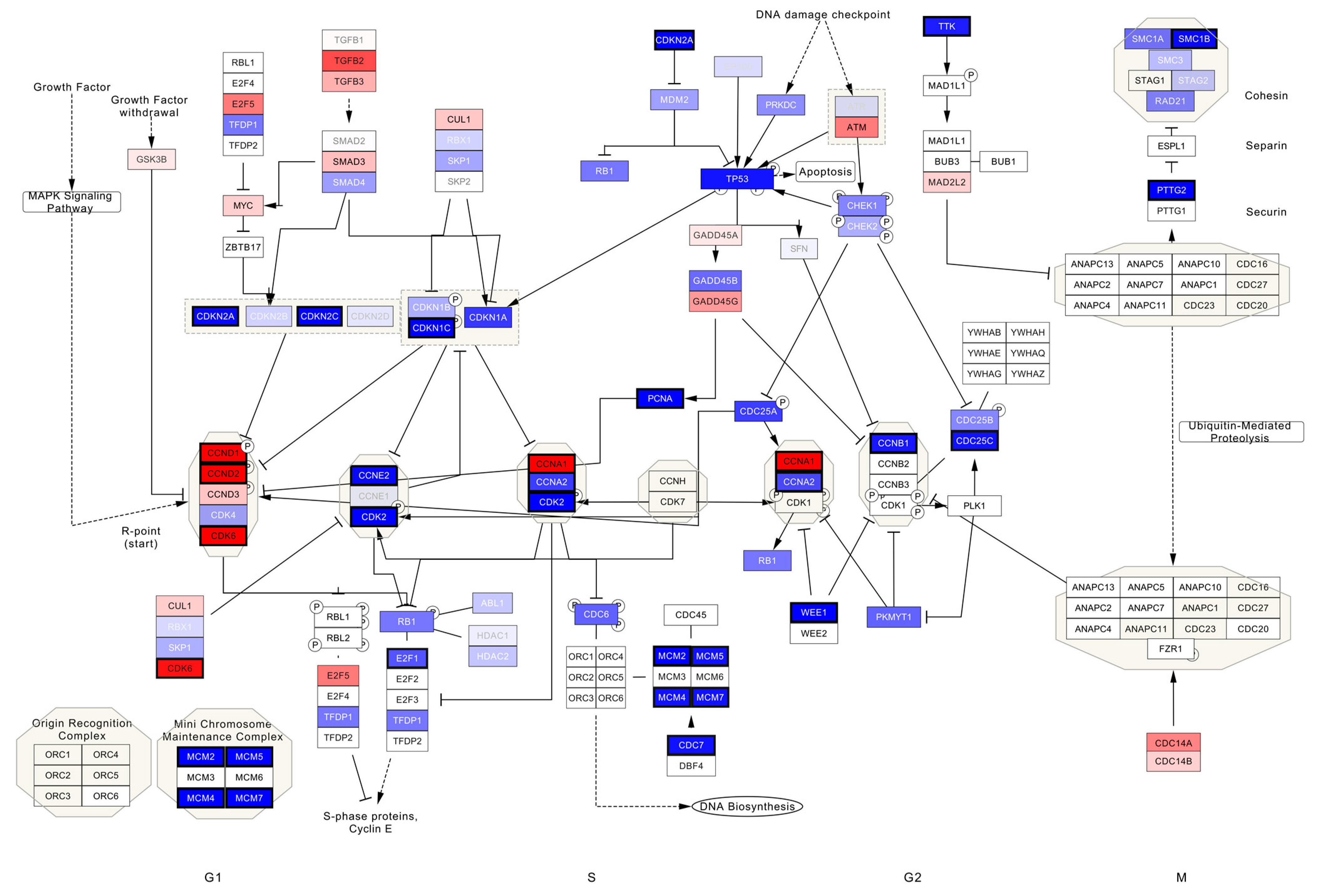
3.2.1. TP53mut VSCC
3.2.2. HPV+ VSCC
3.2.3. “OTHER”
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Buttmann-Schweiger, N.; Klug, S.J.; Luyten, A.; Holleczek, B.; Heitz, F.; Du Bois, A.; Kraywinkel, K. Incidence Patterns and Temporal Trends of Invasive Nonmelanotic Vulvar Tumors in Germany 1999–2011. A Population-Based Cancer Registry Analysis. PLoS ONE 2015, 10, e0128073. [Google Scholar] [CrossRef] [Green Version]
- Holleczek, B.; Sehouli, J.; Barinoff, J. Vulvar cancer in Germany: Increase in incidence and change in tumour biological characteristics from 1974 to 2013. Acta Oncol. 2018, 57, 324–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Martel, C.; Georges, D.; Bray, F.; Ferlay, J.; Clifford, G.M. Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob. Health 2020, 8, e180–e190. [Google Scholar] [CrossRef] [Green Version]
- Woelber, L.; Jaeger, A.; Prieske, K. New treatment standards for vulvar cancer. Curr. Opin. Obstet. Gynecol. 2020, 32, 9–14. [Google Scholar] [CrossRef]
- Prieske, K.; Alawi, M.; Oliveira-Ferrer, L.; Jaeger, A.; Eylmann, K.; Burandt, E.; Schmalfeldt, B.; Joosse, S.A.; Woelber, L. Genomic characterization of vulvar squamous cell carcinoma. Gynecol. Oncol. 2020, 158, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Schuurman, M.; Einden, L.V.D.; Massuger, L.; Kiemeney, L.; van der Aa, M.; de Hullu, J. Trends in incidence and survival of Dutch women with vulvar squamous cell carcinoma. Eur. J. Cancer 2013, 49, 3872–3880. [Google Scholar] [CrossRef]
- Woelber, L.; Prieske, K.; Eulenburg, C.; Oliveira-Ferrer, L.; de Gregorio, N.; Klapdor, R.; Kalder, M.; Braicu, I.; Fuerst, S.; Klar, M.; et al. p53 and p16 expression profiles in vulvar cancer—A translational analysis by the AGO-CaRE- study group. Am. J. Obstet. Gynecol. 2021, 224, 595.e1–595.e11. [Google Scholar] [CrossRef] [PubMed]
- Nooij, L.S.; Ter Haar, N.T.; Ruano, D.; Rakislova, N.; Van Wezel, T.; Smit, V.T.; Trimbos, B.J.; Ordi, J.; Van Poelgeest, M.I.; Bosse, T. Genomic Characterization of Vulvar (Pre)cancers Identifies Distinct Molecular Subtypes with Prognostic Significance. Clin. Cancer Res. 2017, 23, 6781–6789. [Google Scholar] [CrossRef] [Green Version]
- Mahner, S.; Jueckstock, J.; Hilpert, F.; Neuser, P.; Harter, P.; de Gregorio, N.; Hasenburg, A.; Sehouli, J.; Habermann, A.; Hillemanns, P.; et al. Adjuvant Therapy in Lymph Node–Positive Vulvar Cancer: The AGO-CaRE-1 Study. J. Natl. Cancer Inst. 2015, 107, dju426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prieske, K.; Haeringer, N.; Grimm, D.; Trillsch, F.; Eulenburg, C.; Burandt, E.; Schmalfeldt, B.; Mahner, S.; Mueller, V.; Woelber, L. Patterns of distant metastases in vulvar cancer. Gynecol. Oncol. 2016, 142, 427–434. [Google Scholar] [CrossRef]
- Chung, H.; Ros, W.; Delord, J.-P.; Perets, R.; Italiano, A.; Shapira-Frommer, R.; Manzuk, L.; Piha-Paul, S.; Xu, L.; Zeigenfuss, S.; et al. Efficacy and Safety of Pembrolizumab in Previously Treated Advanced Cervical Cancer: Results From the Phase II KEYNOTE-158 Study. J. Clin. Oncol. 2019, 37, 1470–1478. [Google Scholar] [CrossRef]
- Tewari, K.S.; Sill, M.W.; Long, H.J., 3rd; Penson, R.T.; Huang, H.; Ramondetta, L.M.; Landrum, L.M.; Oaknin, A.; Reid, T.J.; Leitao, M.M.; et al. Improved Survival with Bevacizumab in Advanced Cervical Cancer. N. Engl. J. Med. 2014, 370, 734–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabatini, M.E.; Chiocca, S. Human papillomavirus as a driver of head and neck cancers. Br. J. Cancer 2019, 122, 306–314. [Google Scholar] [CrossRef]
- Trietsch, M.D.; Nooij, L.S.; Gaarenstroom, K.; van Poelgeest, M.I. Genetic and epigenetic changes in vulvar squamous cell carcinoma and its precursor lesions: A review of the current literature. Gynecol. Oncol. 2015, 136, 143–157. [Google Scholar] [CrossRef]
- Zięba, S.; Kowalik, A.; Zalewski, K.; Rusetska, N.; Goryca, K.; Piaścik, A.; Misiek, M.; Bakuła-Zalewska, E.; Kopczyński, J.; Kowalski, K.; et al. Somatic mutation profiling of vulvar cancer: Exploring therapeutic targets. Gynecol. Oncol. 2018, 150, 552–561. [Google Scholar] [CrossRef]
- Weberpals, J.I.; Lo, B.; Duciaume, M.M.; Spaans, J.N.; Clancy, A.; Dimitroulakos, J.; Goss, G.D.; Sekhon, H.S. Vulvar Squamous Cell Carcinoma (VSCC) as Two Diseases: HPV Status Identifies Distinct Mutational Profiles Including Oncogenic Fibroblast Growth Factor Receptor. Clin. Cancer Res. 2017, 23, 4501–4510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, E.A.; Werth, A.J.; Sharaf, R.; Montesion, M.; Sokol, E.S.; Pavlick, D.C.; McLaughlin-Drubin, M.; Erlich, R.; Toma, H.; Williams, K.J.; et al. Vulvar Squamous Cell Carcinoma: Comprehensive Genomic Profiling of HPV+ Versus HPV– Forms Reveals Distinct Sets of Potentially Actionable Molecular Targets. JCO Precis. Oncol. 2020, 4, 647–661. [Google Scholar] [CrossRef]
- Han, M.-R.; Shin, S.; Park, H.-C.; Kim, M.S.; Lee, S.H.; Jung, S.H.; Song, S.Y.; Lee, S.H.; Chung, Y.-J. Mutational signatures and chromosome alteration profiles of squamous cell carcinomas of the vulva. Exp. Mol. Med. 2018, 50, e442. [Google Scholar] [CrossRef]
- Dasgupta, S.; Ewing-Graham, P.; Swagemakers, S.; Bosch, T.V.D.; Atmodimedjo, P.; Verbiest, M.; de Haan, M.; van Doorn, H.; van der Spek, P.; Koljenović, S.; et al. Exploring Differentially Methylated Genes in Vulvar Squamous Cell Carcinoma. Cancers 2021, 13, 3580. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Cancer Genome Atlas Research Network. Integrated genomic and molecular characterization of cervical cancer. Nature 2017, 543, 378–384. [Google Scholar] [CrossRef]
- Pappa, K.I.; Jacob-Hirsch, J.; Vlachos, G.D.; Christodoulou, I.; Partsinevelos, G.; Amariglio, N.; Markaki, S.; Antsaklis, A.; Anagnou, N.P. Expression Profiling of Vulvar Carcinoma: Clues for Deranged Extracellular Matrix Remodeling and Effects on Multiple Signaling Pathways Combined with Discrete Patient Subsets. Transl. Oncol. 2011, 4, 301-IN6. [Google Scholar] [CrossRef] [Green Version]
- Kolitz, E.; Lucas, E.; Hosler, G.A.; Kim, J.; Hammer, S.; Lewis, C.; Xu, L.; Day, A.T.; Mauskar, M.; Lea, J.S.; et al. Human Papillomavirus-Positive and -Negative Vulvar Squamous Cell Carcinoma Are Biologically but Not Clinically Distinct. J. Investig. Dermatol. 2021. [Google Scholar] [CrossRef]
- Beller, U.; Quinn, M.A.; Benedet, J.L.; Creasman, W.T.; Ngan, H.Y.S.; Maisonneuve, P.; Pecorelli, S.; Odicino, F.; Heintz, A. Carcinoma of the Vulva. Int. J. Gynecol. Obstet. 2006, 95, S7–S27. [Google Scholar] [CrossRef]
- Benedet, J.L.; Bender, H.; Jones, H., 3rd; Ngan, H.Y.; Pecorelli, S. FIGO staging classifications and clinical practice guidelines in the management of gynecologic cancers. FIGO Committee on Gynecologic Oncology. Int. J. Gynaecol. Obstet. 2000, 70, 209–262. [Google Scholar]
- Risso, D.; Ngai, J.; Speed, T.P.; Dudoit, S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014, 32, 896–902. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, A.; Hamilton, A.M.; Furberg, H.; Pietzak, E.; Purdue, M.P.; Troester, M.A.; Hoadley, K.A.; Love, M.I. An approach for normalization and quality control for NanoString RNA expression data. Brief. Bioinform. 2021, 22, bbaa163. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Lein, E.S.; Hawrylycz, M.J.; Ao, N.; Ayres, M.; Bensinger, A.; Bernard, A.; Boe, A.F.; Boguski, M.S.; Brockway, K.S.; Byrnes, E.J.; et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 2007, 445, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Joosse, S.A. In-Silico Online (Version 2.1.2). 2019. Available online: http://in-silico.online (accessed on 16 March 2021).
- Weiss, D.; Basel, T.; Sachse, F.; Braeuninger, A.; Rudack, C. Promoter methylation of cyclin A1 is associated with human papillomavirus 16 induced head and neck squamous cell carcinoma independently of p53 mutation. Mol. Carcinog. 2011, 50, 680–688. [Google Scholar] [CrossRef] [PubMed]
- Chalertpet, K.; Pakdeechaidan, W.; Patel, V.; Mutirangura, A.; Yanatatsaneejit, P. Human papillomavirus type 16 E7 oncoprotein mediates CCNA1 promoter methylation. Cancer Sci. 2015, 106, 1333–1340. [Google Scholar] [CrossRef] [Green Version]
- Malumbres, M.; Barbacid, M. To cycle or not to cycle: A critical decision in cancer. Nat. Rev. Cancer 2001, 1, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Musgrove, E.A.; Caldon, C.E.; Barraclough, J.; Stone, A.; Sutherland, R.L. Cyclin D as a therapeutic target in cancer. Nat. Rev. Cancer 2011, 11, 558–572. [Google Scholar] [CrossRef] [PubMed]
- Saâda-Bouzid, E.; Defaucheux, C.; Karabajakian, A.; Coloma, V.P.; Servois, V.; Paoletti, X.; Even, C.; Fayette, J.; Guigay, J.; Loirat, D.; et al. Hyperprogression during anti-PD-1/PD-L1 therapy in patients with recurrent and/or metastatic head and neck squamous cell carcinoma. Ann. Oncol. 2017, 28, 1605–1611. [Google Scholar] [CrossRef]
- Yu, J.; Yan, J.; Guo, Q.; Chi, Z.; Tang, B.; Zheng, B.; Yu, J.; Yin, T.; Cheng, Z.; Wu, X.; et al. Genetic Aberrations in the CDK4 Pathway Are Associated with Innate Resistance to PD-1 Blockade in Chinese Patients with Non-Cutaneous Melanoma. Clin. Cancer Res. 2019, 25, 6511–6523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Huang, Y.; Gao, X.; Li, Y.; Lin, J.; Chen, L.; Chang, L.; Chen, G.; Guan, Y.; Pan, L.K.; et al. CCND1 Amplification Contributes to Immunosuppression and Is Associated With a Poor Prognosis to Immune Checkpoint Inhibitors in Solid Tumors. Front. Immunol. 2020, 11, 1620. [Google Scholar] [CrossRef]
- Choschzick, M.; Hess, S.; Tennstedt, P.; Holst, F.; Bohlken, H.; Gieseking, F.; Mahner, S.; Woelber, L.; Simon, R.; Sauter, G. Role of cyclin D1 amplification and expression in vulvar carcinomas. Hum. Pathol. 2012, 43, 1386–1393. [Google Scholar] [CrossRef] [PubMed]
- Nooij, L.S.; Dreef, E.J.; Smit, V.T.; van Poelgeest, M.I.; Bosse, T. Stathmin is a highly sensitive and specific biomarker for vulvar high-grade squamous intraepithelial lesions. J. Clin. Pathol. 2016, 69, 1070–1075. [Google Scholar] [CrossRef]
- Thorpe, L.M.; Spangle, J.M.; Ohlson, C.E.; Cheng, H.; Roberts, T.M.; Cantley, L.C.; Zhao, J.J. PI3K-p110α mediates the oncogenic activity induced by loss of the novel tumor suppressor PI3K-p85α. Proc. Natl. Acad. Sci. USA 2017, 114, 7095–7100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeimet, A.G. Molecular characterization of vulvar squamous cell cancer: High time to gain ground. Gynecol. Oncol. 2020, 158, 519–520. [Google Scholar] [CrossRef] [PubMed]
- Moore, G.; Annett, S.; McClements, L.; Robson, T. Top Notch Targeting Strategies in Cancer: A Detailed Overview of Recent Insights and Current Perspectives. Cells 2020, 9, 1503. [Google Scholar] [CrossRef]
- Leonard, S.; Pereira, M.; Fox, R.; Gordon, N.; Yap, J.; Kehoe, S.; Luesley, D.; Woodman, C.; Ganesan, R. Over-expression of DNMT3A predicts the risk of recurrent vulvar squamous cell carcinomas. Gynecol. Oncol. 2016, 143, 414–420. [Google Scholar] [CrossRef] [Green Version]
- Chen, B.-F.; Chan, W.-Y. The de novo DNA methyltransferase DNMT3A in development and cancer. Epigenetics 2014, 9, 669–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, H.Y.; Chen, Z.; Wang, L.; Zhang, Z.-K.; Tan, X.; Liu, S.; Zhang, B.-T.; Lu, A.; Yu, Y.; Zhang, G. Dickkopf-1: A Promising Target for Cancer Immunotherapy. Front. Immunol. 2021, 12, 658097. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.-J.; Hsu, T.-W.; Wu, S.-R.; Lan, S.-W.; Hsiao, T.-F.; Lin, H.-Y.; Tu, H.-F.; Lee, C.-F.; Huang, C.-C.; Chen, M.-J.M.; et al. Inhibition of TMPRSS2 by HAI-2 reduces prostate cancer cell invasion and metastasis. Oncogene 2020, 39, 5950–5963. [Google Scholar] [CrossRef]
- Zhang, L.; Qiang, J.; Yang, X.; Wang, D.; Rehman, A.U.; He, X.; Chen, W.; Sheng, D.; Zhou, L.; Jiang, Y.; et al. IL1R2 Blockade Suppresses Breast Tumorigenesis and Progression by Impairing USP15-Dependent BMI1 Stability. Adv. Sci. 2019, 7, 1901728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luiken, S.; Fraas, A.; Bieg, M.; Sugiyanto, R.; Goeppert, B.; Singer, S.; Ploeger, C.; Warsow, G.; Marquardt, J.U.; Sticht, C.; et al. NOTCH target gene HES5 mediates oncogenic and tumor suppressive functions in hepatocarcinogenesis. Oncogene 2020, 39, 3128–3144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Characteristics | HPV+ N = 5 | HPV+ % | TP53mut N = 15 | TP53mut % |
|---|---|---|---|---|
| Age at FD (yrs) median | 63 | n.a. | 67 | n.a. |
| (range) | (40–81) | (38–84) | ||
| Tumor stage | ||||
| pT1b | 1 | 20 | 3 | 20 |
| pT2 | 4 | 80 | 8 | 53.3 |
| pT3 | 0 | 0 | 1 | 6.7 |
| pT4 | 0 | 0 | 1 | 6.7 |
| unknown | 0 | 0 | 2 | 13.3 |
| Nodal status | ||||
| unilateral groin metastases | 1 | 20 | 1 | 6.7 |
| bilateral groin metastases | 0 | 0 | 1 | 6.7 |
| no groin metastases | 4 | 80 | 10 | 66.7 |
| unknown | 0 | 0 | 3 | 20 |
| Median tumor diameter | ||||
| mm | 49.5 | n.a. | 50 | n.a. |
| (range) | (45–50) | (15–150) | ||
| Median depth of invasion | n.a. | n.a. | ||
| mm | 10 | 7 | ||
| (range) | (8–10) | (5–28) | ||
| Resection status of vulvar primary | ||||
| R0 | 3 | 60 | 11 | 73.3 |
| R1 | 1 | 20 | 2 | 13.3 |
| unknown | 1 | 20 | 2 | 13.3 |
| Grading | ||||
| G1 | 1 | 20 | 1 | 6.7 |
| G2 | 2 | 40 | 5 | 33.3 |
| G3 | 2 | 40 | 6 | 40 |
| unknown | 0 | 0 | 3 | 20 |
| Vulvar surgery | ||||
| Vulvectomy | 3 | 60 | 7 | 46.7 |
| Radical local Excision | 2 | 40 | 6 | 40 |
| No local surgery | 0 | 0 | 1 | 6.7 |
| unknown | 0 | 0 | 1 | 6.7 |
| Groin Surgery | ||||
| Full Groin dissection (unilateral) | 1 | 20 | 2 | 13.3 |
| Full Groin dissection (bilateral) | 4 | 80 | 5 | 33.3 |
| SNL only (bilateral) | 0 | 0 | 3 | 20 |
| SLN+ followed by bilateral groin dissection | 0 | 0 | 0 | 0 |
| No LNE | 0 | 0 | 4 | 26.7 |
| Unknown | 0 | 0 | 1 | 6.7 |
| Pelvic node dissection | 1 | 20 | 1 | 6.7 |
| Adjuvant therapy | ||||
| Radiotherapy | 2 | 40 | 3 | 20 |
| Chemoradiation | 1 | 20 | 0 | 0 |
| Laser | 0 | 0 | 0 | 0 |
| no adjuvant therapy | 2 | 40 | 11 | 73.3 |
| unknown | 0 | 0 | 1 | 6.7 |
| Radiation field | ||||
| Vulva ±.groin ±.pelvis | 3 | 60 | 2 | 13.3 |
| Groin ±.pelvis only | 0 | 0 | 1 | 6.7 |
| unknown | 0 | 0 | 0 | 0 |
| Recurrence | 1 | 20 | 3 | 20 |
| Localization of recurrence (multiples possible) | ||||
| Vulva | 1 | 20 | 3 | 20 |
| Groins | 1 | 20 | 2 | 13.3 |
| Recurrence free | 4 | 80 | 10 | 66.7 |
| Distant metastasis in course of disease | 1 | 20 | 2 | 13.3 |
| Gene | Log2FC | Base Mean | Gene Name |
|---|---|---|---|
| TMPRSS2 | 4.15 | 156.3 | Transmembrane protease serine subtype 2 |
| PLA2G3 | 2.66 | 136.03 | Phospholipase A2 Group III |
| IL20RA | 2.49 | 84.09 | Interleukin 20 Receptor Subunit Alpha |
| RASAL1 | 2.45 | 156.73 | RAS Protein Activator Like 1 |
| IL1R2 | 2.18 | 526.73 | Interleukin 1 Receptor Type 2 |
| PRMT8 | 2.11 | 42.75 | Protein Arginine Methyltransferase 8 |
| MAPK8IP2 | 2.04 | 64.02 | Mitogen-Activated Protein Kinase 8 Interacting Protein 2 |
| PLA2G4A | 1.95 | 127.75 | Phospholipase A2 Group IVA) |
| HES5 | 1.58 | 125.22 | Hes Family BHLH Transcription Factor 5 |
| SYK | 1.40 | 922.43 | Spleen Associated Tyrosine Kinase |
| PTPRR | −3.05 | 55.63 | Protein Tyrosine Phosphatase Receptor Type R |
| TSLP | −2.59 | 78.84 | Thymic stromal lymphopoietin |
| GPC4 | −2.25 | 269.24 | Glypican 4 |
| IGFBP3 | −2.20 | 5798.71 | Insulin-like growth factor-binding protein 3 |
| PRLR | −2.08 | 67.63 | Prolactin Receptor |
| VEGFC | −2.07 | 630.41 | Vascular endothelial growth factor C |
| FN1 | −1.9 | 6497.01 | Fibronectin 1 |
| RET | −1.67 | 75.33 | Rezeptor-Tyrosinkinase |
| DUSP10 | −1.25 | 543.2 | Dual specificity protein phosphatase |
| ITGB3 | −1.1 | 82.5 | (Integrin Subunit Beta 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prieske, K.; Alawi, M.; Jaeger, A.; Wankner, M.C.; Eylmann, K.; Reuter, S.; Lebok, P.; Burandt, E.; Blessin, N.C.; Schmalfeldt, B.; et al. Transcriptome Analysis in Vulvar Squamous Cell Cancer. Cancers 2021, 13, 6372. https://doi.org/10.3390/cancers13246372
Prieske K, Alawi M, Jaeger A, Wankner MC, Eylmann K, Reuter S, Lebok P, Burandt E, Blessin NC, Schmalfeldt B, et al. Transcriptome Analysis in Vulvar Squamous Cell Cancer. Cancers. 2021; 13(24):6372. https://doi.org/10.3390/cancers13246372
Chicago/Turabian StylePrieske, Katharina, Malik Alawi, Anna Jaeger, Maximilian Christian Wankner, Kathrin Eylmann, Susanne Reuter, Patrick Lebok, Eike Burandt, Niclas C. Blessin, Barbara Schmalfeldt, and et al. 2021. "Transcriptome Analysis in Vulvar Squamous Cell Cancer" Cancers 13, no. 24: 6372. https://doi.org/10.3390/cancers13246372
APA StylePrieske, K., Alawi, M., Jaeger, A., Wankner, M. C., Eylmann, K., Reuter, S., Lebok, P., Burandt, E., Blessin, N. C., Schmalfeldt, B., Oliveira-Ferrer, L., Joosse, S. A., & Woelber, L. (2021). Transcriptome Analysis in Vulvar Squamous Cell Cancer. Cancers, 13(24), 6372. https://doi.org/10.3390/cancers13246372






