Role of MicroRNAs in the Development and Progression of the Four Medulloblastoma Subgroups
Abstract
:Simple Summary
Abstract
1. Introduction
2. Medulloblastoma’s Classification
- - Classic, that is the most common subtype;
- - Desmoplastic/nodular;
- - Extensive nodularity, that is predominantly in infants;
- - Anaplastic/large cell.
Diagnosis, Prognosis and Therapy
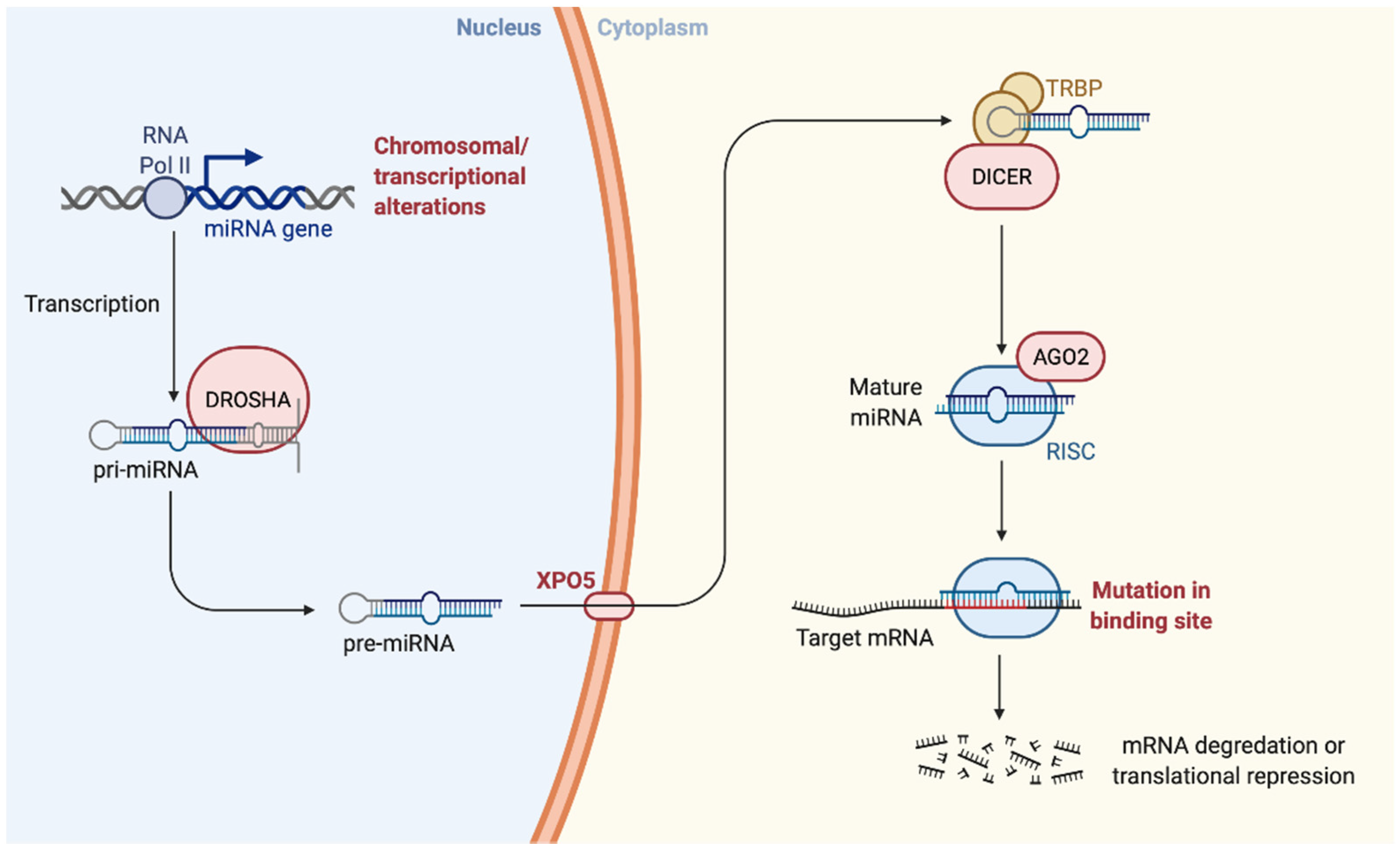
3. miRNAs
3.1. Role of miRNAs in Neuronal Development
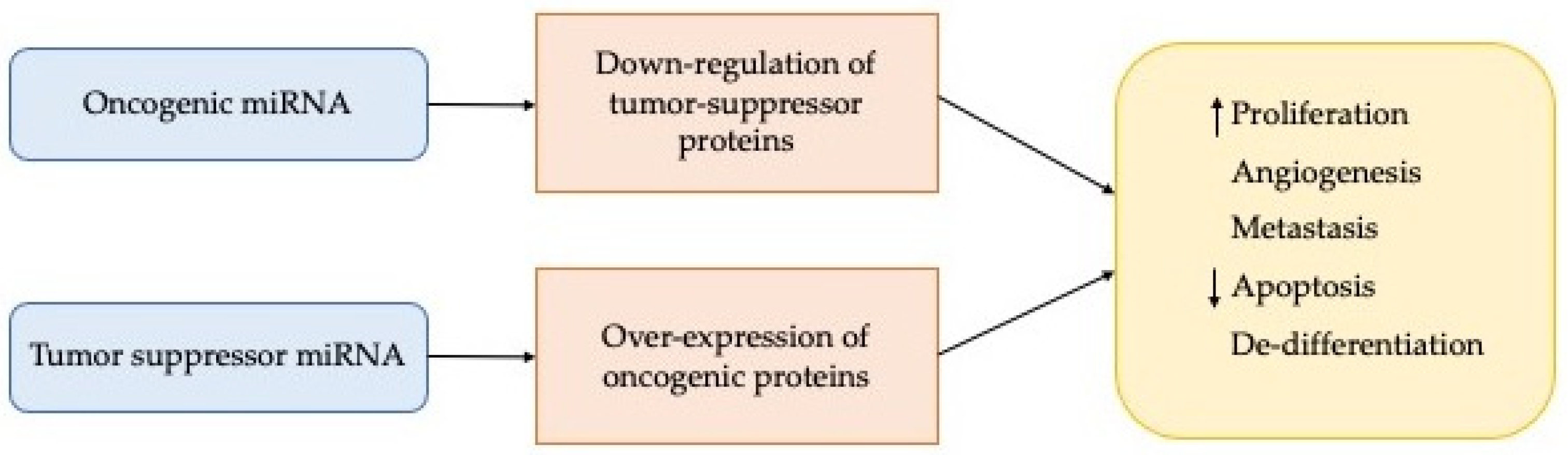
3.2. Role of miRNAs in Cancer
4. miRNAs Involvement in the Different Subgroups of Medulloblastoma
4.1. Wingless (WNT) Subgroup
4.2. Sonic Hedgehog (SHH) Subgroup
4.3. Group 3 (Grp3) Subgroup
4.4. Group 4 (Grp4) Subgroup
5. Role of miRNAs in Medulloblastoma
Clinical Application of miRNAs in Medulloblastoma
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Packer, R.J.; Vezina, G. Management of and prognosis with medulloblastoma: Therapy at a crossroads. Arch. Neurol. 2008, 65, 1419–1424. [Google Scholar] [CrossRef] [PubMed]
- Bondy, M.L.; Scheurer, M.E.; Malmer, B.; Barnholtz-Sloan, J.S.; Davis, F.G.; Il’yasova, D.; Kruchko, C.; McCarthy, B.J.; Rajaraman, P.; Schwartzbaum, J.A.; et al. Brain tumor epidemiology: Consensus from the Brain Tumor Epidemiology Consortium. Cancer 2008, 113, 1953–1968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Group, A.W.; Busco, S.; Buzzoni, C.; Mallone, S.; Trama, A.; Castaing, M.; Bella, F.; Amodio, R.; Bizzoco, S.; Cassetti, T.; et al. Italian cancer figures—Report 2015: The burden of rare cancers in Italy. Epidemiol. Prev. 2016, 40, 1–120. [Google Scholar] [CrossRef]
- Northcott, P.A.; Robinson, G.W.; Kratz, C.P.; Mabbott, D.J.; Pomeroy, S.L.; Clifford, S.C.; Rutkowski, S.; Ellison, D.W.; Malkin, D.; Taylor, M.D.; et al. Medulloblastoma. Nat. Rev. Dis. Primers 2019, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Tailor, J.; Zhen, Q.; Gillmor, A.H.; Miller, M.L.; Weishaupt, H.; Chen, J.; Zheng, T.; Nash, E.K.; McHenry, L.K.; et al. Engineering Genetic Predisposition in Human Neuroepithelial Stem Cells Recapitulates Medulloblastoma Tumorigenesis. Cell Stem Cell 2019, 25, 433–446.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinlan, A.; Rizzolo, D. Understanding medulloblastoma. J. Am. Acad. Phys. Assist. 2017, 30, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Wells, E.M.; Packer, R.J. Pediatric brain tumors. Continuum 2015, 21, 373–396. [Google Scholar] [CrossRef] [PubMed]
- Waszak, S.M.; Northcott, P.A.; Buchhalter, I.; Robinson, G.W.; Sutter, C.; Groebner, S.; Grund, K.B.; Brugieres, L.; Jones, D.T.W.; Pajtler, K.W.; et al. Spectrum and prevalence of genetic predisposition in medulloblastoma: A retrospective genetic study and prospective validation in a clinical trial cohort. Lancet Oncol. 2018, 19, 785–798. [Google Scholar] [CrossRef]
- Kumar, V.; Kumar, V.; McGuire, T.; Coulter, D.W.; Sharp, J.G.; Mahato, R.I. Challenges and Recent Advances in Medulloblastoma Therapy. Trends Pharmacol. Sci. 2017, 38, 1061–1084. [Google Scholar] [CrossRef] [PubMed]
- Buruiana, A.; Florian, S.I.; Florian, A.I.; Timis, T.L.; Mihu, C.M.; Miclaus, M.; Osan, S.; Hrapsa, I.; Cataniciu, R.C.; Farcas, M.; et al. The Roles of miRNA in Glioblastoma Tumor Cell Communication: Diplomatic and Aggressive Negotiations. Int. J. Mol. Sci. 2020, 21, 1950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Archer, T.C.; Mahoney, E.L.; Pomeroy, S.L. Medulloblastoma: Molecular Classification-Based Personal Therapeutics. Neurotherapeutics 2017, 14, 265–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavalli, F.M.G.; Remke, M.; Rampasek, L.; Peacock, J.; Shih, D.J.H.; Luu, B.; Garzia, L.; Torchia, J.; Nor, C.; Morrissy, A.S.; et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 2017, 31, 737–754.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laneve, P.; Caffarelli, E. The Non-coding Side of Medulloblastoma. Front. Cell Dev. Biol. 2020, 8, 275. [Google Scholar] [CrossRef] [PubMed]
- Orr, B.A. Pathology, diagnostics, and classification of medulloblastoma. Brain Pathol. 2020, 30, 664–678. [Google Scholar] [CrossRef] [PubMed]
- Garzia, L.; Kijima, N.; Morrissy, A.S.; De Antonellis, P.; Guerreiro-Stucklin, A.; Holgado, B.L.; Wu, X.; Wang, X.; Parsons, M.; Zayne, K.; et al. A Hematogenous Route for Medulloblastoma Leptomeningeal Metastases. Cell 2018, 172, 1050–1062.e14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khatua, S.; Song, A.; Citla Sridhar, D.; Mack, S.C. Childhood Medulloblastoma: Current Therapies, Emerging Molecular Landscape and Newer Therapeutic Insights. Curr. Neuropharmacol. 2018, 16, 1045–1058. [Google Scholar] [CrossRef] [PubMed]
- Gottardo, N.G.; Gajjar, A. Current therapy for medulloblastoma. Curr. Treat. Options Neurol. 2006, 8, 319–334. [Google Scholar] [CrossRef] [PubMed]
- Frisira, E.; Rashid, F.; Varma, S.N.; Badodi, S.; Benjamin-Ombo, V.A.; Michod, D.; Niklison-Chirou, M.V. NPI-0052 and gamma-radiation induce a synergistic apoptotic effect in medulloblastoma. Cell Death Dis. 2019, 10, 785. [Google Scholar] [CrossRef]
- Ashley, D.M.; Merchant, T.E.; Strother, D.; Zhou, T.; Duffner, P.; Burger, P.C.; Miller, D.C.; Lyon, N.; Bonner, M.J.; Msall, M.; et al. Induction chemotherapy and conformal radiation therapy for very young children with nonmetastatic medulloblastoma: Children’s Oncology Group study P9934. J. Clin. Oncol. 2012, 30, 3181–3186. [Google Scholar] [CrossRef] [Green Version]
- Gupta, T.; Sarkar, C.; Rajshekhar, V.; Chatterjee, S.; Shirsat, N.; Muzumdar, D.; Pungavkar, S.; Chinnaswamy, G.; Jalali, R. Indian Society of Neuro-Oncology consensus guidelines for the contemporary management of medulloblastoma. Neurol. India 2017, 65, 315–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garzon, R.; Calin, G.A.; Croce, C.M. MicroRNAs in Cancer. Annu. Rev. Med. 2009, 60, 167–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaucheret, H.; Vazquez, F.; Crete, P.; Bartel, D.P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004, 18, 1187–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—MicroRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Harfe, B.D. MicroRNAs in vertebrate development. Curr. Opin. Genet. Dev. 2005, 15, 410–415. [Google Scholar] [CrossRef]
- Fabian, M.R.; Sonenberg, N. The mechanics of miRNA-mediated gene silencing: A look under the hood of miRISC. Nat. Struct. Mol. Biol. 2012, 19, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Shalom-Feuerstein, R.; Riley, J.; Zhang, S.D.; Tucci, P.; Agostini, M.; Aberdam, D.; Knight, R.A.; Genchi, G.; Nicotera, P.; et al. miR-7 and miR-214 are specifically expressed during neuroblastoma differentiation, cortical development and embryonic stem cells differentiation, and control neurite outgrowth in vitro. Biochem. Biophys. Res. Commun. 2010, 394, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Feng, Y. MicroRNAs in neural cell development and brain diseases. Sci. China Life Sci. 2011, 54, 1103–1112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bak, M.; Silahtaroglu, A.; Moller, M.; Christensen, M.; Rath, M.F.; Skryabin, B.; Tommerup, N.; Kauppinen, S. MicroRNA expression in the adult mouse central nervous system. RNA 2008, 14, 432–444. [Google Scholar] [CrossRef] [Green Version]
- Agostini, M.; Tucci, P.; Killick, R.; Candi, E.; Sayan, B.S.; Rivetti di Val Cervo, P.; Nicotera, P.; McKeon, F.; Knight, R.A.; Mak, T.W.; et al. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc. Natl. Acad. Sci. USA 2011, 108, 21093–21098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agostini, M.; Tucci, P.; Steinert, J.R.; Shalom-Feuerstein, R.; Rouleau, M.; Aberdam, D.; Forsythe, I.D.; Young, K.W.; Ventura, A.; Concepcion, C.P.; et al. microRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proc. Natl. Acad. Sci. USA 2011, 108, 21099–21104. [Google Scholar] [CrossRef] [Green Version]
- Bonev, B.; Pisco, A.; Papalopulu, N. MicroRNA-9 reveals regional diversity of neural progenitors along the anterior-posterior axis. Dev. Cell 2011, 20, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Makeyev, E.V.; Zhang, J.; Carrasco, M.A.; Maniatis, T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 2007, 27, 435–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Titze-de-Almeida, S.S.; Soto-Sanchez, C.; Fernandez, E.; Koprich, J.B.; Brotchie, J.M.; Titze-de-Almeida, R. The Promise and Challenges of Developing miRNA-Based Therapeutics for Parkinson’s Disease. Cells 2020, 9, 841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gui, Y.; Liu, H.; Zhang, L.; Lv, W.; Hu, X. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 2015, 6, 37043–37053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruce, J.P.; Hui, A.B.; Shi, W.; Perez-Ordonez, B.; Weinreb, I.; Xu, W.; Haibe-Kains, B.; Waggott, D.M.; Boutros, P.C.; O’Sullivan, B.; et al. Identification of a microRNA signature associated with risk of distant metastasis in nasopharyngeal carcinoma. Oncotarget 2015, 6, 4537–4550. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [Green Version]
- Mollashahi, B.; Aghamaleki, F.S.; Movafagh, A. The Roles of miRNAs in Medulloblastoma: A Systematic Review. J. Cancer Prev. 2019, 24, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Fasoulakis, Z.; Daskalakis, G.; Diakosavvas, M.; Papapanagiotou, I.; Theodora, M.; Bourazan, A.; Alatzidou, D.; Pagkalos, A.; Kontomanolis, E.N. MicroRNAs Determining Carcinogenesis by Regulating Oncogenes and Tumor Suppressor Genes During Cell Cycle. Microrna 2020, 9, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Huang, J.; Yang, N.; Greshock, J.; Megraw, M.S.; Giannakakis, A.; Liang, S.; Naylor, T.L.; Barchetti, A.; Ward, M.R.; et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl. Acad. Sci. USA 2006, 103, 9136–9141. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, U.; Hasemeier, B.; Christgen, M.; Muller, M.; Romermann, D.; Langer, F.; Kreipe, H. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J. Pathol. 2008, 214, 17–24. [Google Scholar] [CrossRef]
- Kano, M.; Seki, N.; Kikkawa, N.; Fujimura, L.; Hoshino, I.; Akutsu, Y.; Chiyomaru, T.; Enokida, H.; Nakagawa, M.; Matsubara, H. miR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int. J. Cancer 2010, 127, 2804–2814. [Google Scholar] [CrossRef] [PubMed]
- Boscaino, V.; Fiannaca, A.; La Paglia, L.; La Rosa, M.; Rizzo, R.; Urso, A. MiRNA therapeutics based on logic circuits of biological pathways. BMC Bioinform. 2019, 20, 344. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014, 9, 287–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids—The mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef] [Green Version]
- Iorio, M.V.; Croce, C.M. MicroRNAs in cancer: Small molecules with a huge impact. J. Clin. Oncol. 2009, 27, 5848–5856. [Google Scholar] [CrossRef] [PubMed]
- Gambari, R.; Brognara, E.; Spandidos, D.A.; Fabbri, E. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: Nuew trends in the development of miRNA therapeutic strategies in oncology (Review). Int. J. Oncol. 2016, 49, 5–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Northcott, P.A.; Shih, D.J.; Peacock, J.; Garzia, L.; Morrissy, A.S.; Zichner, T.; Stutz, A.M.; Korshunov, A.; Reimand, J.; Schumacher, S.E.; et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 2012, 488, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.D.; Northcott, P.A.; Korshunov, A.; Remke, M.; Cho, Y.J.; Clifford, S.C.; Eberhart, C.G.; Parsons, D.W.; Rutkowski, S.; Gajjar, A.; et al. Molecular subgroups of medulloblastoma: The current consensus. Acta Neuropathol. 2012, 123, 465–472. [Google Scholar] [CrossRef] [Green Version]
- Ellison, D.W.; Dalton, J.; Kocak, M.; Nicholson, S.L.; Fraga, C.; Neale, G.; Kenney, A.M.; Brat, D.J.; Perry, A.; Yong, W.H.; et al. Medulloblastoma: Clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 2011, 121, 381–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goschzik, T.; Zur Muhlen, A.; Kristiansen, G.; Haberler, C.; Stefanits, H.; Friedrich, C.; von Hoff, K.; Rutkowski, S.; Pfister, S.M.; Pietsch, T. Molecular stratification of medulloblastoma: Comparison of histological and genetic methods to detect Wnt activated tumours. Neuropathol. Appl. Neurobiol. 2015, 41, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Juraschka, K.; Taylor, M.D. Medulloblastoma in the age of molecular subgroups: A review. J. Neurosurg. Pediatrics 2019, 24, 353–363. [Google Scholar] [CrossRef] [Green Version]
- Ramaswamy, V.; Nor, C.; Taylor, M.D. p53 and Meduloblastoma. Cold Spring Harb. Perspect. Med. 2015, 6, a026278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phoenix, T.N.; Patmore, D.M.; Boop, S.; Boulos, N.; Jacus, M.O.; Patel, Y.T.; Roussel, M.F.; Finkelstein, D.; Goumnerova, L.; Perreault, S.; et al. Medulloblastoma Genotype Dictates Blood Brain Barrier Phenotype. Cancer Cell 2016, 29, 508–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manoranjan, B.; Venugopal, C.; Bakhshinyan, D.; Adile, A.A.; Richards, L.; Kameda-Smith, M.M.; Whitley, O.; Dvorkin-Gheva, A.; Subapanditha, M.; Savage, N.; et al. Wnt activation as a therapeutic strategy in medulloblastoma. Nat. Commun. 2020, 11, 4323. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Li, Q.; Bing, Z.; Zhang, Y.; Niu, L.; Yin, H.; Yuan, G.; Pan, Y. Comprehensive analysis of a microRNA expression profile in pediatric medulloblastoma. Mol. Med. Rep. 2017, 15, 4109–4115. [Google Scholar] [CrossRef] [PubMed]
- Panwalkar, P.; Moiyadi, A.; Goel, A.; Shetty, P.; Goel, N.; Sridhar, E.; Shirsat, N. MiR-206, a Cerebellum Enriched miRNA Is Downregulated in All Medulloblastoma Subgroups and Its Overexpression Is Necessary for Growth Inhibition of Medulloblastoma Cells. J. Mol. Neurosci. 2015, 56, 673–680. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Jiang, T.; Shao, L.; Liu, Y.; Zheng, C.; Zhong, Y.; Zhang, J.; Chang, Q. Mir-449a, a potential diagnostic biomarker for WNT group of medulloblastoma. J. Neurooncol. 2016, 129, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Yogi, K.; Sridhar, E.; Goel, N.; Jalali, R.; Goel, A.; Moiyadi, A.; Thorat, R.; Panwalkar, P.; Khire, A.; Dasgupta, A.; et al. MiR-148a, a microRNA upregulated in the WNT subgroup tumors, inhibits invasion and tumorigenic potential of medulloblastoma cells by targeting Neuropilin 1. Oncoscience 2015, 2, 334–348. [Google Scholar] [CrossRef] [Green Version]
- Kool, M.; Jones, D.T.; Jager, N.; Northcott, P.A.; Pugh, T.J.; Hovestadt, V.; Piro, R.M.; Esparza, L.A.; Markant, S.L.; Remke, M.; et al. Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell 2014, 25, 393–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhukova, N.; Ramaswamy, V.; Remke, M.; Pfaff, E.; Shih, D.J.; Martin, D.C.; Castelo-Branco, P.; Baskin, B.; Ray, P.N.; Bouffet, E.; et al. Subgroup-specific prognostic implications of TP53 mutation in medulloblastoma. J. Clin. Oncol. 2013, 31, 2927–2935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rausch, T.; Jones, D.T.; Zapatka, M.; Stutz, A.M.; Zichner, T.; Weischenfeldt, J.; Jager, N.; Remke, M.; Shih, D.; Northcott, P.A.; et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 2012, 148, 59–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Besharat, Z.M.; Sabato, C.; Po, A.; Gianno, F.; Abballe, L.; Napolitano, M.; Miele, E.; Giangaspero, F.; Vacca, A.; Catanzaro, G.; et al. Low Expression of miR-466f-3p Sustains Epithelial to Mesenchymal Transition in Sonic Hedgehog Medulloblastoma Stem Cells Through Vegfa-Nrp2 Signaling Pathway. Front. Pharmacol. 2018, 9, 1281. [Google Scholar] [CrossRef]
- Miele, E.; Po, A.; Begalli, F.; Antonucci, L.; Mastronuzzi, A.; Marras, C.E.; Carai, A.; Cucchi, D.; Abballe, L.; Besharat, Z.M.; et al. beta-arrestin1-mediated acetylation of Gli1 regulates Hedgehog/Gli signaling and modulates self-renewal of SHH medulloblastoma cancer stem cells. BMC Cancer 2017, 17, 488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, B.L.; Obad, S.; Bihannic, L.; Ayrault, O.; Zindy, F.; Kauppinen, S.; Roussel, M.F. Silencing of the miR-17~92 cluster family inhibits medulloblastoma progression. Cancer Res. 2013, 73, 7068–7078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Northcott, P.A.; Fernandez, L.A.; Hagan, J.P.; Ellison, D.W.; Grajkowska, W.; Gillespie, Y.; Grundy, R.; Van Meter, T.; Rutka, J.T.; Croce, C.M.; et al. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009, 69, 3249–3255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tantawy, M.; Elzayat, M.G.; Yehia, D.; Taha, H. Identification of microRNA signature in different pediatric brain tumors. Genet. Mol. Biol. 2018, 41, 27–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pal, R.; Greene, S. microRNA-10b Is Overexpressed and Critical for Cell Survival and Proliferation in Medulloblastoma. PLoS ONE 2015, 10, e0137845. [Google Scholar] [CrossRef]
- Weeraratne, S.D.; Amani, V.; Teider, N.; Pierre-Francois, J.; Winter, D.; Kye, M.J.; Sengupta, S.; Archer, T.; Remke, M.; Bai, A.H.; et al. Pleiotropic effects of miR-183~96~182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta Neuropathol. 2012, 123, 539–552. [Google Scholar] [CrossRef]
- Singh, S.V.; Dakhole, A.N.; Deogharkar, A.; Kazi, S.; Kshirsagar, R.; Goel, A.; Moiyadi, A.; Jalali, R.; Sridhar, E.; Gupta, T.; et al. Restoration of miR-30a expression inhibits growth, tumorigenicity of medulloblastoma cells accompanied by autophagy inhibition. Biochem. Biophys. Res. Commun. 2017, 491, 946–952. [Google Scholar] [CrossRef] [PubMed]
- Kanchan, R.K.; Perumal, N.; Atri, P.; Chirravuri Venkata, R.; Thapa, I.; Klinkebiel, D.L.; Donson, A.M.; Perry, D.; Punsoni, M.; Talmon, G.A.; et al. MiR-1253 exerts tumor-suppressive effects in medulloblastoma via inhibition of CDK6 and CD276 (B7-H3). Brain Pathol. 2020, 30, 732–745. [Google Scholar] [CrossRef]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senfter, D.; Samadaei, M.; Mader, R.M.; Gojo, J.; Peyrl, A.; Krupitza, G.; Kool, M.; Sill, M.; Haberler, C.; Ricken, G.; et al. High impact of miRNA-4521 on FOXM1 expression in medulloblastoma. Cell Death Dis. 2019, 10, 696. [Google Scholar] [CrossRef]
- Kool, M.; Korshunov, A.; Remke, M.; Jones, D.T.; Schlanstein, M.; Northcott, P.A.; Cho, Y.J.; Koster, J.; Schouten-van Meeteren, A.; van Vuurden, D.; et al. Molecular subgroups of medulloblastoma: An international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012, 123, 473–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gershanov, S.; Toledano, H.; Michowiz, S.; Barinfeld, O.; Pinhasov, A.; Goldenberg-Cohen, N.; Salmon-Divon, M. MicroRNA-mRNA expression profiles associated with medulloblastoma subgroup 4. Cancer Manag. Res. 2018, 10, 339–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stromecki, M.; Tatari, N.; Morrison, L.C.; Kaur, R.; Zagozewski, J.; Palidwor, G.; Ramaswamy, V.; Skowron, P.; Wolfl, M.; Milde, T.; et al. Characterization of a novel OTX2-driven stem cell program in Group 3 and Group 4 medulloblastoma. Mol. Oncol. 2018, 12, 495–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, L.; Cheng, Z.; Zhang, J.; Li, R.; Zhao, P.; Fu, Z.; You, Y. hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008, 1236, 185–193. [Google Scholar] [CrossRef]
- Zhao, J.; Lei, T.; Xu, C.; Li, H.; Ma, W.; Yang, Y.; Fan, S.; Liu, Y. MicroRNA-187, down-regulated in clear cell renal cell carcinoma and associated with lower survival, inhibits cell growth and migration though targeting B7-H3. Biochem. Biophys. Res. Commun. 2013, 438, 439–444. [Google Scholar] [CrossRef]
- Mulrane, L.; Madden, S.F.; Brennan, D.J.; Gremel, G.; McGee, S.F.; McNally, S.; Martin, F.; Crown, J.P.; Jirstrom, K.; Higgins, D.G.; et al. miR-187 is an independent prognostic factor in breast cancer and confers increased invasive potential in vitro. Clin. Cancer Res. 2012, 18, 6702–6713. [Google Scholar] [CrossRef] [Green Version]
- Hovestadt, V.; Jones, D.T.; Picelli, S.; Wang, W.; Kool, M.; Northcott, P.A.; Sultan, M.; Stachurski, K.; Ryzhova, M.; Warnatz, H.J.; et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature 2014, 510, 537–541. [Google Scholar] [CrossRef] [Green Version]
- Ferretti, E.; De Smaele, E.; Miele, E.; Laneve, P.; Po, A.; Pelloni, M.; Paganelli, A.; Di Marcotullio, L.; Caffarelli, E.; Screpanti, I.; et al. Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 2008, 27, 2616–2627. [Google Scholar] [CrossRef] [Green Version]
- Tang, B.; Xu, A.; Xu, J.; Huang, H.; Chen, L.; Su, Y.; Zhang, L.; Li, J.; Fan, F.; Deng, J.; et al. MicroRNA-324-5p regulates stemness, pathogenesis and sensitivity to bortezomib in multiple myeloma cells by targeting hedgehog signaling. Int. J. Cancer 2018, 142, 109–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Smaele, E.; Di Marcotullio, L.; Ferretti, E.; Screpanti, I.; Alesse, E.; Gulino, A. Chromosome 17p deletion in human medulloblastoma: A missing checkpoint in the Hedgehog pathway. Cell Cycle 2004, 3, 1263–1266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferretti, E.; De Smaele, E.; Po, A.; Di Marcotullio, L.; Tosi, E.; Espinola, M.S.; Di Rocco, C.; Riccardi, R.; Giangaspero, F.; Farcomeni, A.; et al. MicroRNA profiling in human medulloblastoma. Int. J. Cancer 2009, 124, 568–577. [Google Scholar] [CrossRef]
- Uziel, T.; Karginov, F.V.; Xie, S.; Parker, J.S.; Wang, Y.D.; Gajjar, A.; He, L.; Ellison, D.; Gilbertson, R.J.; Hannon, G.; et al. The miR-17~92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc. Natl. Acad. Sci. USA 2009, 106, 2812–2817. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Ryan, S.L.; Elliott, D.J.; Bignell, G.R.; Futreal, P.A.; Ellison, D.W.; Bailey, S.; Clifford, S.C. Amplification and overexpression of Hsa-miR-30b, Hsa-miR-30d and KHDRBS3 at 8q24.22-q24.23 in medulloblastoma. PLoS ONE 2009, 4, e6159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, A.H.; Milde, T.; Remke, M.; Rolli, C.G.; Hielscher, T.; Cho, Y.J.; Kool, M.; Northcott, P.A.; Jugold, M.; Bazhin, A.V.; et al. MicroRNA-182 promotes leptomeningeal spread of non-sonic hedgehog-medulloblastoma. Acta Neuropathol. 2012, 123, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Weeraratne, S.D.; Amani, V.; Neiss, A.; Teider, N.; Scott, D.K.; Pomeroy, S.L.; Cho, Y.J. miR-34a confers chemosensitivity through modulation of MAGE-A and p53 in medulloblastoma. Neuro Oncol. 2011, 13, 165–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venkataraman, S.; Alimova, I.; Fan, R.; Harris, P.; Foreman, N.; Vibhakar, R. MicroRNA 128a increases intracellular ROS level by targeting Bmi-1 and inhibits medulloblastoma cancer cell growth by promoting senescence. PLoS ONE 2010, 5, e10748. [Google Scholar] [CrossRef] [PubMed]
- Garzia, L.; Andolfo, I.; Cusanelli, E.; Marino, N.; Petrosino, G.; De Martino, D.; Esposito, V.; Galeone, A.; Navas, L.; Esposito, S.; et al. MicroRNA-199b-5p impairs cancer stem cells through negative regulation of HES1 in medulloblastoma. PLoS ONE 2009, 4, e4998. [Google Scholar] [CrossRef] [PubMed]
- Fiaschetti, G.; Abela, L.; Nonoguchi, N.; Dubuc, A.M.; Remke, M.; Boro, A.; Grunder, E.; Siler, U.; Ohgaki, H.; Taylor, M.D.; et al. Epigenetic silencing of miRNA-9 is associated with HES1 oncogenic activity and poor prognosis of medulloblastoma. Br. J. Cancer 2014, 110, 636–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gokhale, A.; Kunder, R.; Goel, A.; Sarin, R.; Moiyadi, A.; Shenoy, A.; Mamidipally, C.; Noronha, S.; Kannan, S.; Shirsat, N.V. Distinctive microRNA signature of medulloblastomas associated with the WNT signaling pathway. J. Cancer Res. Ther. 2010, 6, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, S.; Geng, S.; Ma, S.; Liang, Z.; Jiao, B. Upregulation of microRNA-224 confers a poor prognosis in glioma patients. Clin. Transl. Oncol. 2013, 15, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Rossi, J.J. Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 2007, 8, 173–184. [Google Scholar] [CrossRef] [PubMed]
- De Antonellis, P.; Medaglia, C.; Cusanelli, E.; Andolfo, I.; Liguori, L.; De Vita, G.; Carotenuto, M.; Bello, A.; Formiggini, F.; Galeone, A.; et al. MiR-34a targeting of Notch ligand delta-like 1 impairs CD15+/CD133+ tumor-propagating cells and supports neural differentiation in medulloblastoma. PLoS ONE 2011, 6, e24584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrissy, A.S.; Garzia, L.; Shih, D.J.; Zuyderduyn, S.; Huang, X.; Skowron, P.; Remke, M.; Cavalli, F.M.; Ramaswamy, V.; Lindsay, P.E.; et al. Divergent clonal selection dominates medulloblastoma at recurrence. Nature 2016, 529, 351–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Y.N.; Meley, D.; Pizer, B.; See, V. Mir-34a mimics are potential therapeutic agents for p53-mutated and chemo-resistant brain tumour cells. PLoS ONE 2014, 9, e108514. [Google Scholar] [CrossRef] [Green Version]
- Thor, T.; Kunkele, A.; Pajtler, K.W.; Wefers, A.K.; Stephan, H.; Mestdagh, P.; Heukamp, L.; Hartmann, W.; Vandesompele, J.; Sadowski, N.; et al. MiR-34a deficiency accelerates medulloblastoma formation in vivo. Int. J. Cancer 2015, 136, 2293–2303. [Google Scholar] [CrossRef] [PubMed]
- Visani, M.; Marucci, G.; Biase, D.; Giangaspero, F.; Buttarelli, F.R.; Brandes, A.A.; Franceschi, E.; Acquaviva, G.; Ciarrocchi, A.; Rhoden, K.J.; et al. miR-196B-5P and miR-200B-3P Are Differentially Expressed in Medulloblastomas of Adults and Children. Diagnostics 2020, 10, 265. [Google Scholar] [CrossRef] [PubMed]
- Hemmesi, K.; Squadrito, M.L.; Mestdagh, P.; Conti, V.; Cominelli, M.; Piras, I.S.; Sergi, L.S.; Piccinin, S.; Maestro, R.; Poliani, P.L.; et al. miR-135a Inhibits Cancer Stem Cell-Driven Medulloblastoma Development by Directly Repressing Arhgef6 Expression. Stem Cells 2015, 33, 1377–1389. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, S.; Birks, D.K.; Balakrishnan, I.; Alimova, I.; Harris, P.S.; Patel, P.R.; Handler, M.H.; Dubuc, A.; Taylor, M.D.; Foreman, N.K.; et al. MicroRNA 218 acts as a tumor suppressor by targeting multiple cancer phenotype-associated genes in medulloblastoma. J. Biol. Chem. 2013, 288, 1918–1928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Yang, L.; Wang, T.; Zhang, J.; Guo, X.; Huo, X.; Niu, H. miR-218 is downregulated and directly targets SH3GL1 in childhood medulloblastoma. Mol. Med. Rep. 2013, 8, 1111–1117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.A.; Lu, D.L.; Huang, X.; Tan, W. miR-219 inhibits the proliferation, migration and invasion of medulloblastoma cells by targeting CD164. Int. J. Mol. Med. 2014, 34, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Genovesi, L.A.; Carter, K.W.; Gottardo, N.G.; Giles, K.M.; Dallas, P.B. Integrated analysis of miRNA and mRNA expression in childhood medulloblastoma compared with neural stem cells. PLoS ONE 2011, 6, e23935. [Google Scholar] [CrossRef] [Green Version]
- Bharambe, H.S.; Paul, R.; Panwalkar, P.; Jalali, R.; Sridhar, E.; Gupta, T.; Moiyadi, A.; Shetty, P.; Kazi, S.; Deogharkar, A.; et al. Downregulation of miR-204 expression defines a highly aggressive subset of Group 3/Group 4 medulloblastomas. Acta Neuropathol. Commun. 2019, 7, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Yun, Z.; Zhao, T.; Liu, X.; Ma, X. MiR-495 is a Predictive Biomarker that Downregulates GFI1 Expression in Medulloblastoma. Cell Physiol. Biochem. 2015, 36, 1430–1439. [Google Scholar] [CrossRef] [PubMed]
- Grunder, E.; D’Ambrosio, R.; Fiaschetti, G.; Abela, L.; Arcaro, A.; Zuzak, T.; Ohgaki, H.; Lv, S.Q.; Shalaby, T.; Grotzer, M. MicroRNA-21 suppression impedes medulloblastoma cell migration. Eur. J. Cancer 2011, 47, 2479–2490. [Google Scholar] [CrossRef]
- Jin, Y.; Xiong, A.; Zhang, Z.; Li, S.; Huang, H.; Yu, T.T.; Cao, X.; Cheng, S.Y. MicroRNA-31 suppresses medulloblastoma cell growth by inhibiting DNA replication through minichromosome maintenance 2. Oncotarget 2014, 5, 4821–4833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, H.; Cao, W.; Ding, M. MicroRNA-31 weakens cisplatin resistance of medulloblastoma cells via NF-kappaB and PI3K/AKT pathways. Biofactors 2020, 46, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Gruszka, R.; Zakrzewski, K.; Liberski, P.P.; Zakrzewska, M. mRNA and miRNA Expression Analyses of the MYC/E2F/miR-17-92 Network in the Most Common Pediatric Brain Tumors. Int. J. Mol. Sci. 2021, 22, 543. [Google Scholar] [CrossRef] [PubMed]
- Pierson, J.; Hostager, B.; Fan, R.; Vibhakar, R. Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J. Neurooncol. 2008, 90, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Silber, J.; Hashizume, R.; Felix, T.; Hariono, S.; Yu, M.; Berger, M.S.; Huse, J.T.; VandenBerg, S.R.; James, C.D.; Hodgson, J.G.; et al. Expression of miR-124 inhibits growth of medulloblastoma cells. Neuro Oncol. 2013, 15, 83–90. [Google Scholar] [CrossRef] [Green Version]
- Li, K.K.; Xia, T.; Ma, F.M.; Zhang, R.; Mao, Y.; Wang, Y.; Zhou, L.; Lau, K.M.; Ng, H.K. miR-106b is overexpressed in medulloblastomas and interacts directly with PTEN. Neuropathol. Appl. Neurobiol. 2015, 41, 145–164. [Google Scholar] [CrossRef] [PubMed]
- Kaid, C.; Silva, P.B.; Cortez, B.A.; Rodini, C.O.; Semedo-Kuriki, P.; Okamoto, O.K. miR-367 promotes proliferation and stem-like traits in medulloblastoma cells. Cancer Sci. 2015, 106, 1188–1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andolfo, I.; Liguori, L.; De Antonellis, P.; Cusanelli, E.; Marinaro, F.; Pistollato, F.; Garzia, L.; De Vita, G.; Petrosino, G.; Accordi, B.; et al. The micro-RNA 199b-5p regulatory circuit involves Hes1, CD15, and epigenetic modifications in medulloblastoma. Neuro Oncol. 2012, 14, 596–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunder, R.; Jalali, R.; Sridhar, E.; Moiyadi, A.; Goel, N.; Goel, A.; Gupta, T.; Krishnatry, R.; Kannan, S.; Kurkure, P.; et al. Real-time PCR assay based on the differential expression of microRNAs and protein-coding genes for molecular classification of formalin-fixed paraffin embedded medulloblastomas. Neuro Oncol. 2013, 15, 1644–1651. [Google Scholar] [CrossRef] [Green Version]
- Pan, X.; Wang, Z.; Wan, B.; Zheng, Z. MicroRNA-206 inhibits the viability and migration of medulloblastoma cells by targeting LIM and SH3 protein 1. Exp. Ther. Med. 2017, 14, 3894–3900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.Y.; Zhu, B.; Zhao, X.W.; Zhan, Y.B.; Bao, J.J.; Zhou, J.Q.; Zhang, F.J.; Yu, B.; Liu, J.; Wang, Y.M.; et al. Regulation of UHRF1 by microRNA-378 modulates medulloblastoma cell proliferation and apoptosis. Oncol. Rep. 2017, 38, 3078–3084. [Google Scholar] [CrossRef] [PubMed]
- Li, K.K.; Pang, J.C.; Lau, K.M.; Zhou, L.; Mao, Y.; Wang, Y.; Poon, W.S.; Ng, H.K. MiR-383 is downregulated in medulloblastoma and targets peroxiredoxin 3 (PRDX3). Brain Pathol. 2013, 23, 413–425. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Chen, L.G.; Yao, H.B.; Zhang, J.; Ding, K.F. Upregulation of microRNA-383 inhibits the proliferation, migration and invasion of colon cancer cells. Oncol Lett. 2018, 15, 1184–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pickles, J.C.; Fairchild, A.R.; Stone, T.J.; Brownlee, L.; Merve, A.; Yasin, S.A.; Avery, A.; Ahmed, S.W.; Ogunbiyi, O.; Gonzalez Zapata, J.; et al. DNA methylation-based profiling for paediatric CNS tumour diagnosis and treatment: A population-based study. Lancet Child. Adolesc. Health 2020, 4, 121–130. [Google Scholar] [CrossRef]
- Shalaby, T.; Grotzer, M.A. Tumor-Associated CSF MicroRNAs for the Prediction and Evaluation of CNS Malignancies. Int. J. Mol. Sci. 2015, 16, 29103–29119. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Holgado, B.L.; Ramaswamy, V.; Mack, S.; Zayne, K.; Remke, M.; Wu, X.; Garzia, L.; Daniels, C.; Kenney, A.M.; et al. miR miR on the wall, who’s the most malignant medulloblastoma miR of them all? Neuro Oncol. 2018, 20, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Shih, D.J.; Northcott, P.A.; Remke, M.; Korshunov, A.; Ramaswamy, V.; Kool, M.; Luu, B.; Yao, Y.; Wang, X.; Dubuc, A.M.; et al. Cytogenetic prognostication within medulloblastoma subgroups. J. Clin. Oncol. 2014, 32, 886–896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Po, A.; Ferretti, E.; Miele, E.; De Smaele, E.; Paganelli, A.; Canettieri, G.; Coni, S.; Di Marcotullio, L.; Biffoni, M.; Massimi, L.; et al. Hedgehog controls neural stem cells through p53-independent regulation of Nanog. EMBO J. 2010, 29, 2646–2658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niklison-Chirou, M.V.; Erngren, I.; Engskog, M.; Haglof, J.; Picard, D.; Remke, M.; McPolin, P.H.R.; Selby, M.; Williamson, D.; Clifford, S.C.; et al. TAp73 is a marker of glutamine addiction in medulloblastoma. Genes Dev. 2017, 31, 1738–1753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| SUBGROUP 1: WINGLESS | |||||
|---|---|---|---|---|---|
| miRNAs as Oncogenes | Cellular Function | Ref. | miRNAs as Suppressors | Cellular Function | Ref. |
| miR 30b, miR-30d | N/A | [87] | miR-9 | Antiproliferation Differentiation Pro-apoptosis | [85,92] |
| miR-193a | Metastasis Proliferation | [93] | miR-148a | Antiproliferation Invasion Reduces tumorigenicity | [60,85,93] |
| miR-224 | Proliferation Radiation-sensitivity. Anchorage-independent growth | [93,94] | |||
| SUBGROUP 2: SONIC HEDGEHOG | |||||
| miRNAs as Oncogenes | Cellular Function | Ref. | miRNAs as Suppressors | Cellular Function | Ref. |
| miR-17/92 | N-Myc target | [66,67,89] | miR-let-7 | Chemoresistance | [82,85,95] |
| miR-183/96/182 | Migration | [70] | miR-34a | Antiproliferation Pro-apoptosis Senescence | [89,96,97,98,99] |
| miR-196b-5p, miR-200b-3p | C-Myc target Proliferation Migration Invasion | [100] | miR-125b | Suppressing progenitor and tumor cell growth | [82] |
| miR-128a | Antiproliferation Senescence | [85,90] | |||
| miR-135a | Reduces tumorigenicity | [82,85,101] | |||
| miR-218 | Antiproliferation. Reduces clonogenicity Promotes differentiation | [102,103,104] | |||
| miR-219 | Antiproliferation Invasion Migration [85,104,105] (Ferretti et al., 2009, Genovesi et al., 2011, Shi et al., 2014) | [64,101,102] | |||
| miR-324-5p | Proliferation | [82] | |||
| miR-326 | Reduces clonogenicity | [82] | |||
| SUBGROUP 3 | |||||
| miRNAs as Oncogenes | Cellular Function | Ref. | miRNAs as Suppressors | Cellular Function | Ref. |
| miR-204 | IGF2R and LC3B target Anchorage-indipendent growth Invasion.Autophagy | [106] | |||
| miR-218 | Antiproliferation. Reduces clonogenicity Promotes differentiation | [102,103,104] | |||
| miR-495 | Gfi1 target Cell growth inhibition | [107] | |||
| miR-1253 | Pro-apoptosis Antiproliferation | [72] | |||
| miR-9 | Antiproliferation Differentiation Pro-apoptosis | [85,92] | |||
| SUBGROUP 4 | |||||
| miRNAs as Oncogenes | Cellular Function | Ref. | miRNAs as Suppressors | Cellular Function | Ref. |
| miR-9 | Antiproliferation Differentiation Pro-apoptosis | [85,92] | |||
| miR-204 | IGF2R and LC3B target Anchorage-independent growth Invasion. Autophagy | [106] | |||
| miR-495 | Gfi1 target Cell growth inhibition | [107] | |||
| miR-1253 | Pro-apoptosis Antiproliferation | [72] | |||
| SUBGROUP NOT SPECIFIED | |||||
| miRNAs as Oncogenes | Cellular Function | Ref. | miRNAs as Suppressors | Cellular Function | Ref. |
| miR-21 | Metastasis | [108] | miR-31 | Antiproliferation | [85,109,110] |
| miR-106a/363 | Proliferation Apoptosis Angiogenesis | [111] | miR-124 | Differentiation Antiproliferation Pro-apoptosis | [82,112,113] |
| miR-106b | PTEN target Migration Invasion Tumor-sphere formation | [114] | miR-125a | Antiproliferation | [85] |
| miR-367 | Invasion Proliferation | [115] | miR-199b-5p | Antiproliferation Reduces cancer stem cells | [91,116] |
| miR-206 | Antiproliferation | [58,93,117,118] | |||
| miR-378 | Differentiation Cell growth inhibition | [119] | |||
| miR-383 | Pro-apoptosis | [120,121] | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bevacqua, E.; Farshchi, J.; Niklison-Chirou, M.V.; Tucci, P. Role of MicroRNAs in the Development and Progression of the Four Medulloblastoma Subgroups. Cancers 2021, 13, 6323. https://doi.org/10.3390/cancers13246323
Bevacqua E, Farshchi J, Niklison-Chirou MV, Tucci P. Role of MicroRNAs in the Development and Progression of the Four Medulloblastoma Subgroups. Cancers. 2021; 13(24):6323. https://doi.org/10.3390/cancers13246323
Chicago/Turabian StyleBevacqua, Emilia, Jasmin Farshchi, Maria Victoria Niklison-Chirou, and Paola Tucci. 2021. "Role of MicroRNAs in the Development and Progression of the Four Medulloblastoma Subgroups" Cancers 13, no. 24: 6323. https://doi.org/10.3390/cancers13246323
APA StyleBevacqua, E., Farshchi, J., Niklison-Chirou, M. V., & Tucci, P. (2021). Role of MicroRNAs in the Development and Progression of the Four Medulloblastoma Subgroups. Cancers, 13(24), 6323. https://doi.org/10.3390/cancers13246323






