Loss of SATB2 Occurs More Frequently Than CDX2 Loss in Colorectal Carcinoma and Identifies Particularly Aggressive Cancers in High-Risk Subgroups
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
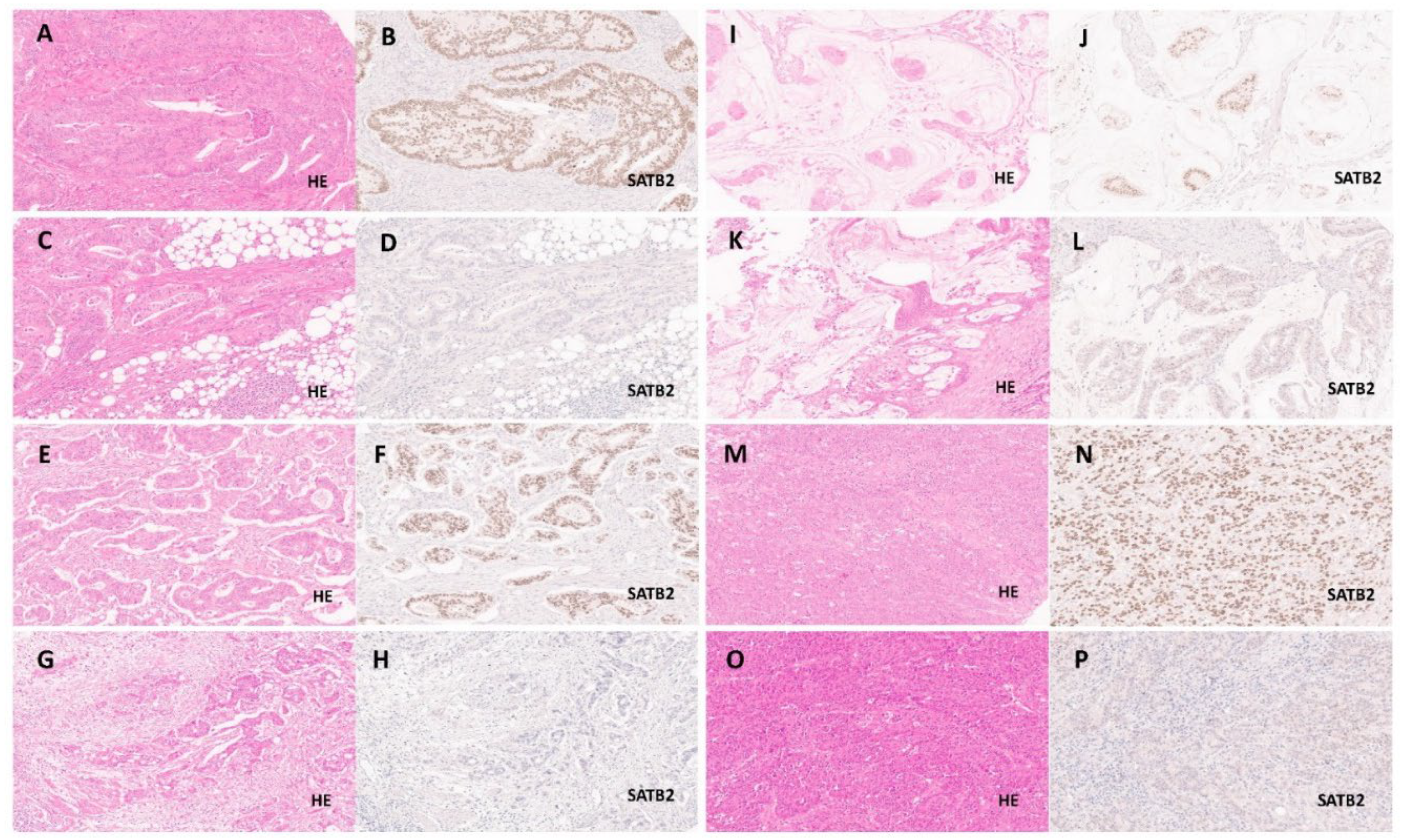
2.2. Evaluation of SATB2 Expression and Clinicopathological Parameters
2.3. Statistics
3. Results
3.1. Clinicopathological Features and Survival
3.1.1. Distribution of SATB2 Expression and Biostatistical Generation of SATB2 Expression Groups
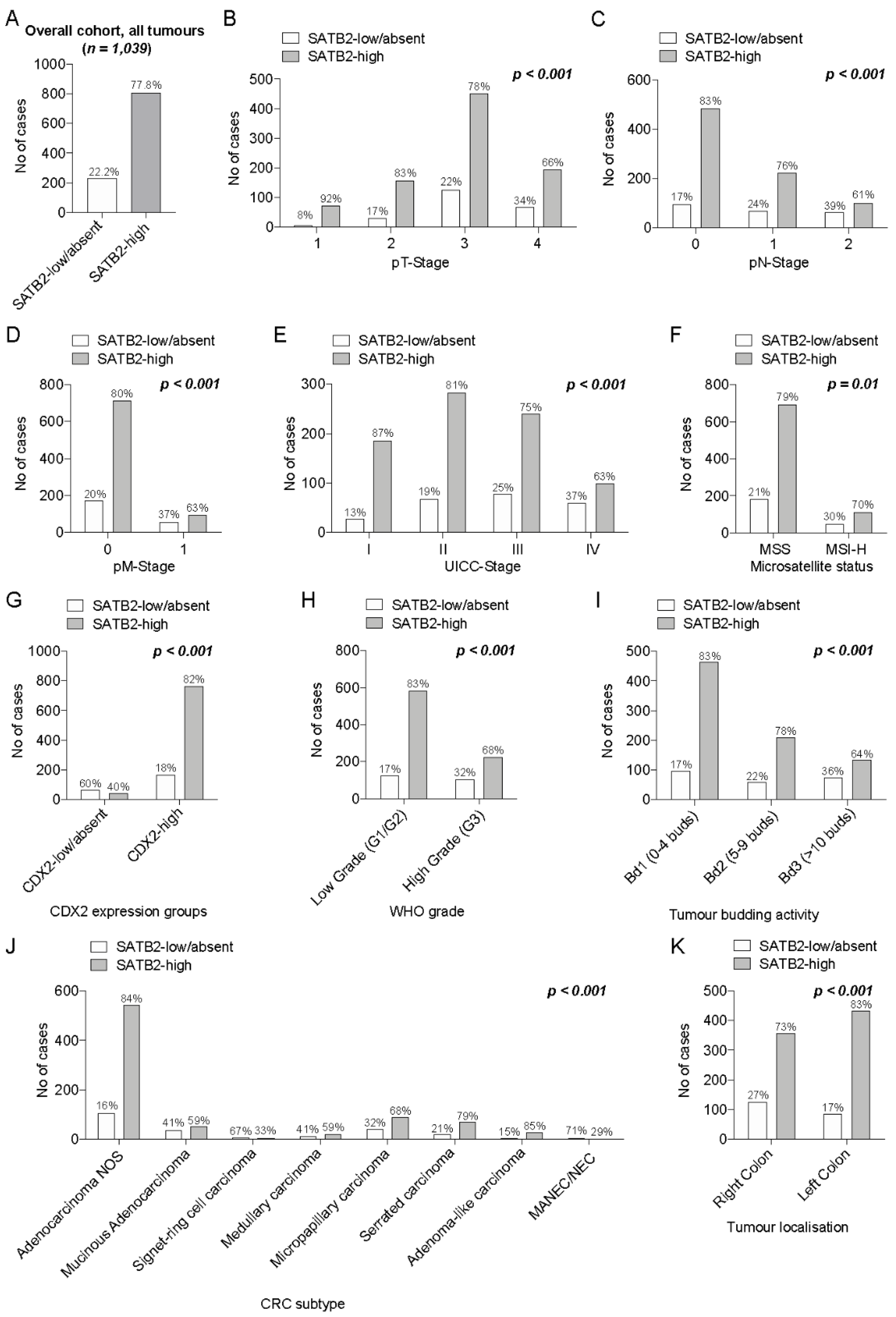
3.1.2. Association of SATB2-Groups with pTNM/UICC Staging, Morphologic Parameters (CRC Subtypes/Tumour Budding/WHO Grade) and Microsatellite Status
3.1.3. Association of SATB2 Expression Groups with CDX2 Expression Groups
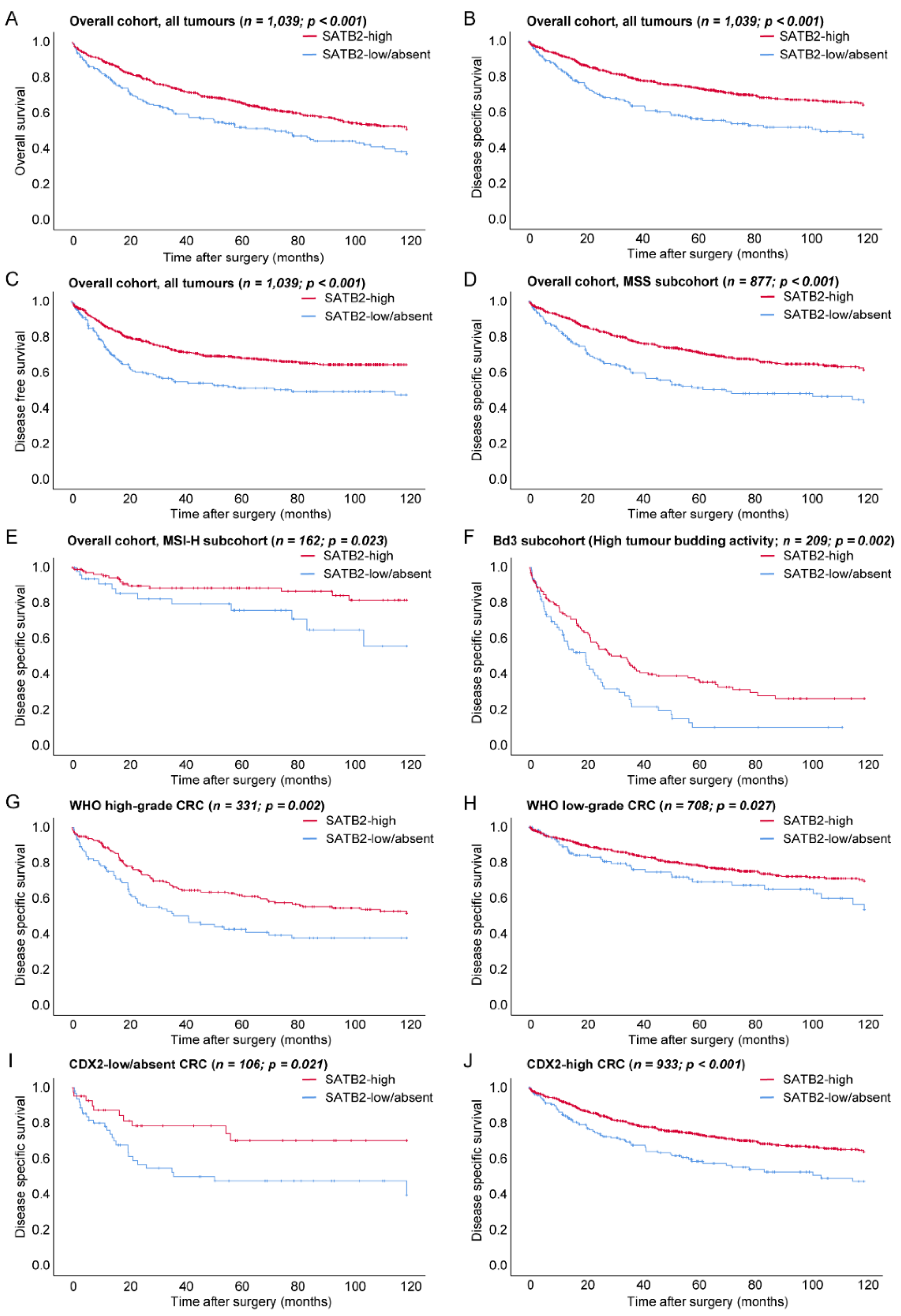
3.2. Prognostic Relevance of SATB2-Groups in the Overall Cohort
3.2.1. Prognostic Relevance of SATB2 in Microsatellite and CDX2 Expression Subgroups
3.2.2. Prognostic Relevance of SATB2 in WHO Grade and Tumour Budding Subgroups
3.2.3. Prognostic Relevance of SATB2 in UICC Stage Subgroups and Right vs. Left-Sided CRCs
3.3. Multivariate Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Zarate, Y.A.; Kaylor, J.; Fish, J. Satb2-associated syndrome. In Genereviews((r)); Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Mirzaa, G., Amemiya, A., Eds.; National Center for Biotechnology Information Bookshelf Site: Seattle, WA, USA, 1993. [Google Scholar]
- FitzPatrick, D.R.; Carr, I.M.; McLaren, L.; Leek, J.P.; Wightman, P.; Williamson, K.; Gautier, P.; McGill, N.; Hayward, C.; Firth, H.; et al. Identification of satb2 as the cleft palate gene on 2q32-q33. Hum. Mol. Genet. 2003, 12, 2491–2501. [Google Scholar] [CrossRef] [Green Version]
- Magnusson, K.; de Wit, M.; Brennan, D.J.; Johnson, L.B.; McGee, S.F.; Lundberg, E.; Naicker, K.; Klinger, R.; Kampf, C.; Asplund, A.; et al. Satb2 in combination with cytokeratin 20 identifies over 95% of all colorectal carcinomas. Am. J. Surg. Pathol. 2011, 35, 937–948. [Google Scholar] [CrossRef]
- Ordonez, N.G. Satb2 is a novel marker of osteoblastic differentiation and colorectal adenocarcinoma. Adv. Anat. Pathol. 2014, 21, 63–67. [Google Scholar] [CrossRef]
- Moh, M.; Krings, G.; Ates, D.; Aysal, A.; Kim, G.E.; Rabban, J.T. Satb2 expression distinguishes ovarian metastases of colorectal and appendiceal origin from primary ovarian tumors of mucinous or endometrioid type. Am. J. Surg. Pathol. 2016, 40, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Conner, J.R.; Hornick, J.L. Metastatic carcinoma of unknown primary: Diagnostic approach using immunohistochemistry. Adv. Anat. Pathol. 2015, 22, 149–167. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Chen, J.W.; He, X.S.; Zhang, H.Z.; Ling, Y.H.; Wen, J.H.; Deng, W.H.; Li, P.; Yun, J.P.; Xie, D.; et al. Satb2 is a promising biomarker for identifying a colorectal origin for liver metastatic adenocarcinomas. EBioMedicine 2018, 28, 62–69. [Google Scholar] [CrossRef] [Green Version]
- Kriegsmann, M.; Harms, A.; Longuespee, R.; Muley, T.; Winter, H.; Kriegsmann, K.; Kazdal, D.; Goeppert, B.; Pathil, A.; Warth, A. Role of conventional immunomarkers, hnf4-alpha and satb2, in the differential diagnosis of pulmonary and colorectal adenocarcinomas. Histopathology 2018, 72, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Dragomir, A.; de Wit, M.; Johansson, C.; Uhlen, M.; Ponten, F. The role of satb2 as a diagnostic marker for tumors of colorectal origin: Results of a pathology-based clinical prospective study. Am. J. Clin. Pathol. 2014, 141, 630–638. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Zhou, J.; Wang, X.Y.; Hao, J.M.; Chen, J.Z.; Zhang, X.M.; Jin, H.; Liu, L.; Zhang, Y.F.; Liu, J.; et al. Down-regulated expression of satb2 is associated with metastasis and poor prognosis in colorectal cancer. J. Pathol. 2009, 219, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Eberhard, J.; Gaber, A.; Wangefjord, S.; Nodin, B.; Uhlen, M.; Ericson Lindquist, K.; Jirstrom, K. A cohort study of the prognostic and treatment predictive value of satb2 expression in colorectal cancer. Br. J. Cancer 2012, 106, 931–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mezheyeuski, A.; Ponten, F.; Edqvist, P.H.; Sundstrom, M.; Thunberg, U.; Qvortrup, C.; Pfeiffer, P.; Sorbye, H.; Glimelius, B.; Dragomir, A. Metastatic colorectal carcinomas with high satb2 expression are associated with better prognosis and response to chemotherapy: A population-based scandinavian study. Acta Oncol. 2020, 59, 284–290. [Google Scholar] [CrossRef] [Green Version]
- Lin, F.; Shi, J.; Zhu, S.; Chen, Z.; Li, A.; Chen, T.; Wang, H.L.; Liu, H. Cadherin-17 and satb2 are sensitive and specific immunomarkers for medullary carcinoma of the large intestine. Arch. Pathol. Lab. Med. 2014, 138, 1015–1026. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Lowenthal, B.M.; Pai, R.K. Satb2 is superior to cdx2 in distinguishing signet ring cell carcinoma of the upper gastrointestinal tract and lower gastrointestinal tract. Am. J. Surg. Pathol. 2018, 42, 1715–1722. [Google Scholar] [CrossRef]
- Jesinghaus, M.; Schmitt, M.; Lang, C.; Reiser, M.; Scheiter, A.; Konukiewitz, B.; Steiger, K.; Silva, M.; Tschurtschenthaler, M.; Lange, S.; et al. Morphology matters: A critical reappraisal of the clinical relevance of morphologic criteria from the 2019 who classification in a large colorectal cancer cohort comprising 1004 cases. Am. J. Surg. Pathol. 2021, 45, 969–978. [Google Scholar] [CrossRef]
- Konukiewitz, B.; Schmitt, S.; Silva, M.; Pohl, J.; Lang, C.; Steiger, K.; Halfter, K.; Engel, J.; Schlitter, A.M.; Boxberg, M.; et al. Loss of cdx2 in colorectal cancer is associated with histopathologic subtypes and microsatellite instability but is prognostically inferior to hematoxylin-eosin based morphologic parameters from the who classification. Br. J. Cancer 2021, in press. [Google Scholar] [CrossRef]
- Konukiewitz, B.; Kasajima, A.; Schmitt, M.; Schwamborn, K.; Groll, T.; Schicktanz, F.; Delbridge, C.; Schutze, L.M.; Wilhelm, D.; Lang, C.; et al. Neuroendocrine differentiation in conventional colorectal adenocarcinomas: Incidental finding or prognostic biomarker? Cancers 2021, 13, 5111. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yuan, J.; Wei, L.; Zhou, L.; Mei, K.; Yue, J.; Gao, H.; Zhang, M.; Jia, L.; Kang, Q.; et al. Satb2 is a sensitive marker for lower gastrointestinal well-differentiated neuroendocrine tumors. Int. J. Clin. Exp. Pathol. 2015, 8, 7072–7082. [Google Scholar]
- Budczies, J.; Klauschen, F.; Sinn, B.V.; Gyorffy, B.; Schmitt, W.D.; Darb-Esfahani, S.; Denkert, C. Cutoff finder: A comprehensive and straightforward web application enabling rapid biomarker cutoff optimization. PLoS ONE 2012, 7, e51862. [Google Scholar] [CrossRef] [Green Version]
- Brierley, J.D.; Gospodarowicz, M.K.; Wittekind, C. Tnm Classification of Malignant Tumours; John Wiley & Sons: Hoboken, NJ, USA, 2017. [Google Scholar]
- Berg, K.B.; Schaeffer, D.F. Satb2 as an immunohistochemical marker for colorectal adenocarcinoma: A concise review of benefits and pitfalls. Arch. Pathol. Lab. Med. 2017, 141, 1428–1433. [Google Scholar] [CrossRef] [Green Version]
- Cigerova, V.; Adamkov, M.; Drahosova, S.; Grendar, M. Immunohistochemical expression and significance of satb2 protein in colorectal cancer. Ann. Diagn. Pathol. 2021, 52, 151731. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Olevian, D.; Miller, C.; Herbst, C.; Jayachandran, P.; Kozak, M.M.; Chang, D.T.; Pai, R.K. Satb2 and cdx2 are prognostic biomarkers in DNA mismatch repair protein deficient colon cancer. Mod. Pathol. 2019, 32, 1217–1231. [Google Scholar] [CrossRef]
- Ma, C.; Olevian, D.C.; Lowenthal, B.M.; Jayachandran, P.; Kozak, M.M.; Chang, D.T.; Pai, R.K. Loss of satb2 expression in colorectal carcinoma is associated with DNA mismatch repair protein deficiency and braf mutation. Am. J. Surg. Pathol. 2018, 42, 1409–1417. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Jiang, D.M.; Hu, S.S.; Zhao, L.; Wang, L.; Yang, M.H.; Ai, M.L.; Jiang, H.J.; Han, Y.; Ding, Y.Q.; et al. Satb2-as1 suppresses colorectal carcinoma aggressiveness by inhibiting satb2-dependent snail transcription and epithelial-mesenchymal transition. Cancer Res. 2019, 79, 3542–3556. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Xu, X.; Pan, B.; Chen, X.; Lin, K.; Zeng, K.; Liu, X.; Xu, T.; Sun, L.; Qin, J.; et al. Lncrna satb2-as1 inhibits tumor metastasis and affects the tumor immune cell microenvironment in colorectal cancer by regulating satb2. Mol. Cancer 2019, 18, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, J.; Wang, G.; Liu, H.; Xiong, C. Satb2 targeted by methylated mir-34c-5p suppresses proliferation and metastasis attenuating the epithelial-mesenchymal transition in colorectal cancer. Cell Prolif. 2018, 51, e12455. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.H.; Yu, J.; Jiang, D.M.; Li, W.L.; Wang, S.; Ding, Y.Q. Microrna-182 targets special at-rich sequence-binding protein 2 to promote colorectal cancer proliferation and metastasis. J. Transl. Med. 2014, 12, 109. [Google Scholar] [CrossRef] [Green Version]
| Variables | Overall n (%) | Median Overall Survival (SE) (Months) | p-Value | Mean Disease Specific Survival (SE) (Months) | p-Value | Mean Disease Free Survival (SE) (Months) | p-Value | |
|---|---|---|---|---|---|---|---|---|
| Age | <0.001 | 0.02 | 0.98 | |||||
| below median | 504 (48.5%) | 86.4 (2.2) | 91.2 (2.1) | 82.4 (2.3) | ||||
| above median | 535 (51.5%) | 72.1 (2.2) | 84.2 (2.3) | 82.7 (2.4) | ||||
| Sex | 0.33 | 0.93 | 0.54 | |||||
| male | 599 (57.7%) | 78.1 (2.1) | 88 (2.0) | 83.6 (2.2) | ||||
| female | 440 (42.3%) | 80.7 (2.4) | 87.4 (2.4) | 81 (2.6) | ||||
| SATB2 Subgroups | <0.001 | <0.001 | <0.001 | |||||
| SATB2-low/absent | 231 (22.2%) | 68.4 (3.6) | 74.2 (3.6) | 68 (3.9) | ||||
| SATB2-high | 808 (77.8%) | 82.2 (1.7) | 91.5 (1.7) | 86.3 (1.8) | ||||
| pT | <0.001 | <0.001 | <0.001 | |||||
| 1 | 79 (7.6%) | 97.7 (4.8) | 115.5 (3.0) | 109.8 (3.6) | ||||
| 2 | 187 (18%) | 92.8 (3.2) | 103.9 (2.8) | 100 (3.1) | ||||
| 3 | 578 (55.6%) | 79.7 (2.1) | 88.3 (2.1) | 82.2 (2.2) | ||||
| 4 | 195 (18.8%) | 57.2 (3.7) | 60.4 (3.8) | 55 (4.0) | ||||
| pN | <0.001 | <0.001 | <0.001 | |||||
| 0 | 580 (55.8%) | 89 (1.9) | 101.2 (1.7) | 99.1 (1.8) | ||||
| 1 | 292 (28.1%) | 75.7 (3.0) | 81 (3.0) | 73.2 (3.2) | ||||
| 2 | 167 (16.1%) | 51.4 (4.0) | 54.6 (4.2) | 42.6 (4.0) | ||||
| pM | <0.001 | <0.001 | <0.001 | |||||
| 0 | 887 (85.4%) | 86.5 (1.6) | 96.3 (1.5) | 91.1 (1.7) | ||||
| 1 | 152 (14.6%) | 40.1 (3.5) | 42.7 (3.7) | 34.4 (3.6) | ||||
| UICC Stage | <0.001 | <0.001 | <0.001 | |||||
| 1 | 213 (20.5%) | 96.6 (2.9) | 111.1 (2.1) | 107.8 (2.4) | ||||
| 2 | 350 (33.7%) | 86 (2.6) | 97 (2.4) | 95.7 (2.6) | ||||
| 3 | 318 (30.6%) | 81.2 (2.8) | 87.2 (2.8) | 76.6 (3.1) | ||||
| 4 | 158 (15.2%) | 39.3 (3.4) | 41.8 (3.6) | 33.3 (3.5) | ||||
| Tumour type (WHO) | <0.001 | <0.001 | <0.001 | |||||
| Adenocarcinoma NOS | 650 (62.6%) | 83.7 (1.9) | 92.5 (1.9) | 87.4 (2.0) | ||||
| Mucinous adenocarcinoma | 88 (8.5%) | 76.5 (5.5) | 87 (5.6) | 78.1 (6.0) | ||||
| Signet-ring cell carcinoma | 9 (0.8%) | 54 (22.5) | 54 (22.5) | 34.4 (18.9) | ||||
| Medullary adenocarcinoma | 32 (3.1%) | 98.6 (7.2) | 116.3 (3.6) | 112.8 (4.9) | ||||
| Micropapillary adenocarcinoma | 129 (12.4%) | 53.6 (4.4) | 56.2 (4.6) | 47.3 (4.5) | ||||
| Serrated adenocarcinoma | 91 (8.7%) | 78.4 (5.6) | 87.7 (5.4) | 84.4 (5.6) | ||||
| Adenoma-like adenocarcinoma | 33 (3.2%) | 98 (6.4) | 115.2 (3.5) | 116.4 (3.5) | ||||
| MANEC/NEC | 7 (0.7%) | 18 (8.2) | 18.0 (8.1) | 15.8 (8.4) | ||||
| WHO grade | <0.001 | <0.001 | <0.001 | |||||
| low-grade | 708 (68.1%) | 86 (1.8) | 95.2 (1.7) | 89.6 (1.9) | ||||
| high-grade | 331 (31.9%) | 65.4 (2.8) | 72.6 (2.9) | 67.8 (3.1) | ||||
| Tumour | <0.001 | <0.001 | <0.001 | |||||
| budding | Bd1 | 560 (53.9%) | 97.8 (1.7) | 109.3 (1.3) | 107 (1.5) | |||
| Bd2 | 270 (26%) | 70.5 (3.1) | 77.6 (3.1) | 66.8 (3.3) | ||||
| Bd3 | 209 (20.1%) | 41 (3.1) | 44 (3.4) | 36.6 (3.4) | ||||
| Resection | <0.001 | <0.001 | <0.001 | |||||
| margin | R0 | 960 (92.4%) | 83.0 (1.6) | 92.4 (1.5) | 87.4 (1.7) | |||
| R1 | 49 (4.7%) | 40.8 (7.2) | 42.3 (7.4) | 29.2 (6.1) | ||||
| R2 | 30 (2.9%) | 25.0 (4.5) | 25 (4.5) | 21.5 (3.7) | ||||
| Lymphatic | <0.001 | <0.001 | <0.001 | |||||
| invasion | not present | 508 (48.9%) | 89.7 (2.0) | 101.8 (1.8) | 100.6 (1.9) | |||
| present | 531 (51.1%) | 69 (1.6) | 74.2 (2.3) | 65.2 (2.5) | ||||
| Venous | <0.001 | <0.001 | <0.001 | |||||
| invasion | not present | 904 (87%) | 83.7 (1.6) | 93.4 (1.6) | 89.2 (1.7) | |||
| present | 135 (13%) | 48.7 (4.3) | 50.9 (4.4) | 38.6 (4.1) | ||||
| Microsatellite | 0.01 | 0.001 | <0.001 | |||||
| status | Microsatellite stable | 877 (84.4%) | 77.6 (1.7) | 85.5 (1.7) | 80 (1.8) | |||
| Microsatellite instable | 162 (15.6%) | 88.7 (3.8) | 101.4 (3.3) | 97.8 (3.7) | ||||
| CDX2 | 0.012 | 0.006 | 0.012 | |||||
| subgroups | CDX2-low/absent | 106 (10.2%) | 67.9 (5.6) | 75.8 (5.7) | 70.4 (5.9) | |||
| CDX2-high | 933 (89.8%) | 80.4 (1.6) | 89.1 (1.6) | 83.7 (1.7) | ||||
| Tumour | 0.26 | 0.83 | 0.93 | |||||
| localization | Right (Coec/Asc/Trans) | 503 (48.4%) | 77.2 (2.3) | 87.1 (2.3) | 82.7 (2.4) | |||
| Left (Desc/Sigm/Rect) | 536 (51.6%) | 81.1 (2.1) | 88.4 (2.1) | 82.3 (2.3) | ||||
| Variables | Total | p-Value | |||||
|---|---|---|---|---|---|---|---|
| A | SATB2 expression group | ||||||
| CDX2 expression group | low/absent | high | |||||
| low/absent | 64 | 42 | 106 | p < 0.001 | |||
| high | 167 | 766 | 933 | ||||
| total | 231 | 808 | 1039 | ||||
| B | CDX2 staining pattern | ||||||
| SATB2 staining pattern | Absent | heterogenous | diffuse | ||||
| absent | 2 | 19 | 39 | 60 | p < 0.001 | ||
| heterogenous | 7 | 70 | 263 | 340 | |||
| diffuse | 4 | 32 | 603 | 639 | |||
| total | 13 | 121 | 905 | 1039 | |||
| Variables | HR (DSS) | Lower CI (95%) | Upper CI (95%) | p-Value | |
|---|---|---|---|---|---|
| SATB2 subgroups | 0.007 | ||||
| SATB2 high | 1.00 | ||||
| SATB2 Low/absent | 1.95 | 1.20 | 3.16 | ||
| WHO Subtype | 0.026 | ||||
| Adenocarcinoma NOS | 1.00 | ||||
| Mucinous adenocarcinoma | 0.41 | 0.12 | 1.34 | ||
| Signet-ring cell carcinoma | 1.89 | 0.53 | 6.70 | ||
| Medullary carcinoma | 0.17 | 0.02 | 1.40 | ||
| Micropapillary adenocarcinoma | 0.75 | 0.43 | 1.29 | ||
| Serrated adenocarcinoma | 0.82 | 0.40 | 1.68 | ||
| Adenoma-like adenocarcinoma | 0.00 | 0.00 | >30 | ||
| MANEC/NEC | 5.67 | 1.23 | 26.04 | ||
| Tumour budding | <0.001 | ||||
| Bd1 | 1.00 | ||||
| Bd2 | 3.35 | 1.76 | 6.36 | ||
| Bd3 | 5.78 | 2.95 | 11.34 | ||
| WHO-grade | 0.027 | ||||
| Low grade | 1.00 | ||||
| High grade | 1.63 | 1.06 | 2.51 | ||
| Gender | 0.723 | ||||
| female | 1.00 | ||||
| male | 1.08 | 0.69 | 1.69 | ||
| Resection status | 0.001 | ||||
| R0 | 1.00 | ||||
| R1/2 | 2.72 | 1.47 | 5.04 | ||
| Tumour | 0.812 | ||||
| localization | Right colon | 1.00 | |||
| Left colon | 0.94 | 0.59 | 1.52 | ||
| Age group | 0.112 | ||||
| Below median | 1.00 | ||||
| Median and above | 1.43 | 0.92 | 2.34 | ||
| Microsatellite | 0.817 | ||||
| status | Microsatellite instable | 1.00 | |||
| Microsatellite stable | 1.09 | 0.51 | 2.32 | ||
| Variables | HR (DSS) | Lower CI (95%) | Upper CI (95%) | p-Value | |
|---|---|---|---|---|---|
| SATB2 subgroups | 0.01 | ||||
| SATB2 high | 1.00 | ||||
| SATB2 Low/absent | 1.67 | 1.13 | 2.46 | ||
| WHO Subtype | 0.1 | ||||
| Adenocarcinoma NOS | 1.00 | ||||
| Mucinous adenocarcinoma | 1.49 | 0.70 | 3.16 | ||
| Signet-ring cell carcinoma | 1.30 | 0.44 | 3.80 | ||
| Micropapillary adenocarcinoma | 0.80 | 0.54 | 1.19 | ||
| Serrated adenocarcinoma | 0.90 | 0.43 | 1.90 | ||
| MANEC/NEC | 2.31 | 0.85 | 6.25 | ||
| WHO-grade | 0.067 | ||||
| Low grade | 1.00 | ||||
| High grade | 1.43 | 0.97 | 2.09 | ||
| UICC Stage | 0.006 | ||||
| I | 1.00 | ||||
| II | 0.67 | 0.24 | 1.85 | ||
| III | 0.64 | 0.24 | 1.70 | ||
| IV | 1.41 | 0.53 | 3.75 | ||
| Gender | 0.716 | ||||
| female | 1.00 | ||||
| male | 1.07 | 0.74 | 1.55 | ||
| Resection status | 0.013 | ||||
| R0 | 1.00 | ||||
| R1/2 | 1.44 | 1.08 | 1.91 | ||
| Tumour | 0.108 | ||||
| localization | Right colon | 1.00 | |||
| Left colon | 1.34 | 0.94 | 1.93 | ||
| Age group | 0.767 | ||||
| Below median | 1.00 | ||||
| Median and above | 1.06 | 0.72 | 1.57 | ||
| Microsatellite | 0.159 | ||||
| status | Microsatellite instable | 1.00 | |||
| Microsatellite stable | 1.76 | 0.80 | 3.88 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schmitt, M.; Silva, M.; Konukiewitz, B.; Lang, C.; Steiger, K.; Halfter, K.; Engel, J.; Jank, P.; Pfarr, N.; Wilhelm, D.; et al. Loss of SATB2 Occurs More Frequently Than CDX2 Loss in Colorectal Carcinoma and Identifies Particularly Aggressive Cancers in High-Risk Subgroups. Cancers 2021, 13, 6177. https://doi.org/10.3390/cancers13246177
Schmitt M, Silva M, Konukiewitz B, Lang C, Steiger K, Halfter K, Engel J, Jank P, Pfarr N, Wilhelm D, et al. Loss of SATB2 Occurs More Frequently Than CDX2 Loss in Colorectal Carcinoma and Identifies Particularly Aggressive Cancers in High-Risk Subgroups. Cancers. 2021; 13(24):6177. https://doi.org/10.3390/cancers13246177
Chicago/Turabian StyleSchmitt, Maxime, Miguel Silva, Björn Konukiewitz, Corinna Lang, Katja Steiger, Kathrin Halfter, Jutta Engel, Paul Jank, Nicole Pfarr, Dirk Wilhelm, and et al. 2021. "Loss of SATB2 Occurs More Frequently Than CDX2 Loss in Colorectal Carcinoma and Identifies Particularly Aggressive Cancers in High-Risk Subgroups" Cancers 13, no. 24: 6177. https://doi.org/10.3390/cancers13246177
APA StyleSchmitt, M., Silva, M., Konukiewitz, B., Lang, C., Steiger, K., Halfter, K., Engel, J., Jank, P., Pfarr, N., Wilhelm, D., Foersch, S., Denkert, C., Tschurtschenthaler, M., Weichert, W., & Jesinghaus, M. (2021). Loss of SATB2 Occurs More Frequently Than CDX2 Loss in Colorectal Carcinoma and Identifies Particularly Aggressive Cancers in High-Risk Subgroups. Cancers, 13(24), 6177. https://doi.org/10.3390/cancers13246177







