A Comprehensive Survey on Deep-Learning-Based Breast Cancer Diagnosis
Abstract
:Simple Summary
Abstract
1. Introduction
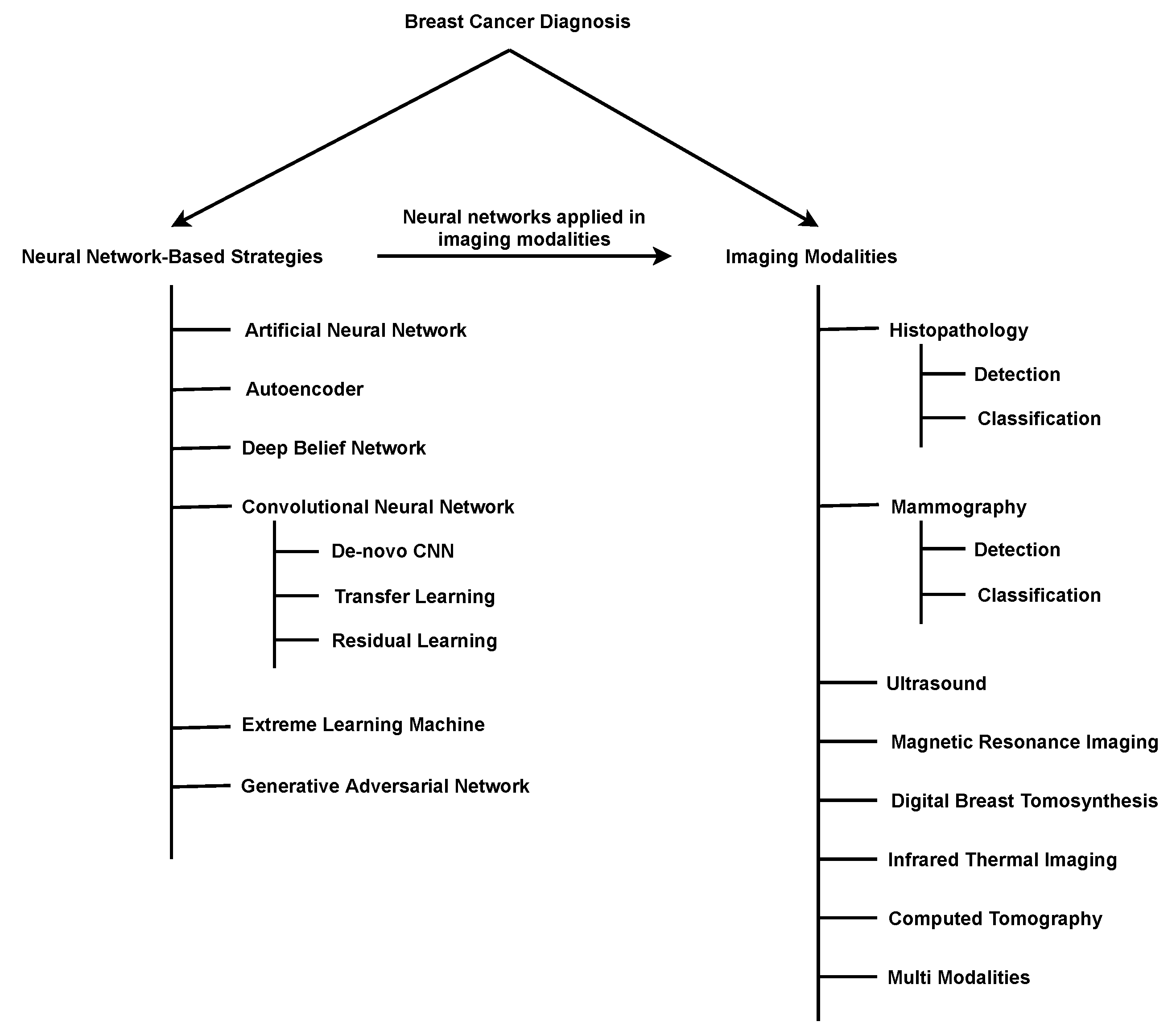
2. Breast-Cancer-Diagnosis Methods Based on Deep Learning
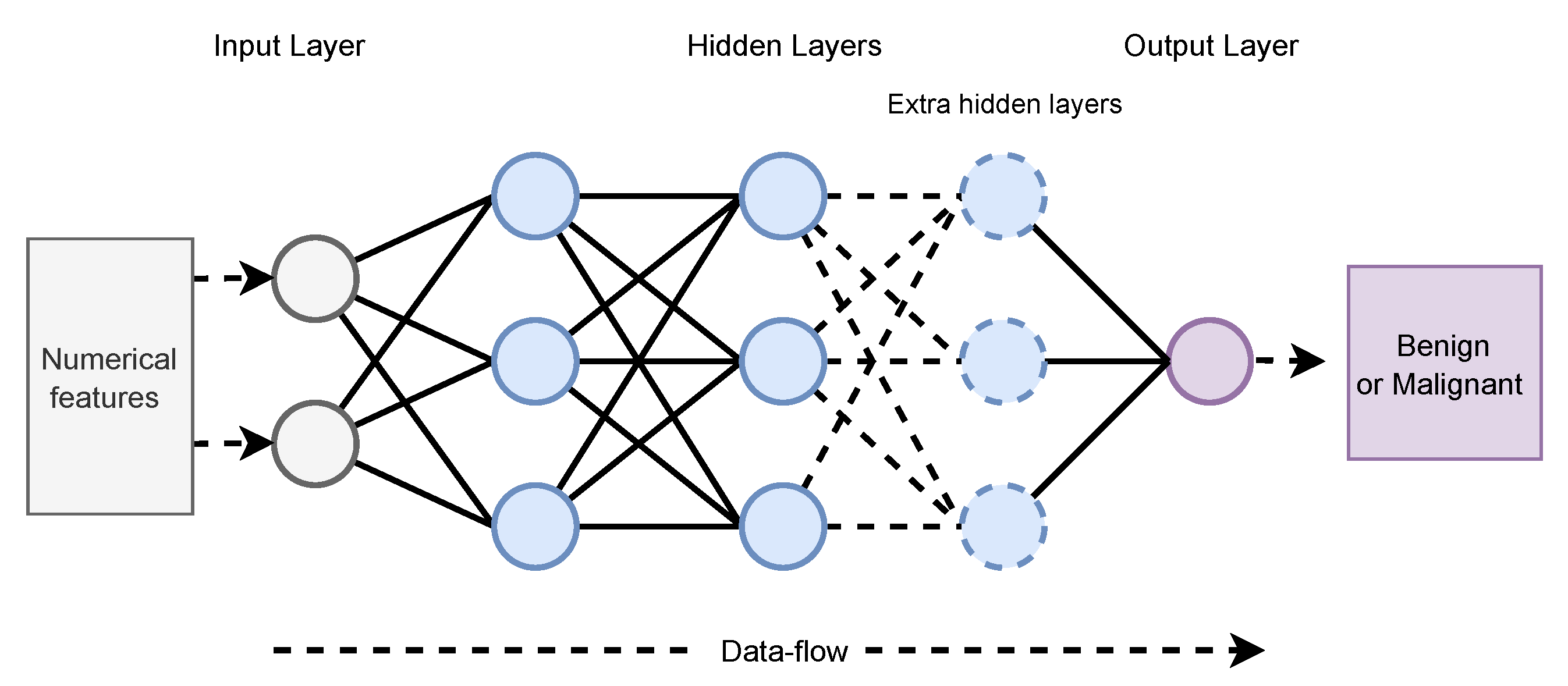
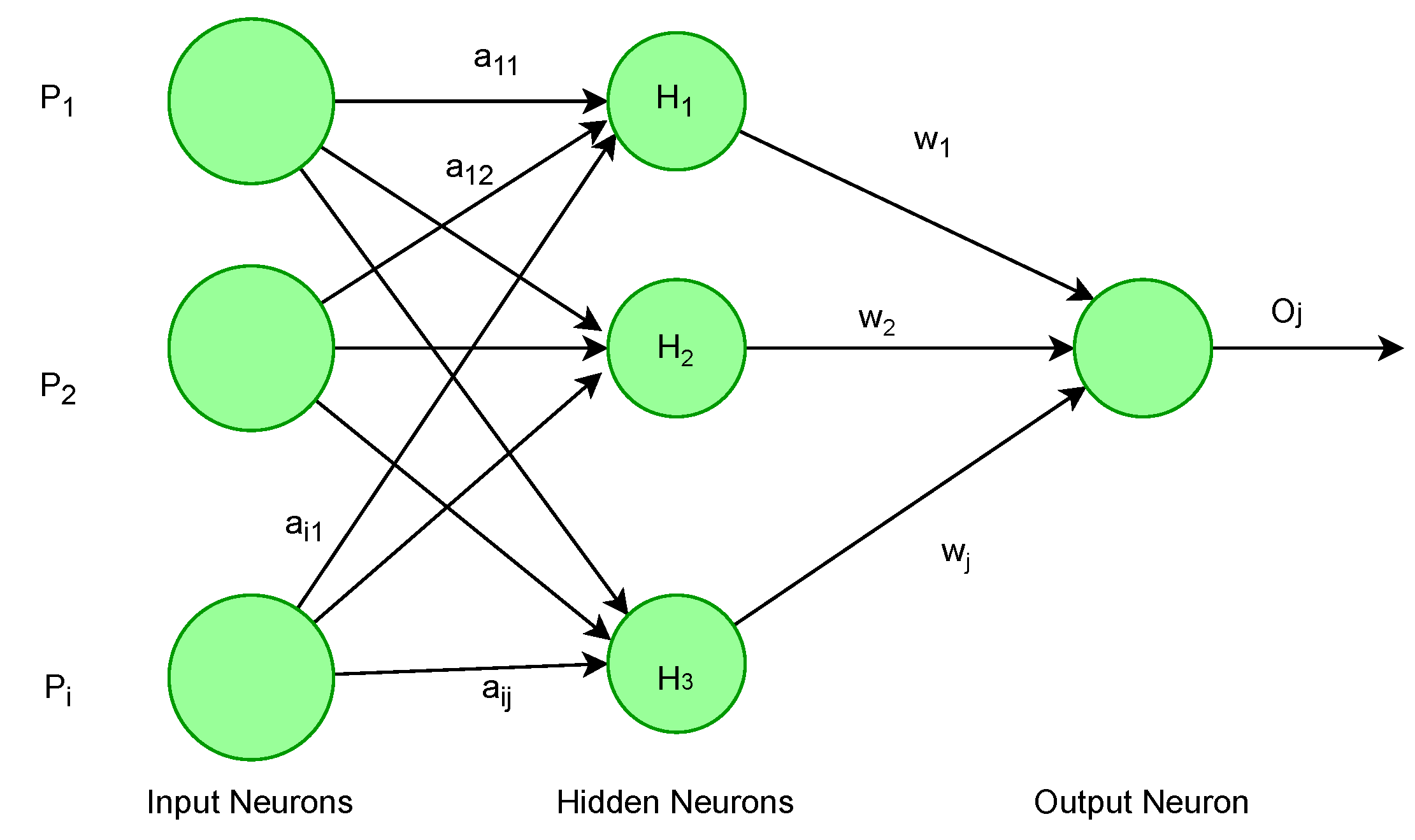
2.1. Artificial Neural Network (ANN)
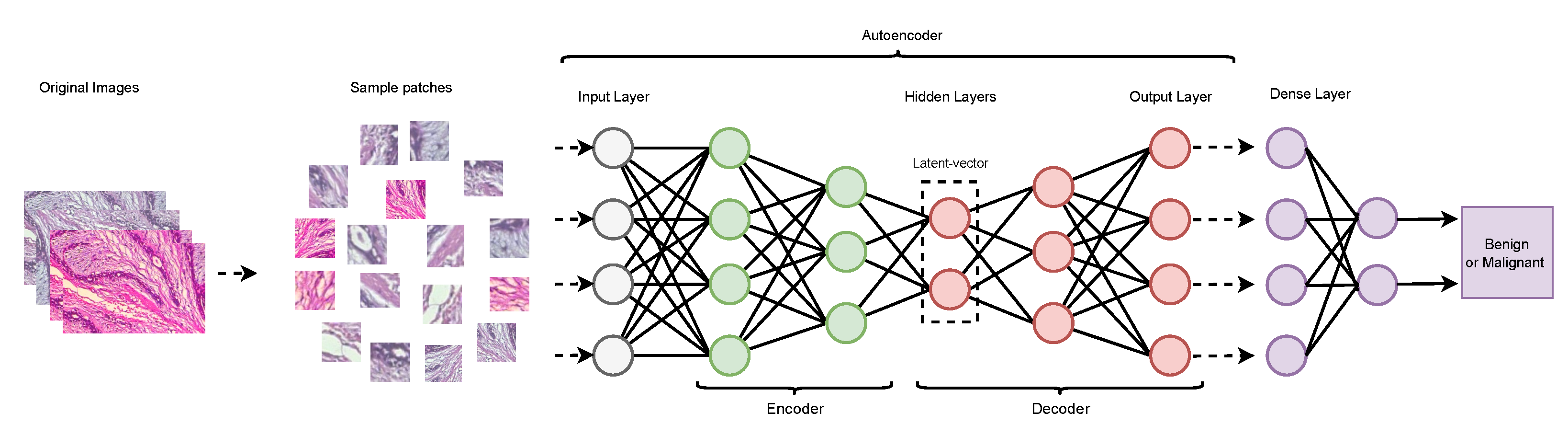
2.2. Autoencoder
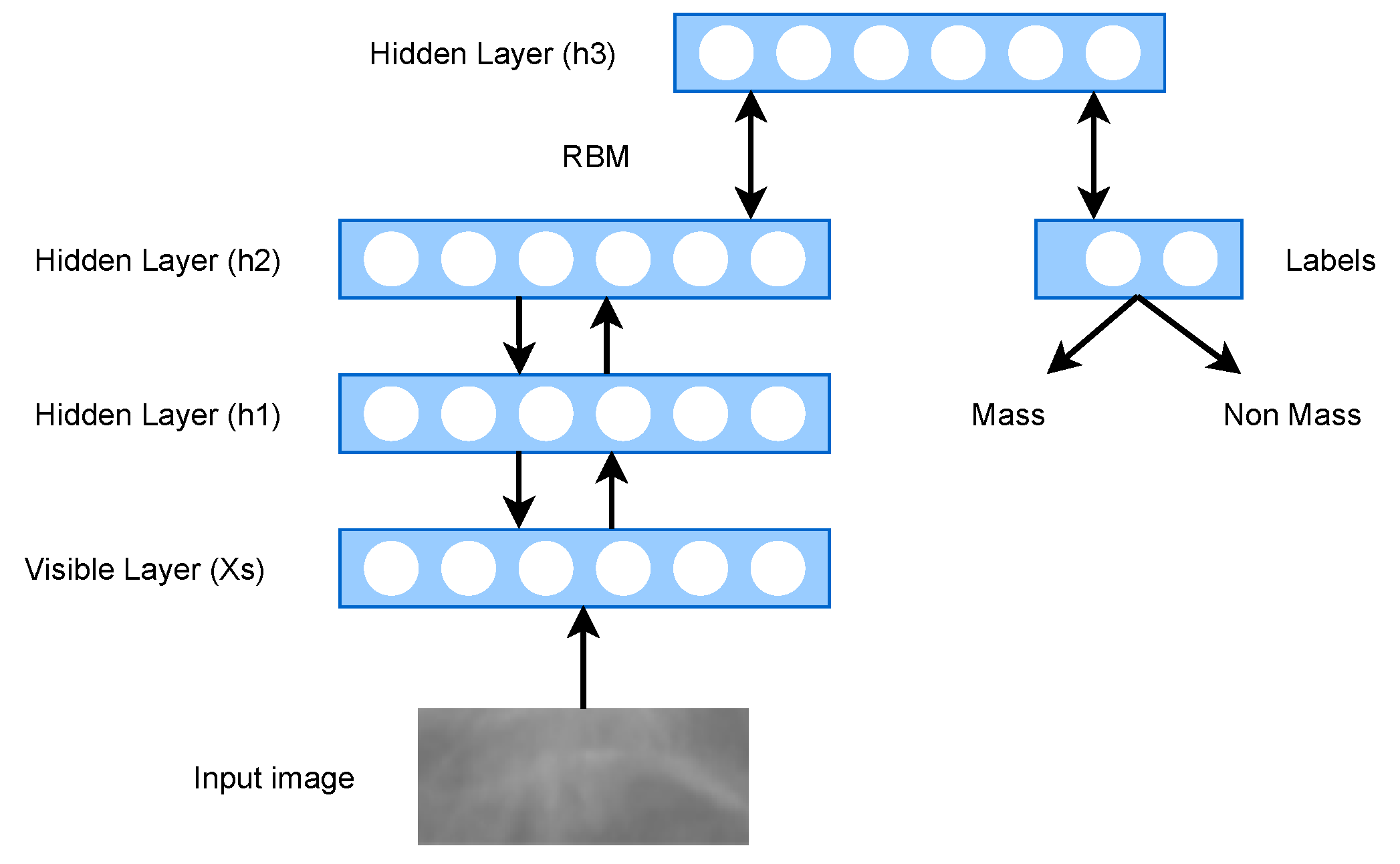
2.3. Deep Belief Network (DBN)
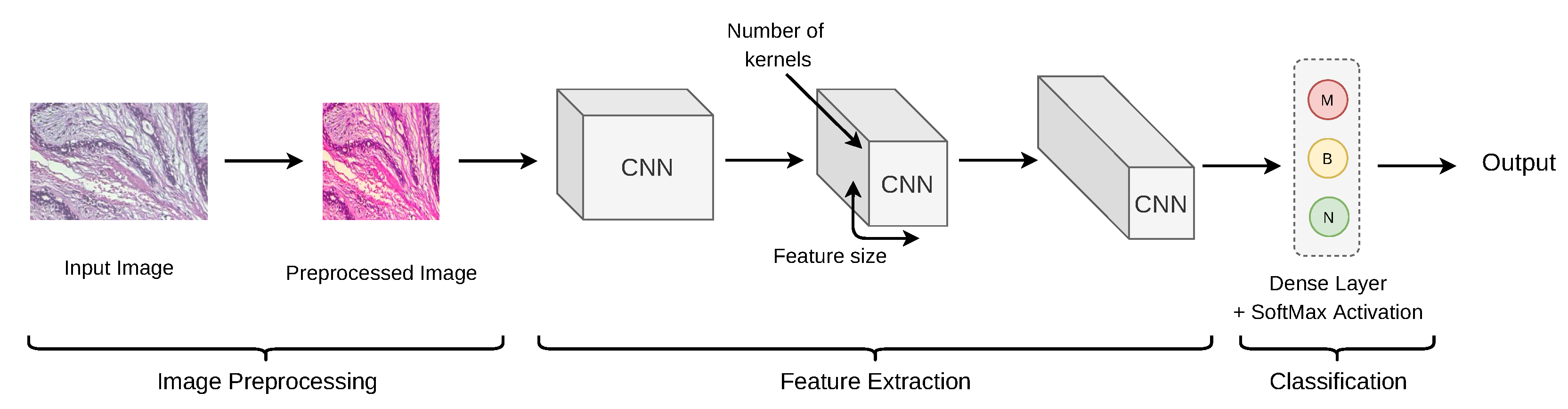
2.4. Convolutional Neural Network (CNN)
2.4.1. De-Novo CNN
2.4.2. Transfer Learning (TL)-Based CNN
2.4.3. Residual Learning (RL)-Based CNN
2.5. Extreme Learning Machine (ELM)
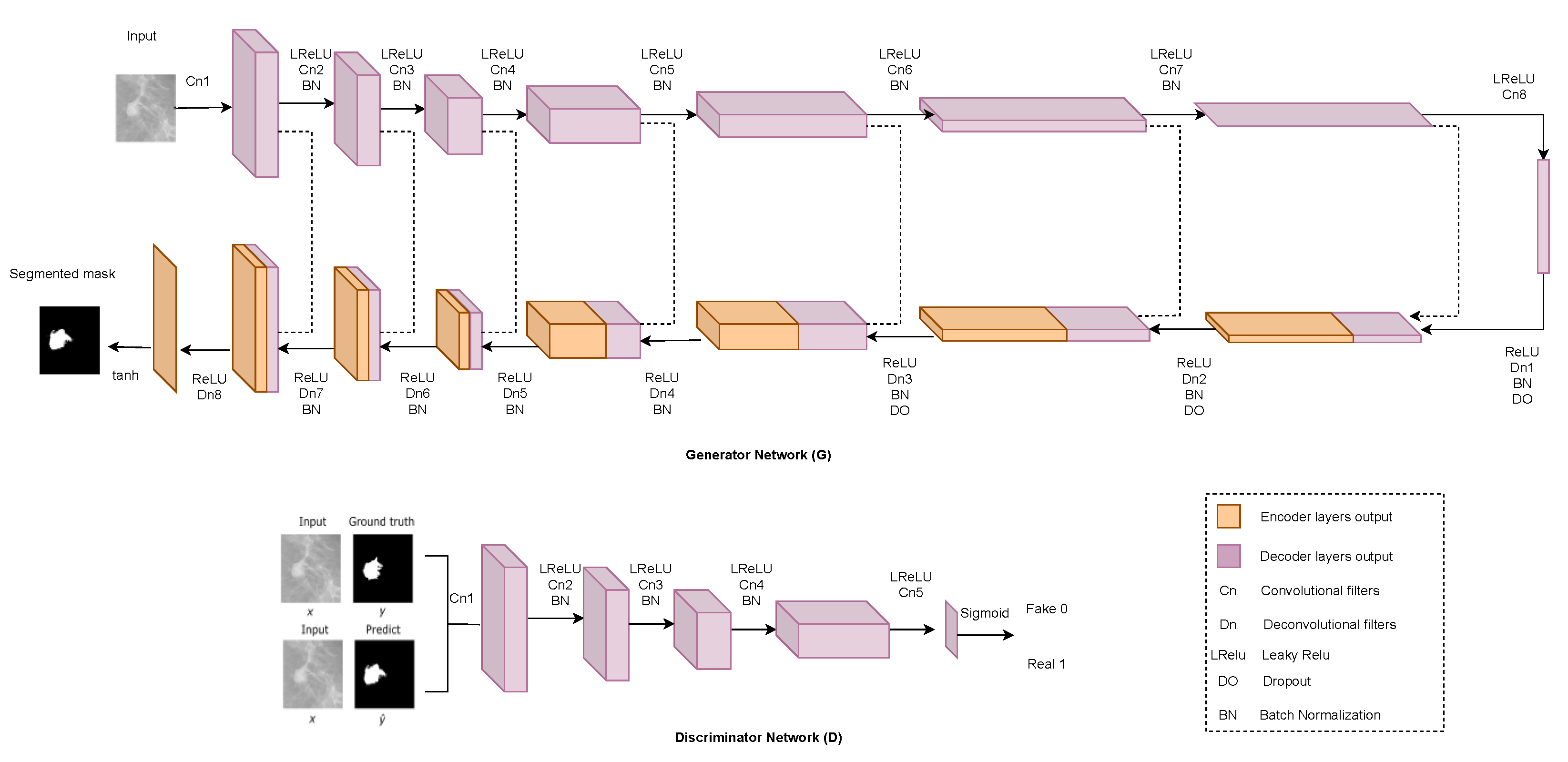
2.6. Generative Adversarial Network (GAN)
3. Datasets and Image Pre-Processing
3.1. Dataset
3.2. Image Pre-Processing
4. Imaging Modalities
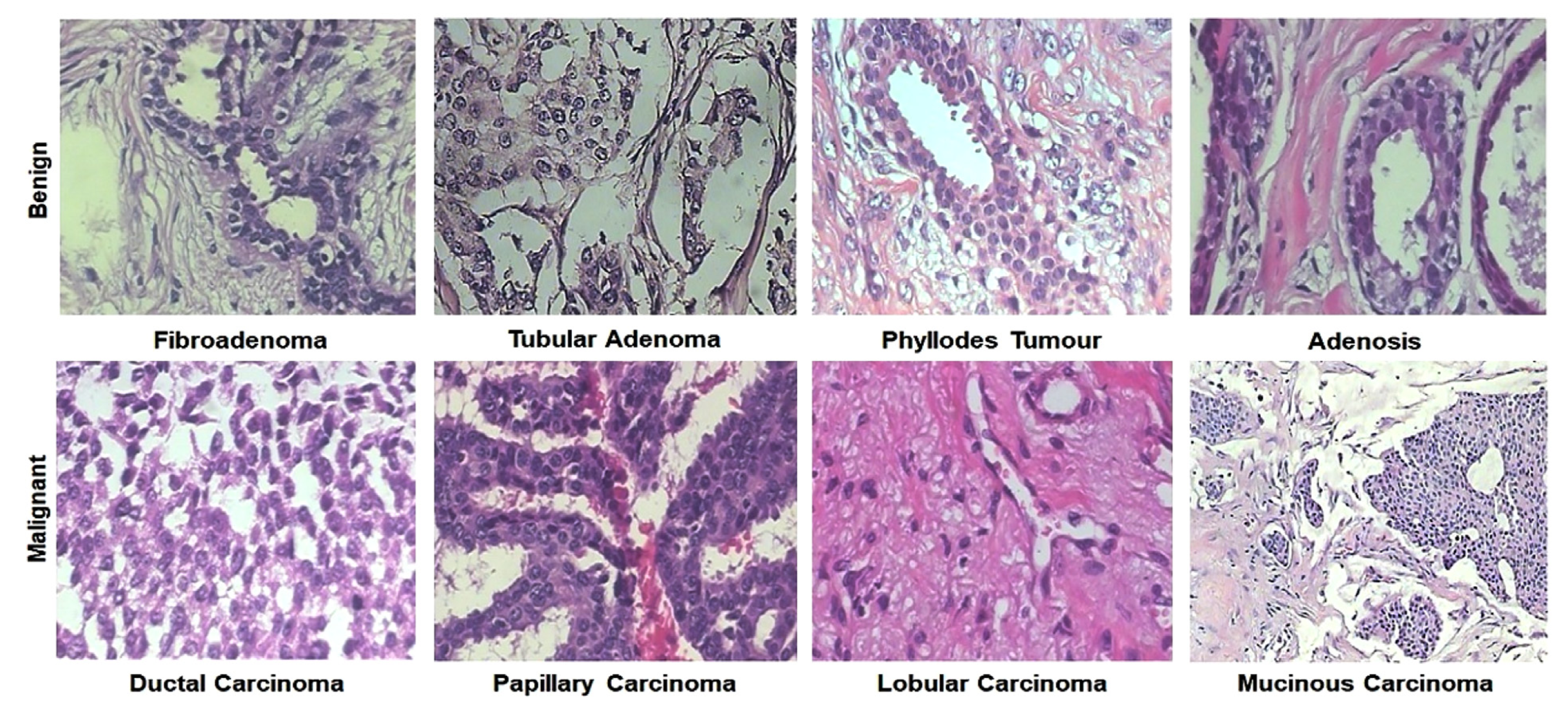
4.1. Histopathology (Hp)
4.1.1. Detection
4.1.2. Classification
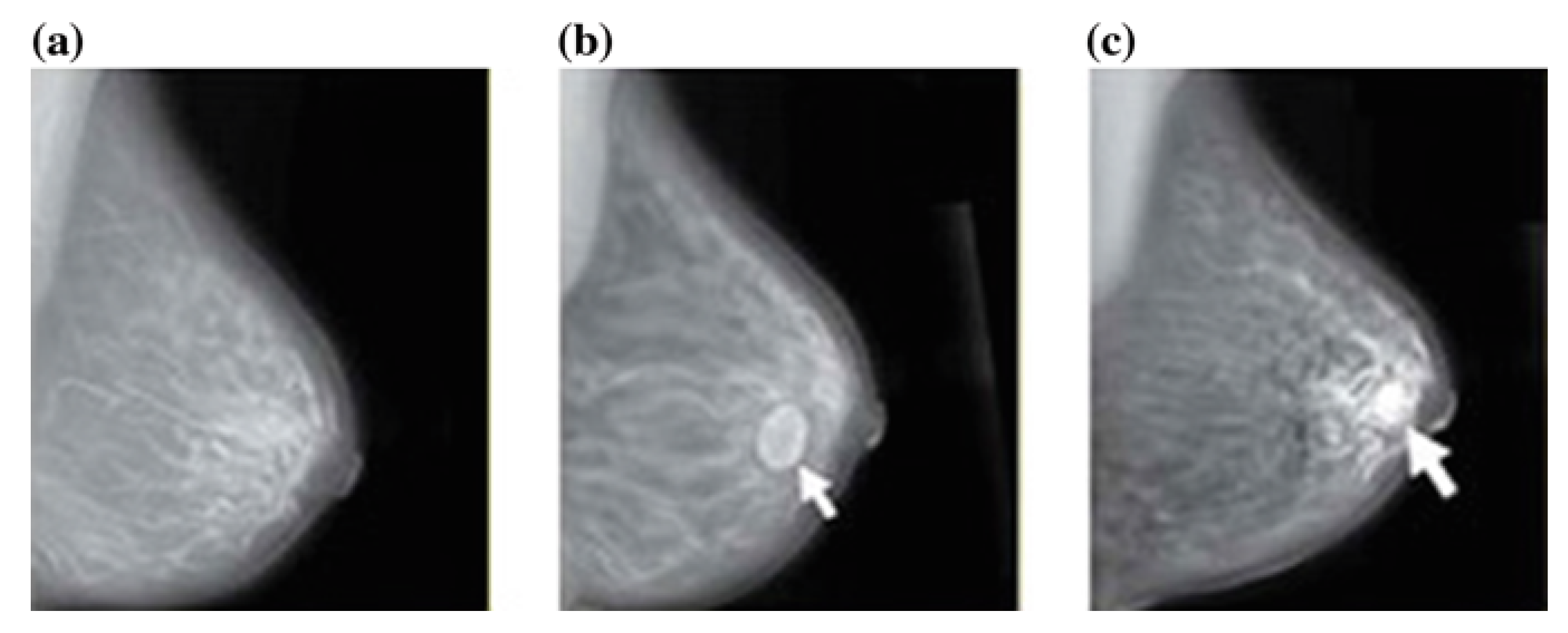
4.2. Mammography (Mg)
4.2.1. Detection
4.2.2. Classification
4.3. Ultrasound (Us)
4.4. Magnetic Resonance Imaging (MRI)
4.5. Digital Breast Tomosynthesis (DBT)
4.6. Infrared Thermal Imaging (ITI)
4.7. Computed Tomography (CT)
4.8. Mass Spectrometry Imaging (MSI)
4.9. Multi-Modalities (MM)
5. Evaluation Metrics and Result Analysis
6. Challenges and Research Directions
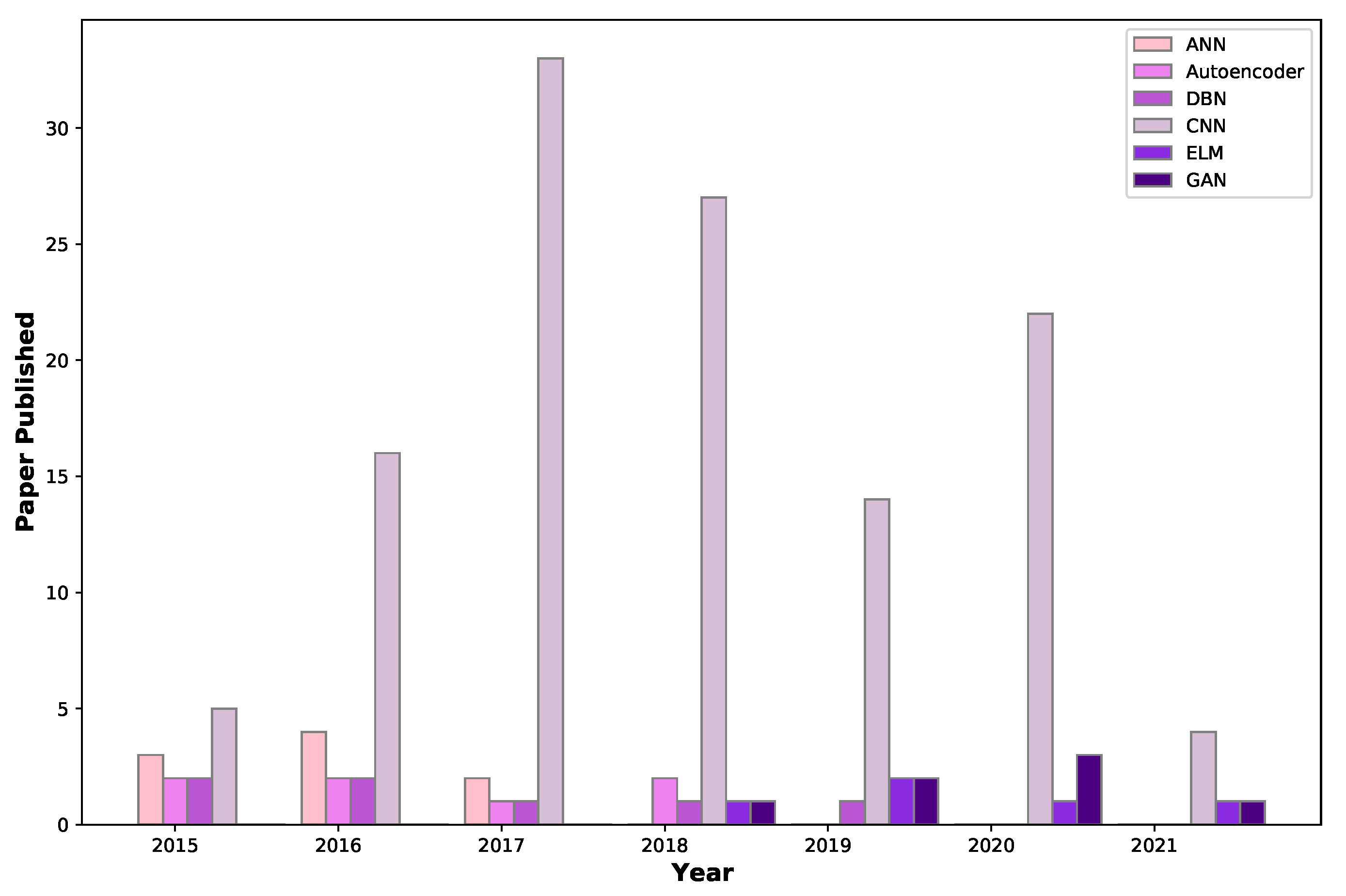
- ANNs, autoencoders, DBNs, and CNNs are currently being used for BrC diagnosis. Deep-learning networks of other kinds, such as RNN, GANs, and clustering should be explored in this field.
- CNN is widely used in BrC classification due to its ability to extract useful features from images. We suggest that strategies based on various CNN architectures, as well as hyperparameter optimization, must be investigated.
- Model efficiency is greatly influenced by feature and classifier selection. Considering the selection of features and classifiers could also boost performance. A study suggested that the alternative-feature-reduction method linear discriminant analysis (LDA) may also be examined [152].
- A study suggested that additional clinical data may boost the performance of the classifier [193]. Increasing data would improve model performance and help to experiment in core model performance more than in handling data scarcity.
- Same-domain transfer learning is a method of training that uses images resembling the target dataset. For example, it trains the model on X-ray images of various breast diseases, and then fine-tunes and trains it on breast-cancer X-ray images for BrC diagnosis. Same-domain transfer learning should be examined as it has achieved promising results recently [199].
- Datasets focused on mammography and histopathology are publicly available. In contrast, datasets based on other imaging modalities such as infrared thermal imaging, computed tomography, and digital breast tomosynthesis are not publicly available. Additionally, studies conducted using such imaging modularities go through unpublished datasets. The research datasets should be published for future knowledge exploration and implementation.
- Future studies may examine the potential of the thermography approach to diagnose cancer levels [186]. The thermography method is currently understood to focus on the metabolic processes of cancerous cells, which generate heat in the affected breast mass areas [200]. A study wants to extend their work by using a 0.5-sensitivity thermal camera to build a 3D structure of the breast to handle the issue of BrC classification [158]. Yet, there are also unanswered questions about what else the deep-learning application can do with thermograms.
- Another field of thermography that requires further investigation is the use of advanced dynamic infrared thermography (DIRT), which allows a non-invasive, painless measurement without the possibility of radiation. The use of different templates, such as CNN, mathematical texture elements, and Gabor coefficient classifiers, in conjunction with the DIRT, can be tested to see if they can detect breast cancer.
- 3D tomosynthesis, a newly emerging breast-imaging technology that creates a video by rebuilding images captured from various viewpoints, has proved to be much more effective than traditional mammograms in manual tumor detection in clinical practice. For BrC classification, mass spectrometry imaging is not employed with deep learning, although it did enable precise classification of distinct-type carcinomas. These imaging technologies need to be investigated in BrC classification.
- Multi-modality research must be investigated. Sutdies have also explored combining mammography and MRI imaging methods. Another intriguing expansion of this work will be to use mammography and MRI-based models as a foundation for analyzing tomosynthesis images, which are increasingly becoming a standard breast-imaging modality.
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Breast Cancer. 2021. Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer (accessed on 19 July 2021).
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- van der Pol, C.C.; Moelans, C.B.; Manson, Q.F.; Batenburg, M.C.; van der Wall, E.; Rinkes, I.B.; Verkooijen, L.; Raman, V.; van Diest, P.J. Cytoplasmic DDX3 as prognosticator in male breast cancer. Virchows Archiv 2021, 479, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Ferdous, S.; Karim-Kos, H.E. Breast cancer in South Asia: A Bangladeshi perspective. Cancer Epidemiol. 2014, 38, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Uddin, A.K.; Khan, Z.J.; Islam, J.; Mahmud, A. Cancer care scenario in Bangladesh. South Asian J. Cancer 2013, 2, 102. [Google Scholar] [CrossRef]
- Bellah, S.F.; Salam, M.A.; Karim, M.R.; Hossain, M.J.; Ashrafudoulla, M. Epidemiology of breast cancer among the female patients in Bangladesh. Orient. Pharm. Exp. Med. 2016, 16, 85–95. [Google Scholar] [CrossRef]
- Arevalo, J.; González, F.A.; Ramos-Pollán, R.; Oliveira, J.L.; Lopez, M.A.G. Convolutional neural networks for mammography mass lesion classification. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milano, Italy, 25–29 August 2015; pp. 797–800. [Google Scholar]
- Yap, M.H.; Pons, G.; Martí, J.; Ganau, S.; Sentís, M.; Zwiggelaar, R.; Davison, A.K.; Martí, R. Automated breast ultrasound lesions detection using convolutional neural networks. IEEE J. Biomed. Health Inform. 2017, 22, 1218–1226. [Google Scholar] [CrossRef] [Green Version]
- Doi, K. Computer-aided diagnosis in medical imaging: Historical review, current status and future potential. Comput. Med. Imaging Graph. 2007, 31, 198–211. [Google Scholar] [CrossRef] [Green Version]
- Brem, R.F.; Rapelyea, J.A.; Zisman, G.; Hoffmeister, J.W.; DeSimio, M.P. Evaluation of breast cancer with a computer-aided detection system by mammographic appearance and histopathology. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2005, 104, 931–935. [Google Scholar] [CrossRef]
- Schneider, M.; Yaffe, M. Better detection: Improving our chances. In Proceedings of the Digital Mammography: 5th International Workshop on Digital Mammography (IWDM), Toronto, ON, Canada, 11–14 June 2000; pp. 3–6. [Google Scholar]
- Huang, D.; Bai, H.; Wang, L.; Hou, Y.; Li, L.; Xia, Y.; Yan, Z.; Chen, W.; Chang, L.; Li, W. The Application and Development of Deep Learning in Radiotherapy: A Systematic Review. Technol. Cancer Res. Treat. 2021, 20, 15330338211016386. [Google Scholar] [CrossRef]
- Munir, K.; Elahi, H.; Farooq, M.U.; Ahmed, S.; Frezza, F.; Rizzi, A. Detection and screening of COVID-19 through chest computed tomography radiographs using deep neural networks. In Data Science for COVID-19; Elsevier: Amsterdam, The Netherlands, 2021; pp. 63–73. [Google Scholar]
- Janowczyk, A.; Madabhushi, A. Deep learning for digital pathology image analysis: A comprehensive tutorial with selected use cases. J. Pathol. Inform. 2016, 7, 29. [Google Scholar] [CrossRef]
- Ghorbani, A.; Ouyang, D.; Abid, A.; He, B.; Chen, J.H.; Harrington, R.A.; Liang, D.H.; Ashley, E.A.; Zou, J.Y. Deep learning interpretation of echocardiograms. NPJ Digit. Med. 2020, 3, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutchinson, L.; Steiert, B.; Soubret, A.; Wagg, J.; Phipps, A.; Peck, R.; Charoin, J.E.; Ribba, B. Models and machines: How deep learning will take clinical pharmacology to the next level. CPT Pharmacomet. Syst. Pharmacol. 2019, 8, 131. [Google Scholar] [CrossRef] [PubMed]
- Munir, K.; Frezza, F.; Rizzi, A. Deep Learning for Brain Tumor Segmentation. In Deep Learning for Cancer Diagnosis; Springer: Berlin/Heidelberg, Germany, 2021; pp. 189–201. [Google Scholar]
- Munir, K.; Frezza, F.; Rizzi, A. Brain Tumor Segmentation Using 2D-UNET Convolutional Neural Network. In Deep Learning for Cancer Diagnosis; Springer: Berlin/Heidelberg, Germany, 2021; pp. 239–248. [Google Scholar]
- Zou, J.; Huss, M.; Abid, A.; Mohammadi, P.; Torkamani, A.; Telenti, A. A primer on deep learning in genomics. Nat. Genet. 2019, 51, 12–18. [Google Scholar] [CrossRef]
- Murthy, N.S.; Bethala, C. Review paper on research direction towards cancer prediction and prognosis using machine learning and deep learning models. J. Ambient. Intell. Humaniz. Comput. 2021, 1–19. [Google Scholar] [CrossRef]
- Aggarwal, R.; Sounderajah, V.; Martin, G.; Ting, D.S.; Karthikesalingam, A.; King, D.; Ashrafian, H.; Darzi, A. Diagnostic accuracy of deep learning in medical imaging: A systematic review and meta-analysis. NPJ Digit. Med. 2021, 4, 65. [Google Scholar] [CrossRef]
- Xie, J.; Liu, R.; Luttrell IV, J.; Zhang, C. Deep learning based analysis of histopathological images of breast cancer. Front. Genet. 2019, 10, 80. [Google Scholar] [CrossRef] [Green Version]
- Lehman, C.D.; Yala, A.; Schuster, T.; Dontchos, B.; Bahl, M.; Swanson, K.; Barzilay, R. Mammographic breast density assessment using deep learning: Clinical implementation. Radiology 2019, 290, 52–58. [Google Scholar] [CrossRef] [Green Version]
- Le, H.; Gupta, R.; Hou, L.; Abousamra, S.; Fassler, D.; Torre-Healy, L.; Moffitt, R.A.; Kurc, T.; Samaras, D.; Batiste, R.; et al. Utilizing Automated Breast Cancer Detection to Identify Spatial Distributions of Tumor Infiltrating Lymphocytes in Invasive Breast Cancer. Am. J. Pathol. 2020, 190, 1491–1504. [Google Scholar] [CrossRef] [Green Version]
- Krithiga, R.; Geetha, P. Breast cancer detection, segmentation and classification on histopathology images analysis: A systematic review. Arch. Comput. Methods Eng. 2021, 28, 2607–2619. [Google Scholar] [CrossRef]
- Bai, J.; Posner, R.; Wang, T.; Yang, C.; Nabavi, S. Applying deep learning in digital breast tomosynthesis for automatic breast cancer detection: A review. Med. Image Anal. 2021, 71, 102049. [Google Scholar] [CrossRef]
- Nahid, A.A.; Kong, Y. Involvement of machine learning for breast cancer image classification: A survey. Comput. Math. Methods Med. 2017, 2017, 3781951. [Google Scholar] [CrossRef] [PubMed]
- Hamidinekoo, A.; Denton, E.; Rampun, A.; Honnor, K.; Zwiggelaar, R. Deep learning in mammography and breast histology, an overview and future trends. Med. Image Anal. 2018, 47, 45–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yassin, N.I.; Omran, S.; El Houby, E.M.; Allam, H. Machine learning techniques for breast cancer computer aided diagnosis using different image modalities: A systematic review. Comput. Methods Programs Biomed. 2018, 156, 25–45. [Google Scholar] [CrossRef]
- Le, E.; Wang, Y.; Huang, Y.; Hickman, S.; Gilbert, F. Artificial intelligence in breast imaging. Clin. Radiol. 2019, 74, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, G.; Shuib, L.; Wahab, A.W.A.; Mujtaba, G.; Nweke, H.F.; Al-garadi, M.A.; Zulfiqar, F.; Raza, G.; Azmi, N.A. Deep learning-based breast cancer classification through medical imaging modalities: State of the art and research challenges. Artif. Intell. Rev. 2019, 53, 1655–1720. [Google Scholar] [CrossRef]
- Munir, K.; Elahi, H.; Ayub, A.; Frezza, F.; Rizzi, A. Cancer diagnosis using deep learning: A bibliographic review. Cancers 2019, 11, 1235. [Google Scholar] [CrossRef] [Green Version]
- Desai, M.; Shah, M. An anatomization on Breast Cancer Detection and Diagnosis employing Multi-layer Perceptron Neural Network (MLP) and Convolutional Neural Network (CNN). Clin. eHealth 2020, 4, 1–11. [Google Scholar] [CrossRef]
- Houssein, E.H.; Emam, M.M.; Ali, A.A.; Suganthan, P.N. Deep and machine learning techniques for medical imaging-based breast cancer: A comprehensive review. Expert Syst. Appl. 2020, 167, 114161. [Google Scholar] [CrossRef]
- Priyanka, K.S. A Review Paper on Breast Cancer Detection Using Deep Learning. In IOP Conference Series: Materials Science and Engineering; IOP Publishing: Bristol, UK, 2021; Volume 1022, p. 012071. [Google Scholar]
- Ayana, G.; Dese, K.; Choe, S.W. Transfer Learning in Breast Cancer Diagnoses via Ultrasound Imaging. Cancers 2021, 13, 738. [Google Scholar] [CrossRef]
- Chugh, G.; Kumar, S.; Singh, N. Survey on Machine Learning and Deep Learning Applications in Breast Cancer Diagnosis. Cogn. Comput. 2021, 1–20. [Google Scholar] [CrossRef]
- Qiu, Y.; Yan, S.; Gundreddy, R.R.; Wang, Y.; Cheng, S.; Liu, H.; Zheng, B. A new approach to develop computer-aided diagnosis scheme of breast mass classification using deep learning technology. J. X-ray Sci. Technol. 2017, 25, 751–763. [Google Scholar] [CrossRef] [Green Version]
- Reardon, S. Rise of robot radiologists. Nature 2019, 576, S54. [Google Scholar]
- Abbass, H.A. An evolutionary artificial neural networks approach for breast cancer diagnosis. Artif. Intell. Med. 2002, 25, 265–281. [Google Scholar] [CrossRef]
- Karabatak, M.; Ince, M.C. An expert system for detection of breast cancer based on association rules and neural network. Expert Syst. Appl. 2009, 36, 3465–3469. [Google Scholar] [CrossRef]
- Jafari-Marandi, R.; Davarzani, S.; Gharibdousti, M.S.; Smith, B.K. An optimum ANN-based breast cancer diagnosis: Bridging gaps between ANN learning and decision-making goals. Appl. Soft Comput. 2018, 72, 108–120. [Google Scholar] [CrossRef]
- Becker, A.S.; Marcon, M.; Ghafoor, S.; Wurnig, M.C.; Frauenfelder, T.; Boss, A. Deep learning in mammography: Diagnostic accuracy of a multipurpose image analysis software in the detection of breast cancer. Investig. Radiol. 2017, 52, 434–440. [Google Scholar] [CrossRef]
- Rouhi, R.; Jafari, M.; Kasaei, S.; Keshavarzian, P. Benign and malignant breast tumors classification based on region growing and CNN segmentation. Expert Syst. Appl. 2015, 42, 990–1002. [Google Scholar] [CrossRef]
- Xu, J.; Xiang, L.; Hang, R.; Wu, J. Stacked Sparse Autoencoder (SSAE) based framework for nuclei patch classification on breast cancer histopathology. In Proceedings of the 2014 IEEE 11th International Symposium on Biomedical Imaging (ISBI), Beijing, China, 29 April–2 May 2014; pp. 999–1002. [Google Scholar]
- Xu, J.; Xiang, L.; Liu, Q.; Gilmore, H.; Wu, J.; Tang, J.; Madabhushi, A. Stacked sparse autoencoder (SSAE) for nuclei detection on breast cancer histopathology images. IEEE Trans. Med Imaging 2015, 35, 119–130. [Google Scholar] [CrossRef] [Green Version]
- Kadam, V.J.; Jadhav, S.M.; Vijayakumar, K. Breast cancer diagnosis using feature ensemble learning based on stacked sparse autoencoders and softmax regression. J. Med. Syst. 2019, 43, 263. [Google Scholar] [CrossRef]
- Toğaçar, M.; Ergen, B.; Cömert, Z. Application of breast cancer diagnosis based on a combination of convolutional neural networks, ridge regression and linear discriminant analysis using invasive breast cancer images processed with autoencoders. Med. Hypotheses 2020, 135, 109503. [Google Scholar] [CrossRef]
- Zhang, D.; Zou, L.; Zhou, X.; He, F. Integrating Feature Selection and Feature Extraction Methods With Deep Learning to Predict Clinical Outcome of Breast Cancer. IEEE Access 2018, 6, 28936–28944. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, L.; Yi, Z. Breast cancer cell nuclei classification in histopathology images using deep neural networks. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.Z.; Ni, D.; Chou, Y.H.; Qin, J.; Tiu, C.M.; Chang, Y.C.; Huang, C.S.; Shen, D.; Chen, C.M. Computer-aided diagnosis with deep learning architecture: Applications to breast lesions in US images and pulmonary nodules in CT scans. Sci. Rep. 2016, 6, 24454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arel, I.; Rose, D.C.; Karnowski, T.P. Deep machine learning-a new frontier in artificial intelligence research [research frontier]. IEEE Comput. Intell. Mag. 2010, 5, 13–18. [Google Scholar] [CrossRef]
- Hinton, G.E.; Osindero, S.; Teh, Y.W. A fast learning algorithm for deep belief nets. Neural Comput. 2006, 18, 1527–1554. [Google Scholar] [CrossRef]
- Abdel-Zaher, A.M.; Eldeib, A.M. Breast cancer classification using deep belief networks. Expert Syst. Appl. 2016, 46, 139–144. [Google Scholar] [CrossRef]
- Khademi, M.; Nedialkov, N.S. Probabilistic Graphical Models and Deep Belief Networks for Prognosis of Breast Cancer. In Proceedings of the 2015 IEEE 14th International Conference on Machine Learning and Applications (ICMLA), Miami, FL, USA, 9–11 December 2015; pp. 727–732. [Google Scholar] [CrossRef]
- Dhungel, N.; Carneiro, G.; Bradley, A.P. Deep structured learning for mass segmentation from mammograms. In Proceedings of the 2015 IEEE International Conference on Image Processing (ICIP), Quebec City, QC, Canada, 27–30 September 2015; pp. 2950–2954. [Google Scholar]
- Dhungel, N.; Carneiro, G.; Bradley, A.P. Deep learning and structured prediction for the segmentation of mass in mammograms. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2015; pp. 605–612. [Google Scholar]
- Al-antari, M.A.; Al-masni, M.A.; Park, S.U.; Park, J.; Metwally, M.K.; Kadah, Y.M.; Han, S.M.; Kim, T.S. An automatic computer-aided diagnosis system for breast cancer in digital mammograms via deep belief network. J. Med. Biol. Eng. 2018, 38, 443–456. [Google Scholar] [CrossRef]
- Zhang, Q.; Xiao, Y.; Dai, W.; Suo, J.; Wang, C.; Shi, J.; Zheng, H. Deep learning based classification of breast tumors with shear-wave elastography. Ultrasonics 2016, 72, 150–157. [Google Scholar] [CrossRef]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. Breast cancer histopathological image classification using convolutional neural networks. In Proceedings of the 2016 International Joint Conference on Neural Networks (IJCNN), Vancouver, BC, Canada, 24–29 July 2016; pp. 2560–2567. [Google Scholar]
- Hu, Q.; Whitney, H.M.; Giger, M.L. A deep learning methodology for improved breast cancer diagnosis using multiparametric MRI. Sci. Rep. 2020, 10, 10536. [Google Scholar] [CrossRef]
- Oyelade, O.N.; Ezugwu, A.E. A deep learning model using data augmentation for detection of architectural distortion in whole and patches of images. Biomed. Signal Process. Control 2021, 65, 102366. [Google Scholar] [CrossRef]
- Huynh, B.Q.; Li, H.; Giger, M.L. Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. J. Med. Imaging 2016, 3, 034501. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Tan, L.; Lin, L.; Cheng, X.; Yi, Z.; Sato, T. Deep-Learning-Empowered Breast Cancer Auxiliary Diagnosis for 5GB Remote E-Health. IEEE Wirel. Commun. 2021, 28, 54–61. [Google Scholar] [CrossRef]
- Xu, J.; Luo, X.; Wang, G.; Gilmore, H.; Madabhushi, A. A deep convolutional neural network for segmenting and classifying epithelial and stromal regions in histopathological images. Neurocomputing 2016, 191, 214–223. [Google Scholar] [CrossRef] [Green Version]
- Kooi, T.; Litjens, G.; Van Ginneken, B.; Gubern-Mérida, A.; Sánchez, C.I.; Mann, R.; den Heeten, A.; Karssemeijer, N. Large scale deep learning for computer aided detection of mammographic lesions. Med. Image Anal. 2017, 35, 303–312. [Google Scholar] [CrossRef]
- Samala, R.K.; Chan, H.P.; Hadjiiski, L.M.; Cha, K.; Helvie, M.A. Deep-learning convolution neural network for computer-aided detection of microcalcifications in digital breast tomosynthesis. In Medical Imaging 2016: Computer-Aided Diagnosis; International Society for Optics and Photonics: Bellingham, WA, USA, 2016; Volume 9785, p. 97850Y. [Google Scholar]
- Yan, R.; Ren, F.; Wang, Z.; Wang, L.; Zhang, T.; Liu, Y.; Rao, X.; Zheng, C.; Zhang, F. Breast cancer histopathological image classification using a hybrid deep neural network. Methods 2020, 173, 52–60. [Google Scholar] [CrossRef]
- Wang, P.; Wang, J.; Li, Y.; Li, P.; Li, L.; Jiang, M. Automatic classification of breast cancer histopathological images based on deep feature fusion and enhanced routing. Biomed. Signal Process. Control 2021, 65, 102341. [Google Scholar] [CrossRef]
- Ting, F.F.; Tan, Y.J.; Sim, K.S. Convolutional neural network improvement for breast cancer classification. Expert Syst. Appl. 2019, 120, 103–115. [Google Scholar] [CrossRef]
- Araújo, T.; Aresta, G.; Castro, E.; Rouco, J.; Aguiar, P.; Eloy, C.; Polónia, A.; Campilho, A. Classification of breast cancer histology images using convolutional neural networks. PLoS ONE 2017, 12, e0177544. [Google Scholar] [CrossRef]
- Albarqouni, S.; Baur, C.; Achilles, F.; Belagiannis, V.; Demirci, S.; Navab, N. Aggnet: Deep learning from crowds for mitosis detection in breast cancer histology images. IEEE Trans. Med. Imaging 2016, 35, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- Samala, R.K.; Chan, H.P.; Hadjiiski, L.M.; Helvie, M.A.; Cha, K.H.; Richter, C.D. Multi-task transfer learning deep convolutional neural network: Application to computer-aided diagnosis of breast cancer on mammograms. Phys. Med. Biol. 2017, 62, 8894. [Google Scholar] [CrossRef] [PubMed]
- Mendel, K.; Li, H.; Sheth, D.; Giger, M. Transfer learning from convolutional neural networks for computer-aided diagnosis: A comparison of digital breast tomosynthesis and full-field digital mammography. Acad. Radiol. 2019, 26, 735–743. [Google Scholar] [CrossRef]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J. Convolutional neural networks for medical image analysis: Full training or fine tuning? IEEE Trans. Med. Imaging 2016, 35, 1299–1312. [Google Scholar] [CrossRef] [Green Version]
- Raghu, M.; Zhang, C.; Kleinberg, J.; Bengio, S. Transfusion: Understanding transfer learning for medical imaging. In Proceedings of the 33rd International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 8–14 December 2019; pp. 3347–3357. [Google Scholar]
- Chougrad, H.; Zouaki, H.; Alheyane, O. Deep convolutional neural networks for breast cancer screening. Comput. Methods Programs Biomed. 2018, 157, 19–30. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, S.K.; Saxena, S.; Lakshmanan, K.; Sangaiah, A.K.; Chauhan, H.; Shrivastava, S.; Singh, R.K. Deep feature learning for histopathological image classification of canine mammary tumors and human breast cancer. Inf. Sci. 2020, 508, 405–421. [Google Scholar] [CrossRef]
- Toğaçar, M.; Özkurt, K.B.; Ergen, B.; Cömert, Z. BreastNet: A novel convolutional neural network model through histopathological images for the diagnosis of breast cancer. Phys. Stat. Mech. Appl. 2020, 545, 123592. [Google Scholar] [CrossRef]
- Gour, M.; Jain, S.; Sunil Kumar, T. Residual learning based CNN for breast cancer histopathological image classification. Int. J. Imaging Syst. Technol. 2020, 30, 621–635. [Google Scholar] [CrossRef]
- Hu, C.; Sun, X.; Yuan, Z.; Wu, Y. Classification of breast cancer histopathological image with deep residual learning. Int. J. Imaging Syst. Technol. 2021, 31, 1583–1594. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference On computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Singh, S.; Matthews, T.P.; Shah, M.; Mombourquette, B.; Tsue, T.; Long, A.; Almohsen, R.; Pedemonte, S.; Su, J. Adaptation of a deep learning malignancy model from full-field digital mammography to digital breast tomosynthesis. In Medical Imaging 2020: Computer-Aided Diagnosis; International Society for Optics and Photonics: Bellingham, WA, USA, 2020; Volume 11314, p. 1131406. [Google Scholar]
- Li, C.; Xu, J.; Liu, Q.; Zhou, Y.; Mou, L.; Pu, Z.; Xia, Y.; Zheng, H.; Wang, S. Multi-View Mammographic Density Classification by Dilated and Attention-Guided Residual Learning. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 18, 1003–1013. [Google Scholar] [CrossRef]
- Lahoura, V.; Singh, H.; Aggarwal, A.; Sharma, B.; Mohammed, M.A.; Damaševičius, R.; Kadry, S.; Cengiz, K. Cloud Computing-Based Framework for Breast Cancer Diagnosis Using Extreme Learning Machine. Diagnostics 2021, 11, 241. [Google Scholar] [CrossRef]
- Toprak, A. Extreme learning machine (elm)-based classification of benign and malignant cells in breast cancer. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018, 24, 6537. [Google Scholar] [CrossRef]
- Nemissi, M.; Salah, H.; Seridi, H. Breast cancer diagnosis using an enhanced Extreme Learning Machine based-Neural Network. In Proceedings of the 2018 International Conference on Signal, Image, Vision and Their Applications (SIVA), Guelma, Algeria, 26–27 November 2018; pp. 1–4. [Google Scholar]
- Wang, P.; Song, Q.; Li, Y.; Lv, S.; Wang, J.; Li, L.; Zhang, H. Cross-task extreme learning machine for breast cancer image classification with deep convolutional features. Biomed. Signal Process. Control 2020, 57, 101789. [Google Scholar] [CrossRef]
- Wang, Z.; Li, M.; Wang, H.; Jiang, H.; Yao, Y.; Zhang, H.; Xin, J. Breast cancer detection using extreme learning machine based on feature fusion with CNN deep features. IEEE Access 2019, 7, 105146–105158. [Google Scholar] [CrossRef]
- Ronoud, S.; Asadi, S. An evolutionary deep belief network extreme learning-based for breast cancer diagnosis. Soft Comput. 2019, 23, 13139–13159. [Google Scholar] [CrossRef]
- Muduli, D.; Dash, R.; Majhi, B. Automated breast cancer detection in digital mammograms: A moth flame optimization based ELM approach. Biomed. Signal Process. Control 2020, 59, 101912. [Google Scholar] [CrossRef]
- Tiwari, S. Ultimate Journey of GANs. 2019. Available online: https://medium.com/analytics-vidhya/ultimate-journey-of-gans-7e5df53b01b (accessed on 19 July 2021).
- Shams, S.; Platania, R.; Zhang, J.; Kim, J.; Lee, K.; Park, S.J. Deep generative breast cancer screening and diagnosis. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2018; pp. 859–867. [Google Scholar]
- Singh, V.K.; Rashwan, H.A.; Romani, S.; Akram, F.; Pandey, N.; Sarker, M.M.K.; Saleh, A.; Arenas, M.; Arquez, M.; Puig, D.; et al. Breast tumor segmentation and shape classification in mammograms using generative adversarial and convolutional neural network. Expert Syst. Appl. 2020, 139, 112855. [Google Scholar] [CrossRef]
- Thuy, M.B.H.; Hoang, V.T. Fusing of deep learning, transfer learning and gan for breast cancer histopathological image classification. In International Conference on Computer Science, Applied Mathematics and Applications; Springer: Cham, Switzerland, 2019; pp. 255–266. [Google Scholar]
- Guan, S.; Loew, M. Breast cancer detection using synthetic mammograms from generative adversarial networks in convolutional neural networks. J. Med. Imaging 2019, 6, 031411. [Google Scholar] [CrossRef]
- Fan, M.; Liu, Z.; Xu, M.; Wang, S.; Zeng, T.; Gao, X.; Li, L. Generative adversarial network-based super-resolution of diffusion-weighted imaging: Application to tumour radiomics in breast cancer. NMR Biomed. 2020, 33, e4345. [Google Scholar] [CrossRef] [PubMed]
- Swiecicki, A.; Buda, M.; Saha, A.; Li, N.; Ghate, S.V.; Walsh, R.; Mazurowski, M.A. Generative adversarial network-based image completion to identify abnormal locations in digital breast tomosynthesis images. In Medical Imaging 2020: Computer-Aided Diagnosis; International Society for Optics and Photonics: Bellingham, WA, USA, 2020; Volume 11314, p. 1131428. [Google Scholar]
- Tien, H.J.; Yang, H.C.; Shueng, P.W.; Chen, J.C. Cone-beam CT image quality improvement using Cycle-Deblur consistent adversarial networks (Cycle-Deblur GAN) for chest CT imaging in breast cancer patients. Sci. Rep. 2021, 11, 1133. [Google Scholar] [CrossRef] [PubMed]
- Wolberg, W.H.; Street, W.N.; Mangasarian, O.L. Breast cytology diagnosis via digital image analysis. Anal. Quant. Cytol. Histol. 1993, 15, 396–404. [Google Scholar] [PubMed]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [Green Version]
- Suckling, J.; Parker, J.; Dance, D.; Astley, S.; Hutt, I.; Boggis, C.; Ricketts, I.; Stamatakis, E.; Cerneaz, N.; Kok, S.; et al. Mammographic Image Analysis Society (MIAS) Database v1.21. 2015. Available online: https://www.repository.cam.ac.uk/handle/1810/250394 (accessed on 19 July 2021).
- Suckling, J.P. The mammographic image analysis society digital mammogram database. Digit. Mammo 1994, 1069, 375–386. [Google Scholar]
- Armato, S.G., III; McLennan, G.; Bidaut, L.; McNitt-Gray, M.F.; Meyer, C.R.; Reeves, A.P.; Zhao, B.; Aberle, D.R.; Henschke, C.I.; Hoffman, E.A.; et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A Completed Reference Database of Lung Nodules on CT Scans. Med. Phys. 2011, 38, 915–931. [Google Scholar] [CrossRef]
- Ramos-Pollán, R.; Guevara-López, M.A.; Suárez-Ortega, C.; Díaz-Herrero, G.; Franco-Valiente, J.M.; Rubio-del Solar, M.; González-de Posada, N.; Vaz, M.A.P.; Loureiro, J.; Ramos, I. Discovering mammography-based machine learning classifiers for breast cancer diagnosis. J. Med. Syst. 2012, 36, 2259–2269. [Google Scholar] [CrossRef] [PubMed]
- Borchartt, T.B.; Conci, A.; Lima, R.C.; Resmini, R.; Sanchez, A. Breast thermography from an image processing viewpoint: A survey. Signal Process. 2013, 93, 2785–2803. [Google Scholar] [CrossRef]
- Lee, R.S.; Gimenez, F.; Hoogi, A.; Miyake, K.K.; Gorovoy, M.; Rubin, D.L. A curated mammography data set for use in computer-aided detection and diagnosis research. Sci. Data 2017, 4, 170177. [Google Scholar] [CrossRef] [PubMed]
- Moreira, I.C.; Amaral, I.; Domingues, I.; Cardoso, A.; Cardoso, M.J.; Cardoso, J.S. Inbreast: Toward a full-field digital mammographic database. Acad. Radiol. 2012, 19, 236–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. A dataset for breast cancer histopathological image classification. IEEE Trans. Biomed. Eng. 2015, 63, 1455–1462. [Google Scholar] [CrossRef]
- Gelasca, E.D.; Byun, J.; Obara, B.; Manjunath, B. Evaluation and Benchmark for Biological Image Segmentation. In Proceedings of the 2008 15th IEEE International Conference on Image Processing, San Diego, CA, USA, 12–15 October 2008. [Google Scholar]
- Aresta, G.; Araújo, T.; Kwok, S.; Chennamsetty, S.S.; Safwan, M.; Alex, V.; Marami, B.; Prastawa, M.; Chan, M.; Donovan, M.; et al. Bach: Grand challenge on breast cancer histology images. Med. Image Anal. 2019, 56, 122–139. [Google Scholar] [CrossRef]
- Veta, M.; Heng, Y.J.; Stathonikos, N.; Bejnordi, B.E.; Beca, F.; Wollmann, T.; Rohr, K.; Shah, M.A.; Wang, D.; Rousson, M.; et al. Predicting breast tumor proliferation from whole-slide images: The TUPAC16 challenge. Med. Image Anal. 2019, 54, 111–121. [Google Scholar] [CrossRef] [Green Version]
- Matheus, B.R.N.; Schiabel, H. Online mammographic images database for development and comparison of CAD schemes. J. Digit. Imaging 2011, 24, 500–506. [Google Scholar] [CrossRef] [Green Version]
- Dua, D.; Graff, C. UCI Machine Learning Repository; University of California: Irvine, CA, USA, 2017. [Google Scholar]
- Sert, E.; Ertekin, S.; Halici, U. Ensemble of convolutional neural networks for classification of breast microcalcification from mammograms. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju, Korea, 11–15 July 2017; pp. 689–692. [Google Scholar]
- Bevilacqua, V.; Brunetti, A.; Triggiani, M.; Magaletti, D.; Telegrafo, M.; Moschetta, M. An optimized feed-forward artificial neural network topology to support radiologists in breast lesions classification. In Proceedings of the 2016 on Genetic and Evolutionary Computation Conference Companion, Denver, CO, USA, 20–24 July 2016; pp. 1385–1392. [Google Scholar]
- Hadad, O.; Bakalo, R.; Ben-Ari, R.; Hashoul, S.; Amit, G. Classification of breast lesions using cross-modal deep learning. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, VIC, Australia, 18–21April 2017; pp. 109–112. [Google Scholar]
- Kim, D.H.; Kim, S.T.; Ro, Y.M. Latent feature representation with 3-D multi-view deep convolutional neural network for bilateral analysis in digital breast tomosynthesis. In Proceedings of the 2016 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Shanghai, China, 20–25 March 2016; pp. 927–931. [Google Scholar]
- Kumar, I.; Bhadauria, H.; Virmani, J.; Thakur, S. A classification framework for prediction of breast density using an ensemble of neural network classifiers. Biocybern. Biomed. Eng. 2017, 37, 217–228. [Google Scholar] [CrossRef]
- Duraisamy, S.; Emperumal, S. Computer-aided mammogram diagnosis system using deep learning convolutional fully complex-valued relaxation neural network classifier. IET Comput. Vis. 2017, 11, 656–662. [Google Scholar] [CrossRef]
- Gandomkar, Z.; Brennan, P.C.; Mello-Thoms, C. MuDeRN: Multi-category classification of breast histopathological image using deep residual networks. Artif. Intell. Med. 2018, 88, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Yu, J.; Han, T.; Chang, H.J.; Park, E. A method for classifying medical images using transfer learning: A pilot study on histopathology of breast cancer. In Proceedings of the 2017 IEEE 19th International Conference on e-Health Networking, Applications and Services (Healthcom), Dalian, China, 12–15 October 2017; pp. 1–4. [Google Scholar]
- Zhang, X.; Zhang, Y.; Han, E.Y.; Jacobs, N.; Han, Q.; Wang, X.; Liu, J. Whole mammogram image classification with convolutional neural networks. In Proceedings of the 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Kansas City, MO, USA, 13–16 November 2017; pp. 700–704. [Google Scholar]
- Samala, R.K.; Chan, H.P.; Hadjiiski, L.M.; Helvie, M.A.; Richter, C.; Cha, K. Evolutionary pruning of transfer learned deep convolutional neural network for breast cancer diagnosis in digital breast tomosynthesis. Phys. Med. Biol. 2018, 63, 095005. [Google Scholar] [CrossRef]
- Jaffar, M.A. Deep learning based computer aided diagnosis system for breast mammograms. Int. J. Adv. Comput. Sci. Appl. 2017, 7, 286–290. [Google Scholar]
- Han, Z.; Wei, B.; Zheng, Y.; Yin, Y.; Li, K.; Li, S. Breast cancer multi-classification from histopathological images with structured deep learning model. Sci. Rep. 2017, 7, 4172. [Google Scholar] [CrossRef]
- Fathy, W.E.; Ghoneim, A.S. A Deep Learning Approach for Breast Cancer Mass Detection. Int. J. Adv. Comput. Sci. Appl. 2019, 10, 175–182. [Google Scholar] [CrossRef]
- Khan, S.; Islam, N.; Jan, Z.; Din, I.U.; Rodrigues, J.J.C. A novel deep learning based framework for the detection and classification of breast cancer using transfer learning. Pattern Recognit. Lett. 2019, 125, 1–6. [Google Scholar] [CrossRef]
- Vesal, S.; Ravikumar, N.; Davari, A.; Ellmann, S.; Maier, A. Classification of breast cancer histology images using transfer learning. In International Conference Image Analysis and Recognition; Springer: Cham, Switzerland, 2018; pp. 812–819. [Google Scholar]
- Vo, D.M.; Nguyen, N.Q.; Lee, S.W. Classification of breast cancer histology images using incremental boosting convolution networks. Inf. Sci. 2019, 482, 123–138. [Google Scholar] [CrossRef]
- Fujioka, T.; Yashima, Y.; Oyama, J.; Mori, M.; Kubota, K.; Katsuta, L.; Kimura, K.; Yamaga, E.; Oda, G.; Nakagawa, T.; et al. Deep-learning approach with convolutional neural network for classification of maximum intensity projections of dynamic contrast-enhanced breast magnetic resonance imaging. Magn. Reson. Imaging 2021, 75, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, C.D.L.; Silva, S.D.d.S.; Silva, T.A.d.; Pereira, W.C.d.A.; Costa, M.G.F.; Costa Filho, C.F.F. Breast tumor classification in ultrasound images using support vector machines and neural networks. Res. Biomed. Eng. 2016, 32, 283–292. [Google Scholar] [CrossRef] [Green Version]
- Arefan, D.; Talebpour, A.; Ahmadinejhad, N.; Asl, A.K. Automatic breast density classification using neural network. J. Instrum. 2015, 10, T12002. [Google Scholar] [CrossRef]
- Khan, M.H.M. Automated breast cancer diagnosis using artificial neural network (ANN). In Proceedings of the 2017 3rd Iranian Conference on Intelligent Systems and Signal Processing (ICSPIS), Shahrood, Iran, 20–21 December 2017; pp. 54–58. [Google Scholar]
- Wan, T.; Cao, J.; Chen, J.; Qin, Z. Automated grading of breast cancer histopathology using cascaded ensemble with combination of multi-level image features. Neurocomputing 2017, 229, 34–44. [Google Scholar] [CrossRef]
- Fonseca, P.; Mendoza, J.; Wainer, J.; Ferrer, J.; Pinto, J.; Guerrero, J.; Castaneda, B. Automatic breast density classification using a convolutional neural network architecture search procedure. In Medical Imaging 2015: Computer-Aided Diagnosis; International Society for Optics and Photonics: Bellingham, WA, USA, 2015; Volume 9414, p. 941428. [Google Scholar]
- Kumar, D.; Kumar, C.; Shao, M. Cross-database mammographic image analysis through unsupervised domain adaptation. In Proceedings of the 2017 IEEE International Conference on Big Data (Big Data), Boston, MA, USA, 11–14 December 2017; pp. 4035–4042. [Google Scholar]
- Benzebouchi, N.E.; Azizi, N.; Ayadi, K. A computer-aided diagnosis system for breast cancer using deep convolutional neural networks. In Computational Intelligence in Data Mining; Springer: Singapore, 2019; pp. 583–593. [Google Scholar]
- Sha, Z.; Hu, L.; Rouyendegh, B.D. Deep learning and optimization algorithms for automatic breast cancer detection. Int. J. Imaging Syst. Technol. 2020, 30, 495–506. [Google Scholar] [CrossRef]
- Cui, C.; Fan, S.; Lei, H.; Qu, X.; Zheng, D. Deep learning-based research on the influence of training data size for breast cancer pathology detection. J. Eng. 2019, 2019, 8729–8732. [Google Scholar] [CrossRef]
- Fan, K.; Wen, S.; Deng, Z. Deep Learning for Detecting Breast Cancer Metastases on WSI. In Innovation in Medicine and Healthcare Systems, and Multimedia; Springer: Singapore, 2019; pp. 137–145. [Google Scholar]
- Mambou, S.J.; Maresova, P.; Krejcar, O.; Selamat, A.; Kuca, K. Breast cancer detection using infrared thermal imaging and a deep learning model. Sensors 2018, 18, 2799. [Google Scholar] [CrossRef] [Green Version]
- Sadeghi, B.; Karimi, M.; Mazaheri, S. Automatic suspicions lesions segmentation based on variable-size windows in mammography images. Health Technol. 2021, 11, 99–110. [Google Scholar] [CrossRef]
- Dhungel, N.; Carneiro, G.; Bradley, A.P. A deep learning approach for the analysis of masses in mammograms with minimal user intervention. Med. Image Anal. 2017, 37, 114–128. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Luo, L.Y.; Dou, Q.; Chen, H.; Chen, C.; Li, G.J.; Jiang, Z.F.; Heng, P.A. Weakly supervised 3D deep learning for breast cancer classification and localization of the lesions in MR images. J. Magn. Reson. Imaging 2019, 50, 1144–1151. [Google Scholar] [CrossRef]
- Nebbia, G.; Mohamed, A.; Chai, R.; Zheng, B.; Zuley, M.; Wu, S. Deep learning of sub-regional breast parenchyma in mammograms for localized breast cancer risk prediction. In Medical Imaging 2019: Computer-Aided Diagnosis; International Society for Optics and Photonics: Bellingham, WA, USA, 2019; Volume 10950, p. 109502P. [Google Scholar]
- Divyashree, B.; Kumar, G.H. Breast Cancer Mass Detection in Mammograms Using Gray Difference Weight and MSER Detector. SN Comput. Sci. 2021, 2, 63. [Google Scholar] [CrossRef]
- Arul Edwin Raj, A.M.; Sundaram, M.; Jaya, T. Thermography based breast cancer detection using self-adaptive gray level histogram equalization color enhancement method. Int. J. Imaging Syst. Technol. 2020, 31, 854–873. [Google Scholar] [CrossRef]
- Suh, Y.J.; Jung, J.; Cho, B.J. Automated Breast Cancer Detection in Digital Mammograms of Various Densities via Deep Learning. J. Pers. Med. 2020, 10, 211. [Google Scholar] [CrossRef]
- Supriya, M.; Deepa, A.; Mythili, C. Mamographic image for breast cancer detection and identification of stages of cancer using MFFC and optimized ANFIS. J. Ambient. Intell. Humaniz. Comput. 2021, 12, 8731–8745. [Google Scholar] [CrossRef]
- Ashiba, H. A proposed framework for diagnosis and breast cancer detection. Multimed. Tools Appl. 2021, 80, 9333–9369. [Google Scholar] [CrossRef]
- Ragab, D.A.; Attallah, O.; Sharkas, M.; Ren, J.; Marshall, S. A framework for breast cancer classification using Multi-DCNNs. Comput. Biol. Med. 2021, 131, 104245. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Qin, Z.; Jing, J.; Chen, J.; Wan, T. An automatic breast cancer grading method in histopathological images based on pixel-, object-, and semantic-level features. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 1151–1154. [Google Scholar]
- McCann, M.T.; Ozolek, J.A.; Castro, C.A.; Parvin, B.; Kovacevic, J. Automated histology analysis: Opportunities for signal processing. IEEE Signal Process. Mag. 2014, 32, 78–87. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, H.; Li, Y.; Wang, S.; Cheng, L.; Li, J. 3D multi-view tumor detection in automated whole breast ultrasound using deep convolutional neural network. Expert Syst. Appl. 2021, 168, 114410. [Google Scholar] [CrossRef]
- Parekh, V.S.; Macura, K.J.; Harvey, S.C.; Kamel, I.R.; EI-Khouli, R.; Bluemke, D.A.; Jacobs, M.A. Multiparametric deep learning tissue signatures for a radiological biomarker of breast cancer: Preliminary results. Med. Phys. 2020, 47, 75–88. [Google Scholar] [CrossRef]
- Ochoa-Albiztegui, R.E.; Sevilimedu, V.; Horvat, J.V.; Thakur, S.B.; Helbich, T.H.; Trattnig, S.; Morris, E.A.; Reiner, J.S.; Pinker, K. Pharmacokinetic Analysis of Dynamic Contrast-Enhanced Magnetic Resonance Imaging at 7T for Breast Cancer Diagnosis and Characterization. Cancers 2020, 12, 3763. [Google Scholar] [CrossRef]
- Mambou, S.; Krejcar, O.; Maresova, P.; Selamat, A.; Kuca, K. Novel four stages classification of breast cancer using infrared thermal imaging and a deep learning model. In International Work-Conference on Bioinformatics and Biomedical Engineering; Springer: Cham, Switzerland, 2019; pp. 63–74. [Google Scholar]
- Litjens, G.; Sánchez, C.I.; Timofeeva, N.; Hermsen, M.; Nagtegaal, I.; Kovacs, I.; Hulsbergen-Van De Kaa, C.; Bult, P.; Van Ginneken, B.; Van Der Laak, J. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci. Rep. 2016, 6, 26286. [Google Scholar] [CrossRef] [Green Version]
- Cireşan, D.C.; Giusti, A.; Gambardella, L.M.; Schmidhuber, J. Mitosis detection in breast cancer histology images with deep neural networks. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2013; pp. 411–418. [Google Scholar]
- Chen, H.; Dou, Q.; Wang, X.; Qin, J.; Heng, P.A. Mitosis detection in breast cancer histology images via deep cascaded networks. In Proceedings of the Thirtieth AAAI Conference on Artificial Intelligence, Phoenix, AZ, USA, 12–17 February 2016. [Google Scholar]
- Saha, M.; Chakraborty, C.; Racoceanu, D. Efficient deep learning model for mitosis detection using breast histopathology images. Comput. Med. Imaging Graph. 2018, 64, 29–40. [Google Scholar] [CrossRef]
- Balkenhol, M.C.; Tellez, D.; Vreuls, W.; Clahsen, P.C.; Pinckaers, H.; Ciompi, F.; Bult, P.; van der Laak, J.A. Deep learning assisted mitotic counting for breast cancer. Lab. Investig. 2019, 99, 1596–1606. [Google Scholar] [CrossRef] [PubMed]
- Elmore, J.G.; Jackson, S.L.; Abraham, L.; Miglioretti, D.L.; Carney, P.A.; Geller, B.M.; Yankaskas, B.C.; Kerlikowske, K.; Onega, T.; Rosenberg, R.D.; et al. Variability in interpretive performance at screening mammography and radiologists’ characteristics associated with accuracy. Radiology 2009, 253, 641–651. [Google Scholar] [CrossRef]
- Suzuki, S.; Zhang, X.; Homma, N.; Ichiji, K.; Sugita, N.; Kawasumi, Y.; Ishibashi, T.; Yoshizawa, M. Mass detection using deep convolutional neural network for mammographic computer-aided diagnosis. In Proceedings of the 2016 55th Annual Conference of the Society of Instrument and Control Engineers of Japan (SICE)., Tsukuba, Japan, 20–23 September 2016; pp. 1382–1386. [Google Scholar]
- Akselrod-Ballin, A.; Karlinsky, L.; Hazan, A.; Bakalo, R.; Horesh, A.B.; Shoshan, Y.; Barkan, E. Deep learning for automatic detection of abnormal findings in breast mammography. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support; Springer: Cham, Switzerland, 2017; pp. 321–329. [Google Scholar]
- Akselrod-Ballin, A.; Chorev, M.; Shoshan, Y.; Spiro, A.; Hazan, A.; Melamed, R.; Barkan, E.; Herzel, E.; Naor, S.; Karavani, E.; et al. Predicting breast cancer by applying deep learning to linked health records and mammograms. Radiology 2019, 292, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Arevalo, J.; González, F.A.; Ramos-Pollán, R.; Oliveira, J.L.; Lopez, M.A.G. Representation learning for mammography mass lesion classification with convolutional neural networks. Comput. Methods Programs Biomed. 2016, 127, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthy, S.S.; Rajaguru, H. Automatic Detection and Classification of Mammograms Using Improved Extreme Learning Machine with Deep Learning. IRBM 2021, in press. [Google Scholar] [CrossRef]
- Dembrower, K.; Liu, Y.; Azizpour, H.; Eklund, M.; Smith, K.; Lindholm, P.; Strand, F. Comparison of a deep learning risk score and standard mammographic density score for breast cancer risk prediction. Radiology 2019, 294, 265–272. [Google Scholar] [CrossRef]
- Al-Masni, M.A.; Al-Antari, M.A.; Park, J.M.; Gi, G.; Kim, T.Y.; Rivera, P.; Valarezo, E.; Choi, M.T.; Han, S.M.; Kim, T.S. Simultaneous detection and classification of breast masses in digital mammograms via a deep learning YOLO-based CAD system. Comput. Methods Programs Biomed. 2018, 157, 85–94. [Google Scholar] [CrossRef]
- Ribli, D.; Horváth, A.; Unger, Z.; Pollner, P.; Csabai, I. Detecting and classifying lesions in mammograms with deep learning. Sci. Rep. 2018, 8, 4165. [Google Scholar] [CrossRef] [Green Version]
- Cao, Z.; Duan, L.; Yang, G.; Yue, T.; Chen, Q.; Fu, H.; Xu, Y. Breast tumor detection in ultrasound images using deep learning. In International Workshop on Patch-based Techniques in Medical Imaging; Springer: Cham, Switzerland, 2017; pp. 121–128. [Google Scholar]
- Wang, N.; Bian, C.; Wang, Y.; Xu, M.; Qin, C.; Yang, X.; Wang, T.; Li, A.; Shen, D.; Ni, D. Densely deep supervised networks with threshold loss for cancer detection in automated breast ultrasound. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2018; pp. 641–648. [Google Scholar]
- Liu, J.; Li, W.; Zhao, N.; Cao, K.; Yin, Y.; Song, Q.; Chen, H.; Gong, X. Integrate domain knowledge in training CNN for ultrasonography breast cancer diagnosis. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2018; pp. 868–875. [Google Scholar]
- Tessa S, K.J. The Difference between an MRI and CT Scan. 2018. Available online: https://www.healthline.com/health/ct-scan-vs-mri (accessed on 12 July 2021).
- Zhang, L.; Luo, Z.; Chai, R.; Arefan, D.; Sumkin, J.; Wu, S. Deep-learning method for tumor segmentation in breast DCE-MRI. In Medical Imaging 2019: Imaging Informatics for Healthcare, Research, and Applications; International Society for Optics and Photonics: Bellingham, WA, USA, 2019; Volume 10954, p. 109540F. [Google Scholar]
- Zhang, L.; Mohamed, A.A.; Chai, R.; Guo, Y.; Zheng, B.; Wu, S. Automated deep learning method for whole-breast segmentation in diffusion-weighted breast MRI. J. Magn. Reson. Imaging 2020, 51, 635–643. [Google Scholar] [CrossRef]
- Rasti, R.; Teshnehlab, M.; Phung, S.L. Breast cancer diagnosis in DCE-MRI using mixture ensemble of convolutional neural networks. Pattern Recognit. 2017, 72, 381–390. [Google Scholar] [CrossRef] [Green Version]
- Amit, G.; Ben-Ari, R.; Hadad, O.; Monovich, E.; Granot, N.; Hashoul, S. Classification of breast MRI lesions using small-size training sets: Comparison of deep learning approaches. In Medical Imaging 2017: Computer-Aided Diagnosis; International Society for Optics and Photonics: Bellingham, WA, USA, 2017; Volume 10134, p. 101341H. [Google Scholar]
- Redman, A.; Lowes, S.; Leaver, A. Imaging techniques in breast cancer. Surgery 2016, 34, 8–18. [Google Scholar] [CrossRef]
- Doganay, E.; Li, P.; Luo, Y.; Chai, R.; Guo, Y.; Wu, S. Breast cancer classification from digital breast tomosynthesis using 3D multi-subvolume approach. In Medical Imaging 2020: Imaging Informatics for Healthcare, Research, and Applications; International Society for Optics and Photonics: Bellingham, WA, USA, 2020; Volume 11318, p. 113180D. [Google Scholar]
- Conant, E.; Toledano, A.Y.; Periaswamy, S.; Fotin, S.; Go, J.; Pike, J.; Hoffmeister, J. Case examples to demonstrate positive and negative impacts of a deep learning based concurrent artificial intelligence system for digital breast tomosynthesis. In Proceedings of the European Congress of Radiology 2019, Vienna, Austria, 27 February–3 March 2019. [Google Scholar]
- Doganay, E.; Luo, Y.; Gao, L.; Li, P.; Berg, W.; Wu, S. Performance comparison of different loss functions for digital breast tomosynthesis classification using 3D deep learning model. In Medical Imaging 2020: Computer-Aided Diagnosis; International Society for Optics and Photonics: Bellingham, WA, USA, 2020; Volume 11314, p. 113141A. [Google Scholar]
- Sakai, A.; Onishi, Y.; Matsui, M.; Adachi, H.; Teramoto, A.; Saito, K.; Fujita, H. A method for the automated classification of benign and malignant masses on digital breast tomosynthesis images using machine learning and radiomic features. Radiol. Phys. Technol. 2020, 13, 27–36. [Google Scholar] [CrossRef]
- Singh, D.; Singh, A.K. Role of image thermography in early breast cancer detection-Past, present and future. Comput. Methods Programs Biomed. 2020, 183, 105074. [Google Scholar] [CrossRef]
- Ekici, S.; Jawzal, H. Breast cancer diagnosis using thermography and convolutional neural networks. Med. Hypotheses 2020, 137, 109542. [Google Scholar] [CrossRef]
- Cong, W.; Shan, H.; Zhang, X.; Liu, S.; Ning, R.; Wang, G. Deep-learning-based breast CT for radiation dose reduction. In Developments in X-ray Tomography XII; International Society for Optics and Photonics: Bellingham, WA, USA, 2019; Volume 11113, p. 111131L. [Google Scholar]
- De Buck, S.; Bertels, J.; Vanbilsen, C.; Dewaele, T.; Van Ongeval, C.; Bosmans, H.; Vandevenne, J.; Suetens, P. Automated breast cancer risk estimation on routine CT thorax scans by deep learning segmentation. In Medical Imaging 2020: Computer-Aided Diagnosis; International Society for Optics and Photonics: Bellingham, WA, USA, 2020; Volume 11314, p. 1131423. [Google Scholar]
- Silva, A.A.; Cardoso, M.R.; Rezende, L.M.; Lin, J.Q.; Guimaraes, F.; Silva, G.R.P.; Murgu, M.; Priolli, D.G.; Eberlin, M.N.; Tata, A.; et al. Multiplatform investigation of plasma and tissue lipid signatures of breast cancer using mass spectrometry tools. Int. J. Mol. Sci. 2020, 21, 3611. [Google Scholar] [CrossRef]
- Al-Wajeeh, A.S.; Salhimi, S.M.; Al-Mansoub, M.A.; Khalid, I.A.; Harvey, T.M.; Latiff, A.; Ismail, M.N. Comparative proteomic analysis of different stages of breast cancer tissues using ultra high performance liquid chromatography tandem mass spectrometer. PLoS ONE 2020, 15, e0227404. [Google Scholar] [CrossRef] [Green Version]
- Behrmann, J.; Etmann, C.; Boskamp, T.; Casadonte, R.; Kriegsmann, J.; Maaβ, P. Deep learning for tumor classification in imaging mass spectrometry. Bioinformatics 2018, 34, 1215–1223. [Google Scholar] [CrossRef] [PubMed]
- Hofvind, S.; Hovda, T.; Holen, Å.S.; Lee, C.I.; Albertsen, J.; Bjørndal, H.; Brandal, S.H.; Gullien, R.; Lømo, J.; Park, D.; et al. Digital breast tomosynthesis and synthetic 2D mammography versus digital mammography: Evaluation in a population-based screening program. Radiology 2018, 287, 787–794. [Google Scholar] [CrossRef]
- Wu, J.; Shi, J.; Li, Y.; Suo, J.; Zhang, Q. Histopathological image classification using random binary hashing based PCANet and bilinear classifier. In Proceedings of the 2016 24th European Signal Processing Conference (EUSIPCO), Budapest, Hungary, 29 August–2 September 2016; pp. 2050–2054. [Google Scholar]
- Mandala, S.; Di, T.C. ECG parameters for malignant ventricular arrhythmias: A comprehensive review. J. Med. Biol. Eng. 2017, 37, 441–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalmış, M.U.; Litjens, G.; Holland, K.; Setio, A.; Mann, R.; Karssemeijer, N.; Gubern-Mérida, A. Using deep learning to segment breast and fibroglandular tissue in MRI volumes. Med. Phys. 2017, 44, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Halicek, M.; Ortega, S.; Fabelo, H.; Lopez, C.; Lejeune, M.; Callico, G.M.; Fei, B. Conditional generative adversarial network for synthesizing hyperspectral images of breast cancer cells from digitized histology. In Medical Imaging 2020: Digital Pathology; International Society for Optics and Photonics: Bellingham, WA, USA, 2020; Volume 11320, p. 113200U. [Google Scholar]
- Yala, A.; Lehman, C.; Schuster, T.; Portnoi, T.; Barzilay, R. A deep learning mammography-based model for improved breast cancer risk prediction. Radiology 2019, 292, 60–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alzubaidi, L.; Fadhel, M.A.; Al-Shamma, O.; Zhang, J.; Santamaría, J.; Duan, Y.; R Oleiwi, S. Towards a better understanding of transfer learning for medical imaging: A case study. Appl. Sci. 2020, 10, 4523. [Google Scholar] [CrossRef]
- Bezerra, L.; Ribeiro, R.; Lyra, P.; Lima, R. An empirical correlation to estimate thermal properties of the breast and of the breast nodule using thermographic images and optimization techniques. Int. J. Heat Mass Transf. 2020, 149, 119215. [Google Scholar] [CrossRef]
| Survey | Taxonomy | Datasets | Imaging Modalities | Evaluation Metrics | Challenges | Deep-Learning Architectures | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ref. | Year | ANN | Autoencoder | DBN | CNN | ELM | GAN | |||||
| [27] | 2017 | ✗ | ✓ | ✗ | ✓ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ |
| [28] | 2018 | ✗ | ✓ | ✗ | ✗ | ✓ | ✓ | ✓ | ✓ | ✓ | ✗ | ✗ |
| [29] | 2018 | ✗ | ✓ | ✓ | ✓ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ |
| [30] | 2019 | ✗ | ✗ | ✓ | ✗ | ✓ | ✓ | ✗ | ✗ | ✓ | ✗ | ✗ |
| [31] | 2019 | ✗ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✗ | ✗ |
| [32] | 2019 | ✗ | ✗ | ✗ | ✓ | ✗ | ✗ | ✓ | ✓ | ✓ | ✗ | ✓ |
| [33] | 2020 | ✗ | ✗ | ✗ | ✗ | ✓ | ✓ | ✗ | ✗ | ✓ | ✗ | ✗ |
| [34] | 2020 | ✗ | ✓ | ✓ | ✗ | ✓ | ✓ | ✗ | ✗ | ✓ | ✗ | ✗ |
| [25] | 2020 | ✗ | ✗ | ✗ | ✗ | ✓ | ✗ | ✓ | ✗ | ✓ | ✗ | ✗ |
| [35] | 2020 | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✓ | ✗ | ✗ |
| [36] | 2021 | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✗ | ✓ | ✗ | ✗ |
| [37] | 2021 | ✗ | ✓ | ✓ | ✓ | ✓ | ✗ | ✗ | ✗ | ✓ | ✗ | ✗ |
| [26] | 2021 | ✗ | ✗ | ✗ | ✓ | ✗ | ✗ | ✓ | ✗ | ✓ | ✗ | ✓ |
| Ours | - | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Reference | Dataset | Architecture | Category | Strength | Limitation |
|---|---|---|---|---|---|
| Abbass [40] | WBCD | MPANN | BrC diagnosis | Better generalization | Absence of feature engineering |
| Karabatak and Ince [41] | WBCD | AR+NN | BrC diagnosis | Reducing feature dimensions | Inadequate model evaluation |
| Rouhi et al. [44] | MIAS, DDSM | ANN | Mammography, image segmentation | Correctly identify small mass lesion | Insufficient images |
| Jafari-Marandi et al. [42] | WDBC | LS-SOED | BrC diagnosis | Driven to better decision-making | Inclusion of the dataset’s missing values |
| Becker et al. [43] | BCDR | ANN | Mammography, BrC detection | Correctly identify small mass lesion | Insufficient images |
| Reference | Dataset | Architecture | Category | Strength | Limitation |
|---|---|---|---|---|---|
| Xu et al. [45] | PD | SSAE + Softmax | Histopathology, nuclei patch classification, unsupervised learning | High-level feature learning | Limited images |
| Xu et al. [46] | PLOS 2018 | SSAE + Softmax | Histopathology, nuclei detection, unsupervised Learning | Lower computation time | Imbalanced data |
| Cheng et al. [51] | PD | SDAE | Ultrasound, supervised learning, breast-lesion classification | Adequate model evaluation | Absence of model comparison |
| Kadam et al. [47] | WDBC | FE-SSAE-SM | Feature ensemble learning, BrC classification | Adequate evaluation | Absence of data-preprocessing techniques |
| Feng et al. [50] | BCC | SDAE + Softmax | Histopathology, nuclei classification, unsupervised feature learning | Utilizing robust features of breast cancer nuclei | Insufficient images |
| Reference | Dataset | Architecture | Category | Strength | Limitation |
|---|---|---|---|---|---|
| Abdel-Zaher and Eldeib [54] | WBCD | DBN-NN | Unsupervised learning, supervised learning, BrC classification | Tested on several train-test partition | May suffer from overfitting issue |
| Zhang et al. [59] | PD | PGBM | Ultrasound (shear-wave elastography (SWE)), feature extraction, classifying breast tumor | Utilized a different ultrasound technique | Higher training time |
| Dhungel et al. [56] | DDSM-BCRP, INbreast | DBN | Mammography, segmentation of masses, structured learning | Can learn complex features | Inadequate model evaluation |
| Dhungel et al. [57] | DDSM-BCRP, INbreast | CRF | Mammography, segmentation of masses, structured output learning | Significantly faster model | Inadequate model evaluation |
| Al-antari et al. [58] | DDSM | DBN | Mammography, automatic mass detection | Feature engineering | Higher error rate for confusing benign with malignant |
| Khademi and Nedialkov [55] | WDBC, WOBC | DBN | Breast cancer diagnosis | The integration of microarray and clinical data | Comparison with ML models instead of other DL models |
| Reference | Dataset | Architecture | Category | Strength | Limitation |
|---|---|---|---|---|---|
| Arevalo et al. [7] | BCDR | CNN (UDM) | Mammography, mass lesion classification | omparison with a pre-trained model | Simple architecture |
| Spanhol et al. [60] | BreakHis | CNN (UDM) | Histopathology, image classification | Used high-resolution histopathological images | For training, only small patches of the images are used. |
| Albarqouni et al. [72] | Crowdsourcing | AggNet | Histopathology, mitosis Detection | Tested with a benchmark dataset | unreliable (crowd) annotations |
| Xu et al. [65] | PD | DCNN (COM) | Histopathology, image segmentation and classification | Can learn complex features | Insufficient images |
| Kooi et al. [66] | PD | CNN | Mammography, breast mass lesion classification | Focused on the detection of solid, malignant lesions including architectural distortions | Absence of benign lesions in training set |
| Araújo et al. [71] | BICBH | CNN (UDM) | Histopathology, image-wise classification, patch-wise classification | Multi-class classification | Limited images |
| Samala et al. [67] | PD | DLCNN (UDM) | Digital breast tomosynthesis, recognition of microcalcification | Learns complex patterns | Limited images |
| Ting et al. [70] | MIAS | CNNI-BCC (UDM) | Mammography, breast-lesion classification | Feature-wise data augmentation | Limited images |
| Yan et al. [68] | PD | CNN+RNN | Histopathology, pathological image classification | Released a larger and more diverse dataset | Lack of data pre-processing |
| Wang et al. [69] | BreakHis | CNN (UDM) | Histopathology, BrC binary classification, deep feature fusion, and enhanced routing | Classification is conducted for different magnification factors | Absence of image pre-processing |
| Reference | Dataset | Architecture | Category | Strength | Limitation |
|---|---|---|---|---|---|
| Huynh et al. [63] | PD | AlexNet | Mammography, feature extraction, breast-mass classification | Automatic lesion segmentation | Inadequate model evaluation |
| Samala et al. [73] | DDSM | CNN (FTM-ML) | Mammography, mass classification | Multi-task transfer learning | Absence of model comparison |
| Chougrad et al. [77] | DDSM, BCDR, INbreast | VGG16, ResNet50 and Inception v3 (FTM-ML) | Mammography, mass-lesion classification | Merged three datasets | Inadequate model evaluation |
| Xie et al. [22] | BreakHis | CNN (FTM-LL) | Histopatology, multi-class classififaction, clustering analysis | Solved the unbalanced distribution of samples | Lack of image pre-processing |
| Mendel et al. [74] | PD | CNN (FTM-ML) | Mammography, digital breast tomosynthesis, classification | Leave-one-out step-wise feature selection was used to eliminate redundant features. | Lack of training data |
| Kumar et al. [78] | BreakHis | VGGNet-16 (FTM-ML) | Histopathology, feature extraction, image classification | Analysis of effects of image pre-processing | Accuracy is influenced by magnification |
| Yu et al. [64] | PD | CNN (FTM-ML) | Histopathology, image classification | Images are collected via the internet. | The quality of the images could be inadequate. |
| Hu et al. [61] | PD | CNN (FTM-ML) | MRI, feature extraction | Pre-processing, large dataset, and extended training times are not required | Issue of class imbalance |
| Reference | Dataset | Architecture | Category | Strength | Limitation |
|---|---|---|---|---|---|
| Toğaçar et al. [79] | BreakHis | BreastNet | Histopathology, BrC diagnosis | Can be used in all microscopic images at different magnification rates | Absence of data pre-processing |
| Gour et al. [80] | BreakHis | ResHist | Histopathology, lassification of benign and malignant | Preserves the global information of histopathological images | Consumes a lot of processing power |
| Hu et al. [81] | BreakHis | myResNet-34 | Histopathology, malignancy-and-benign classification | Automatic target image generation | Significant rate of misclassification |
| Singh et al. [83] | PD | ResNet | Mammography, digital breast tomosynthesis, multi-class classification | The approach is simple and can be applied in different imaging | Only patch-level images are used to train the model. |
| Li et al. [84] | PD, INbreast | ResNet50 | Mammographic density classification | Combination of deep residual networks with integrated dilated convolutions and attention methods | Imbalance classes |
| Reference | Dataset | Architecture | Category | Strength | Limitation |
|---|---|---|---|---|---|
| Lahoura et al. [85] | WBCD | ELM | Feature selection, cloud environment, BrC diagnosis | Consideration of feature engineering | Absence of image pre-processing technique |
| Wang et al. [89] | PD (Mamograms) | ELM | Mass detection, feature extraction, Clustering | Feature fusion | Insufficient data |
| NEMISSI et al. [87] | WBCD | ELM | BrC diagnosis, genetic algorithm | Higher generalization performance | Inadequate evaluation |
| Ronoud and Asadi [90] | WDBC | ELM (DBN+ELM+BP) | BrC diagnosis, ensemble approach | Parameter tuning | |
| Wang et al. [88] | BreaKHis, ImageNet | ELM (ICELM) | Feature extraction, double-step deep transfer learning, BrC diagnosis | A novel method | Not an end-to-end architecture |
| Toprak [86] | WBCD | ELM | Detection and characterization of benign and malignant types | ELM is superior to other methods in performance and speed | Imbalance classes |
| Muduli et al. [91] | WBCD | ELM | Classification of breast masses, feature extraction and reduction | The generalization performance is improved | There is a possibility of data loss |
| Reference | Dataset | Architecture | Category | Strength | Limitation |
|---|---|---|---|---|---|
| Guan and Loew [96] | DDSM | GAN | Image augmentation, BrC diagnosis | Sufficient images | GAN is only used as image generator |
| Shams et al. [93] | WBCD | GAN (DiaGRAM) | BrC diagnosis | Enhanced feature learning | |
| Thuy and Hoang [95] | BreaKHis | GAN (styleGAN, Pix2Pix) | Image augmentation | Feature extraction with VGG16 and VGG19 | Generated images contain noise and affected the classifiers accuracy |
| Singh et al. [94] | DDSM, INbreast | GAN (cGAN) | Breast-tumor segmentation | Works well on limited training samples | Tumor segmentation from full-mammograms has a low accuracy |
| Fan et al. [97] | PD (DCE-MRI images) | GAN (SRGAN) | Image augmentation, BrC diagnosis | Generated super resolution ADC images | There is no conventional medical process that uses ADC images |
| Swiecicki et al. [98] | PD | GAN | Digital breast tomosynthesis, image completion, abnormality detection | Able to identify suspicious regions without the need for training images containing abnormalities | Inadequate model evaluation |
| Tien et al. [99] | PD | GAN | Computed tomography image-quality improvement | Can convert blurred images into clear images | Only for chest region |
| SL | Dataset Name | Category | No. of Images | Classes | Image Format | Resolution | URL |
|---|---|---|---|---|---|---|---|
| 1 | DDSM [107] | Mammograms | 10,480 | Benign, cancer, normal, benign without callback (bwc) | .JPEG | 16-bit | http://www.eng.usf.edu/cvprg/Mammography/Database.html (accessed on 1 October 2021) |
| 2 | MIAS [102] | Mammograms | 322 | Benign, malignant, normal | .PGM | 8 bit | https://www.repository.cam.ac.uk/handle/1810/250394 (accessed on 1 October 2021) |
| 3 | mini-MIAS [103] | Mammograms | 322 | Calcification, circumscribed masses, spiculated masses, other/ill-defined masses, architectural distortion, asymmetry, normal | .PGM | 1024 × 1024 pixels | http://peipa.essex.ac.uk/info/mias.html (accessed on 1 October 2021) |
| 4 | CBIS-DDSM [101] | Mammograms | 1644 | Normal, benign, and malignant | .DICOM | 16-bit | https://wiki.cancerimagingarchive.net/display/Public/CBIS-DDSM (accessed on 1 October 2021) |
| 5 | INBreast [108] | Mammograms | 410 | Benign, malignant, normal | .DICOM | 14-bit | http://medicalresearch.inescporto.pt/breastresearch/index.php/Get_INbreast_Database (accessed on 1 October 2021) |
| 6 | UPMC | Tomography and mamograms | - - | Hamartoma, invasive ductal carcinoma (IDC), asymmetry, lobular carcinoma, papilloma, calcifications | .DICOM | - | https://www.dclunie.com/pixelmedimagearchive/upmcdigitalmammotomocollection/index.html (accessed on 7 October 2021) |
| 7 | BICBH [71] | Histology images | 259 | normal, benign, in situ carconima and invasive carcinoma | .TIFF | - - | https://rdm.inesctec.pt/dataset/nis-2017-003 (accessed on 7 October 2021) |
| http://www.bioimaging2015.ineb.up.pt/dataset.html (accessed on 7 October 2021) | |||||||
| 8 | BreakHis [109] | Histology images | 7909 | Benign and malignant | .PNG | 8-bit | https://web.inf.ufpr.br/vri/databases/breast-cancer-histopathological-database-breakhis/ (accessed on 10 October 2021) |
| 9 | BCC [110] | Histology images | 58 | Malignant, benignant | .TIFF | 896 × 768 pixels, 768 × 512 pixels | http://bioimage.ucsb.edu/research/bio-segmentation (accessed on 10 October 2021) |
| 10 | BACH [111] | Histology images | 400 | Normal, benign, in situ carcinoma, invasive carcinoma | . TIFF | 2048 × 1536 pixels | https://iciar2018-challenge.grand-challenge.org/Dataset/ (accessed on 10 October 2021) |
| 11 | TUPAC16 [112] | Histology images | 500 | - - | .SVS | 50 k × 50 k pixels | https://tupac.grand-challenge.org/Dataset/ (accessed on 10 October 2021) |
| 12 | IDC [14] | Histology images | 162 | Invasive ductal carcinoma (IDC), non-IDC | .PNG | - - | https://www.kaggle.com/paultimothymooney/breast-histopathology-images (accessed on 14 October 2021) |
| 13 | MITOS-ATYPIA 14 | Histology images | - | Mitosis and nuclear atypia | .TIFF | 1539 × 1376 pixels, 1663 × 1485 pixels | https://mitos-atypia-14.grand-challenge.org/dataset/ (accessed on 14 October 2021) |
| 14 | DMR-IR | Infrared Images | - - | - - | - - | 640 × 480 pixels | http://visual.ic.uff.br/en/proeng/thiagoelias/ (accessed on 15 October 2021) |
| 15 | BCDR [105] | Mammograms and ultrasound | - - | Benign, malignant, normal | .DICOM | 720 × 1167 | https://bcdr.ceta-ciemat.es/information/about (accessed on 15 October 2021) |
| 16 | TCGA | Mammograms | 88 | - - | .DICOM | - - | http://cancergenome.nih.gov/ (accessed on 24 October 2021) |
| 17 | BancoWeb LAPIMO [113] | Mammograms | 1473 | Benign, malignant, normal | .TIFF | 12 bits | http://lapimo.sel.eesc.usp.br/bancoweb/ (accessed on 24 October 2021) |
| 18 | PLOS 2018 | Histology images | 537 | Nuclear, non-nuclear | .TIFF | 2200 × 2200 pixels | https://engineering.case.edu/centers/ccipd/data (accessed on 27 October 2021) |
| 19 | WBCD or WBCO [100] | Multivariate | 699 data | Benign, malignant | - - | - - | https://archive.ics.uci.edu/ml/datasets/breast+cancer+wisconsin+(original) (accessed on 3 October 2021) |
| 20 | WDBC [114] | Multivariate | 569 | Malignant, benign | - - | - - | https://archive.ics.uci.edu/ml/datasets/Breast+Cancer+Wisconsin+(Diagnostic)/ (accessed on 3 October 2021) |
| 21 | Histopathological images [68] | Histology images | 3771 | Normal, benign, in situ carcinoma and invasive carcinoma | - - | 2048 × 1536 pixels | http://ear.ict.ac.cn/?page_id=1616 (accessed on 3 October 2021) |
| Pre-Processing Method | Methodology | Advantages | References |
|---|---|---|---|
| Image augmentation | Geometric transformations such as rotation and flipping | To prevent the problem of overfitting. To address the issue of class imbalance in training. For improved interpretation of HP images, the network can learn lesions from several perspectives, much like a pathologist does in real life. | [7,23,50,65,68,70,71,73,78,80,115,116,117,118,119,120,121,122,123,124,125,126,127,128,129,130,131] |
| Insert noise/distortion (Gaussian noise, barrel or pin cushion transforms) | Allows for the robust training of NN It can predict with greater accuracy even when images are noisy | ||
| Patch-creation methods (patches with 50% overlapping, no overlapping, or randomly selected patches) | It can retain the image aspect ratio, architecture, or shape of the lesion, as well as subjective information. It improves the classifier’s performance while decreasing the likelihood of false negatives. It can decrease the possibility of information loss. | ||
| Synthetic minority over-sampling technique (SMOTE) | To solve the class imbalance problem before training NNs, this method increases the number of samples to the minority class. | ||
| ROI extraction | Methods such as region growing, nuclei segmentation, the Otsu method, and the Markov random model were utilized. | Increases the amount of positive and negative image samples available. Assists the neural network (NN) in learning better representations of abnormal areas and decreases the likelihood of overfitting. Reduces calculation time and resource use. | [7,44,50,51,70,73,116,118,119,120,121,124,127,132,133,134,135,136,137,138,139,140,141,142,143] |
| Scaling | Gaussian pyramid, bi-cubic interpolation, bilinear interpolation | The image must be resized before being provided as input to the NN. Carefully chosen interpolation algorithms can prevent information loss while mapping to the new pixel grid. Along with resizing, the Gaussian pyramid can assist to increase the number of images. | [51,59,65,118,119,120,121,122,125,126,133,135,136,144,125] |
| Normalization and enhancement | Histogram equalization, adaptive mean, median filters, log transforms, CLAHE method, Wiener filter, multi-threshold peripheral equalization algorithm. | Normalize the image’s low-value and high-value intensity/contrast. Adaptive filters reduce noise by taking into account mean, variation, and spatial correlations. Reduces the effects of image blurring and impulsive noise in ultrasound images. Multi-threshold peripheral equalization enhances and removes irrelevant information from mammograms. On the normalized image, ANN typically performs better. It aids in the reduction in loss during backpropagation. | [7,44,58,62,89,115,120,122,125,131,133,134,139,143,145,146,147,148,149,150,151,152] |
| Remove artifacts | Using binary images and thresholding the pixel intensity, cropping border, extracting larger regions, using geometric parabola around rib cage. | Non-breast areas (labels, wages, white strips/borders, opaque markers, lungs, thorax, chest wall, and pectoral muscle) in mammograms, US, and MRI can be reduced. | [51,121,127,133,153] |
| Stain normalization or removal | Stain normalization | To make variable color (due to H&E staining of histology images) uniform across all images for certain patients. As a consequence, NN will not be distracted by variations in brightness and color staining and will produce superior classification results for multiclass BrC. The contrast, intensity, and color characteristics of the source images are almost identical to those of the reference image. | [78,115,116,121,129,130,154] |
| Color deconvolution | To extract hematoxylin-eosin (H&E) staining intensities from HP images and to transform them into optical density space images without being considerably affected. It decreases image dimensionality, consumes fewer resources, and improves classification performance. It maintains textural information in histology images that is related with stain colors. |
| Dataset | Reference | Evaluation Metrics | Methods | Accuracy |
|---|---|---|---|---|
| MIAS, DDSM | Rouhi et al. [44] | Sensitivity, specificity, accuracy, AUC | ANN | 90.16%, 96.47% |
| DDSM | Kumar et al. [119] | Accuracy | ANN | 100% |
| BCC | Feng et al. [50] | Precision, recall, F1 Score, accuracy, mean execution time | autoencoder | 98.27% |
| - | Wu et al. [194] | Accuracy, sensitivity, specificity, and autoencoder | 95.45% | |
| WBCO, WDBC | Ronoud and Asadi [90] | Accuracy, sensitivity, specificity, NPV, PPV, | DBN+ELM | 99.75% |
| DDSM | Mandala and Di [195] | Sensitivity, specifity, accuracy | DBN | 93% |
| DDSM, PD (DBT) | Samala et al. [67] | ROC, AUC | CNN (UDM) | 0.93 (AUC) |
| BreakHis | Spanhol et al. [60] | Accuracy, patient score, patient-recognition rate, image-recognition rate | CNN(UDM) | 90% |
| MIAS | Ting et al. [70] | Accuracy, sensitivity, specificity | CNN (UDM) | 90.50% |
| PD | Yan et al. [68] | Accuracy, sensitivity, AUC | CNN (UDM) | 91.3% |
| BreakHis | Han et al. [126] | Recognition rates, accuracy | CNN(COM) | 96.9% |
| BreakHis | Kumar et al. [78] | Accuracy, F1-score | CNN (TL) | 97.01% |
| BreakHis | Toğaçar et al. [79] | Accuracy, sensitivity, specificity, precision, and F1-score | CNN (RL) | 98.70% |
| BreaKHis | Hu et al. [81] | Precision, recall, accuracy, F1-Score | CNN (RL) | 91% |
| BreaKHis | Wang et al. [88] | Accuracy, sensitivity | ELM | 98.18% |
| WBCD | Lahoura et al. [85] | Accuracy, recall, precision, F1-score | ELM | 98.68% |
| DDSM, INbreast | Shams et al. [93] | Accuracy, AUC | GAN+CNN | 89%, 93.5% |
| DDSM, INbreast | Singh et al. [94] | Accuracy | cGAN | 80% |
| Architecture/Policy | Architecture Strength | Architecture Limitation | Research Direction |
|---|---|---|---|
| ANN | Driven to better decision-making. | Not suitable for extracting spatial information | Architectures suitable for extracting spatial information are required. |
| Autoencoder | Excellent for condensing feature information. | Requires separate feature classification system, along with fine-tuned multi-stage training strategies. | Implementation of a single-stage training platform is required. |
| DBN | Requires low data on training. | DBN is not incredibly optimized for image-recognition processes. | Requires stronger fusion with convolutional architectures. |
| Transfer learning | Strong weight initialization results in achieving better accuracy from minimal training data. | Requires intuition of feature relation between pre-trained dataset and target dataset. | Transfer learning strategies should be implemented based on data relevancy. |
| Residual learning | Enables generalization of deeper architectures by auto-calibration on unnecessary features. | Requires batch-normalization resulting in adding extra computation complexity. | Unnecessary and heavy residuals should be avoided. |
| ELM | Faster learning capability with the advantage of avoiding vanishing gradients. | Hard to solve underfitting and overfitting issues. Additionally, ELM is not great at image-classification tasks. | Need to move towards deep-learning strategies or extensively improve architectural perspectives. |
| GAN | Excellent for data distribution learning and generating synthetic data. | Training sometimes falls into local-minima. Additionally, it may cause excessive focus on fixed data patterns. | Should be implemented for generating synthetic datasets and increasing the capability of cancer classifier models. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mridha, M.F.; Hamid, M.A.; Monowar, M.M.; Keya, A.J.; Ohi, A.Q.; Islam, M.R.; Kim, J.-M. A Comprehensive Survey on Deep-Learning-Based Breast Cancer Diagnosis. Cancers 2021, 13, 6116. https://doi.org/10.3390/cancers13236116
Mridha MF, Hamid MA, Monowar MM, Keya AJ, Ohi AQ, Islam MR, Kim J-M. A Comprehensive Survey on Deep-Learning-Based Breast Cancer Diagnosis. Cancers. 2021; 13(23):6116. https://doi.org/10.3390/cancers13236116
Chicago/Turabian StyleMridha, Muhammad Firoz, Md. Abdul Hamid, Muhammad Mostafa Monowar, Ashfia Jannat Keya, Abu Quwsar Ohi, Md. Rashedul Islam, and Jong-Myon Kim. 2021. "A Comprehensive Survey on Deep-Learning-Based Breast Cancer Diagnosis" Cancers 13, no. 23: 6116. https://doi.org/10.3390/cancers13236116
APA StyleMridha, M. F., Hamid, M. A., Monowar, M. M., Keya, A. J., Ohi, A. Q., Islam, M. R., & Kim, J.-M. (2021). A Comprehensive Survey on Deep-Learning-Based Breast Cancer Diagnosis. Cancers, 13(23), 6116. https://doi.org/10.3390/cancers13236116










