Simple Summary
Neuroblastoma is a pediatric cancer that arises in the sympathetic nervous system. High-risk neuroblastoma is clinically challenging and identification of novel therapies, particularly those that offer a reduction in morbidity for these patients, is a high priority. Combining genetic analyses with investigation of molecular mechanisms, while considering recent advances in our understanding of key developmental events, provides avenues for future treatment. Here we review and highlight several recently published articles that address novel molecular mechanisms arising from chromosome 1p, 2p, and 11q aberrations, which likely contribute to high-risk neuroblastoma, and discusses their potential impact on treatment options.
Abstract
Neuroblastoma is the most common extracranial solid pediatric tumor, with around 15% childhood cancer-related mortality. High-risk neuroblastomas exhibit a range of genetic, morphological, and clinical heterogeneities, which add complexity to diagnosis and treatment with existing modalities. Identification of novel therapies is a high priority in high-risk neuroblastoma, and the combination of genetic analysis with increased mechanistic understanding—including identification of key signaling and developmental events—provides optimism for the future. This focused review highlights several recent findings concerning chromosomes 1p, 2p, and 11q, which link genetic aberrations with aberrant molecular signaling output. These novel molecular insights contribute important knowledge towards more effective treatment strategies for neuroblastoma.
1. Introduction
Neuroblastoma is the most common extracranial solid tumor in children, accounting for approximately 8% of all malignancies in children and 15% of all cancer-related deaths in this population [1]. The median age at diagnosis is around 17 months, with 90% of patients younger than 5 years and diagnosis rare after 10 years of age [2]. Neuroblastoma is a heterogeneous cancer with different prognosis depending on criteria such as age at presentation, spread of disease, and biological and genetic characteristics at diagnosis [3,4,5].
Neuroblastoma is considered to originate from undifferentiated neural crest cells and often presents as tumors located in or in close proximity to the adrenal glands or sympathetic ganglia [6,7]. Developmentally, the chromaffin cells of the adrenal medulla arise from migratory neural crest cell derived sympathoadrenal precursor cells (Figure 1). These cells originate from the dorsal neuroepithelium and migrate to the vicinity of the dorsal aorta. Here, neural crest cells differentiate to give rise to sympathetic neurons and chromaffin cells of the adrenal medulla. Lineage-tracing experiments in mice have revealed that chromaffin cells in the adrenal gland arise mostly from embryonic nerve-associated cells of neural crest origin named Schwann cell precursors [8]. Single-cell RNA sequencing of the transcriptome from developing human and mouse adrenal glands, as well as from low versus high risk neuroblastoma samples, has further increased our knowledge of developmental trajectories and cellular states in neuroblastoma [8,9,10,11]. During the developmental transition from Schwann cells, cells pass through a transient cellular state where they exhibit a transcriptional identity known as the ‘bridge cell signature’. Many of the genes found in this ‘bridge cell signature’ are known to be important for differentiation, such as ASCL1, which is a regulator of neuronal differentiation. Recently it was reported that the non-phosphorylated form of ASCL1 mediates mitotic exit and neuronal differentiation [12]. Another ‘bridge cell’ gene is PHOX2B, which is a transcription factor involved in the development of noradrenergic neuron populations, and a known predisposition gene for neuroblastoma. One way in which PHOX2B might contribute to pathogenesis includes via regulation of ALK, as it has been shown to increase ALK expression in vitro. PHOX2B is currently employed as a marker for peripheral neuroblastic tumors ([13,14] and refs therein).
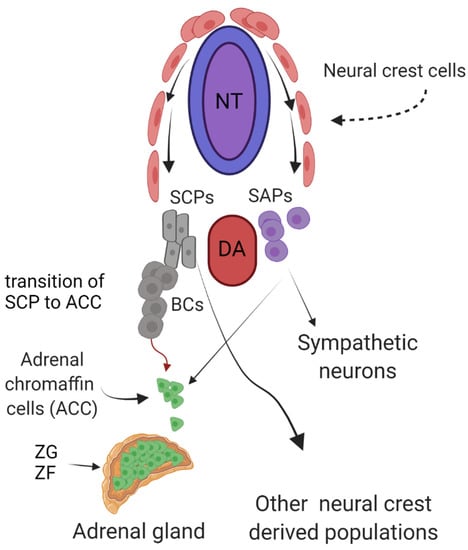
Figure 1.
Development of adrenal chromaffin cells (ACCs) from sympathoadrenal precursors (SAPs) and Schwann cell precursors (SCPs). Neural crest cells migrate from the neural tube (NT) to the dorsal aorta (DA). At the DA, SAPs differentiate into sympathetic neurons and ACCs. SCPs migrate on axons to preganglionic neurons that innervate the adrenal gland or differentiate to other neural crest derived populations, such as endoneurial fibroblast, melanocytes, parasympathetic neurons, tooth pulp cells, odontoblasts, chondrocytes, osteoblast, and enteric neurons [15]. ZG, zona glomerulosa; ZF, zona fasciculata; BCs, bridge cells.
Additionally, other important genetic markers for neuroblastoma are mutations in ALK, PTPN11, TERT, ATRX, and amplification of MYCN. In relapsed neuroblastoma cases, mutations are more frequently observed in ALK, as well as other well-known oncogenes and tumor suppressor genes, such as NF1, RAS, and RAF, and also PTPN11 and FGFR [13,16]. Furthermore, telomere maintenance mechanisms are adopted by neuroblastoma cells, favoring unlimited proliferation in the presence of genome instability and the chromosomal end replication crises. Telomerase activation is commonly observed in high-risk neuroblastoma, such as MYCN-amplified tumors and TERT-rearranged tumors. Furthermore, somatic mutations in ATRX have recently been reported as being connected with alternative lengthening of telomeres, which has recently been summarized [17]. While genetic mutations are rare in neuroblastoma, tumors often harbor many chromosomal aberrations that add to the complexity of the genetic landscape. Neuroblastoma was first suggested as a gene-dosage disorder in the 90s [18,19], and today the International Neuroblastoma Risk Group (INRG) uses a classification system based on several key criteria that include age, stage, tumor histology, MYCN amplification (MNA), 11q-deletion, and ploidy to define very low-, low-, intermediate- and high-risk groups according to 5-year event-free survival (EFS) [20,21]. Low-risk neuroblastomas are mostly observed in children less than 1.5 years old, and often undergo spontaneous differentiation or regression with little or no intervention. Intermediate neuroblastomas are usually treated with chemotherapy and surgery and have a better prognosis than cases that are classified as high-risk neuroblastoma, which also tend to present after 1.5 years [20]. Treatment of high-risk patients includes immunotherapy, radiotherapy, surgery, high dose chemotherapy, autologous stem cell transplantation, and combination treatments. Despite improved clinical treatments, the long-term survival of children and the 5-year survival with high-risk neuroblastoma is less than 50 percent (Table 1) [2,21,22].

Table 1.
Five years survival after diagnosis based on genomic profile. Overview of commonly employed genetic aberrations in neuroblastoma. Data is accumulated from >400 Swedish neuroblastoma patients [21]. MNA, MYCN-amplified; 11q-, 11q deletion; 17q+, 17q gain.
High risk neuroblastoma exhibits genetic features that include deletion of chromosome arm 1p, gain of parts of 17q, aneuploidy and amplification of the proto-oncogene MYCN, and deletion of parts of chromosome arm 11q (Figure 2). At this point, two high-risk neuroblastoma groups are commonly considered: the MYCN amplified (20–25%) group and the more common 11q-deletion (35–45%) group, which together represent currently therapeutically challenging cases. Generally, 11q-deletion and MYCN amplification do not co-exist in the same tumor, although rare cases that harbor both 11q deletion and MYCN amplification have been described and constitute a very high-risk group of neuroblastoma patients (Table 1 [21]).
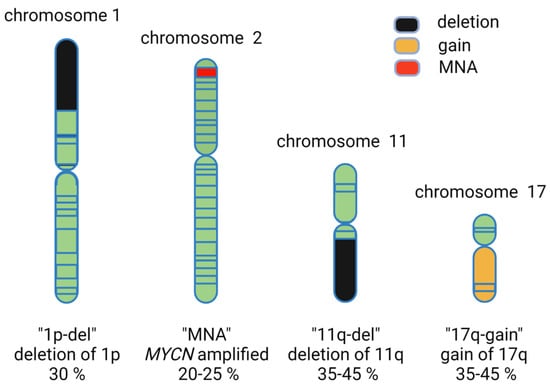
Figure 2.
Common known genetic features of neuroblastoma including deletion of chromosome arm 1p (‘1p-del’), gain of parts of 17q (‘17q-gain’), amplification of MYCN (‘MNA’), and deletion of 11q (‘11q-del’).
While MYCN amplification is conceptually easier to understand at the molecular level, the mechanisms underlying 11q-deletion in neuroblastoma have been elusive. Genetic analyses have given weight to the hypothesis that the contribution of 11q-deletion to neuroblastoma pathogenesis seems to be a result of imbalanced and aberrant cell signaling, which affects key processes—such as cell cycle, growth, differentiation, survival and likely the DNA damage response (DDR) [23]. Understanding these underlying molecular mechanisms, and ultimately placing them in a developmental context is important to understand how neuroblastoma arises and persists despite current aggressive treatment regimes. Here, we discuss recently published reports regarding molecular mechanisms in the 11q-deletion and MYCN-amplified high-risk neuroblastoma groups.
2. Novel Mechanisms Underlying the Contribution of 1p36 Deletion to MYCN Neuroblastoma
The MYCN transcription factor is a known oncoprotein, which forms complex interactions with other proteins and regulates many different target gene regulatory elements to activate or repress gene expression [24,25]. MYCN controls the expression of genes that regulate cell proliferation or cell cycle progression, maintenance of pluripotency of cells and is significantly involved in organogenesis during embryonic development [26]. MYCN is located on chromosome 2p24.3 and is amplified in 20–25% of neuroblastoma cases [27,28,29,30,31]. MYCN amplification drives neuroblastoma cell proliferation and promotes an undifferentiated neuroblastoma phenotype, which strongly correlates with worse prognosis [20,22,30,32,33]. MYCN-amplification is genetically defined as anything from 9 copies up to 500 copies, which are typically localized in double minutes or homogenously stained regions [34] High-risk MYCN-amplified neuroblastomas commonly exhibit loss of heterozygosity at chromosome 1p as well as gain of 17q, with 70% of MYCN amplified neuroblastoma associated with 1p36 deletions [24,35].
Previous genetic studies have identified candidate neuroblastoma tumor suppressor genes, including KIF1Bb, CHD5, miR-34a, ARID1A, and CAMTA1 located at 1p36 [36,37,38]. Convincing data have been presented previously for many of these loci in neuroblastoma and will not be discussed further here [39]. Two independent reports recently reassessed the impact of 1p loss of heterozygosity on the development of MYCN-driven neuroblastoma [40,41]. In one study, García-López et al. genetically deleted the 1p36 syntenic region in mouse neural crest cells (NCCs), developing a mouse model that recapitulates high-risk 1p36 loss of heterozygosity/MYCN amplified neuroblastoma [40]. This 1p36 syntenic deletion, which included the Arid1a and Chd5 tumor suppressors, resulted in increased anchorage independent growth and defects in differentiation. They went on to examine the effects of 1p36 deletion in an elegant approach that asked which genetic deletion event at 1p36 was selected for when Mycn was overexpressed in NCCs. They observed that while loss of the 1p36 syntenic region did not affect tumor penetrance, it did decrease the time to tumor onset on Mycn overexpression [40]. Interestingly, they noted that 1p36 deletion resulted in a less differentiated tumor when compared with Mycn overexpression alone. Their analysis of different candidate 1p36 tumor suppressor genes, such as Arid1a, Chd5, Camta1, and Kif1b, identified Arid1a as the gene with strongest correlation between expression levels and time to tumor onset. Overall, this work teased apart some of the challenges associated with understanding 1p36 loss of heterozygosity in MYCN-driven neuroblastoma, identifying the SWI/SNF chromatin remodeling complex subunit Arid1a as a MYCN collaborator that accelerates neuroblastoma onset [40] (Figure 2).
An independent study of the role of ARID1A in zebrafish resulted in similar conclusions [41]. Here, Shi et al. focused directly on ARID1A due to its high mutation rate across human cancers, including neuroblastoma [42,43]. They showed that deletion of the zebrafish ARID1A homologs, arid1aa and arid1ab, resulted in accelerated onset and increased penetrance of MYCN-driven neuroblastoma in a transgenic zebrafish model [41]. They further interrogated the role of ARID1A in human neuroblastoma cells, showing that genetic removal of ARID1A did not affect proliferation, but did promote cell invasion and migration. Transcriptomic analyses further allowed the authors to identify an adrenergic-to-mesenchymal transition, with neuroblastoma cells lacking ARID1A exhibiting an enriched mesenchymal gene signature [41].
Taken together, these reports support and suggest a role for the 1p36 locus and ARID1A, in neuroblastoma tumor development, where loss of ARID1A modulates onset and invasiveness and induces a mesenchymal tumor phenotype that might increase resistance to chemotherapy (Figure 2). Thus, ARID1A now joins the list of candidate tumor suppressor genes present at 1p36 that have potential to impact on neuroblastoma initiation and progression.
3. Working Alone or in Cooperation? 2p-gain and the ALKAL2, MYCN, and ALK Troika
In vertebrates, activation of the Anaplastic Lymphoma Kinase (ALK) Receptor Tyrosine Kinase (RTK) is regulated by the ALKAL ligands, which drive downstream signal transduction that includes activation of PI3K/AKT and RAS/MAPK pathways [44,45,46,47,48]. In neuroblastoma, activating mutations in ALK are found in both familial and sporadic forms, where they are now appreciated to be present in around 10% of primary cases [49,50,51,52,53]. Activating ALK mutations are associated with worse prognosis in the intermediate- and high-risk groups, and are also more commonly observed in relapsed cases [3,16,54,55]. Several groups have shown that ALK gain-of-function mutations are unable to drive neuroblastoma alone when ALK is expressed at endogenous levels [46,56,57,58]. However, one report, in which ALK is driven in the mouse neural crest by either DBHiCre or TH-IRES-Cre, resulted in tumor development, highlighting the need to consider ALK expression levels carefully in a temporal and spatial context in neuroblastoma [59]. While neither mice nor zebrafish models in which Alk has been activated by in locus knock-in events develop neuroblastoma, they do exhibit phenotypes in neural crest derived structures. For example, an enlarged sympathetic ganglia phenotype is observed with a constitutively active Alk in mice [46,57,58], while in zebrafish, activation of the ALK family RTK Ltk (Ltkmoonstone), results in production of ectopic neural crest derived iridiphore pigment cells [45,56].
It is now well accepted that ALK and MYCN cooperate to drive neuroblastoma. Mechanistically, we know that MYCN transcription is regulated by ALK signaling activity, and conversely, MYCN has been shown to drive ALK expression [60,61]. Several groups have reported that MYCN and ALK gain-of-function mutations, either in locus or overexpressed, cooperate to drive transformation, and result in increased neuroblastoma penetrance in both cell line—mouse and zebrafish—models [46,57,58,59,60,62,63]. This is in keeping with previous analyses of neuroblastoma cases, which identified a correlation between coexistence of ALK activating mutations and MYCN amplification in high-risk neuroblastoma with poor prognosis [3].
One outstanding question is the potential for misregulation of the ALKAL ligands in the development of neuroblastoma, since ligands for several of the RTKs are known to impact on tumorigenesis, a prime example being the PDGF ligands in human cancer [64]. The original reports of ALKAL activation of ALK noted that ALKAL2 was expressed in the adrenal glands and that its expression was observed in neuroblastoma patient samples [44,45,46,48,65]. Two recent studies have further addressed a potential function of ALKAL2 in neuroblastoma [46,66]. Javanmardi et al. homed in on the observation that ALKAL2 is located on chromosome 2p25.3 near the end of the chromosome, in close proximity to ALK and MYCN on chromosome 2p (Figure 3). Thus, all three loci are in the area of ‘2p-gain’, which genetically has been reported to predict poor EFS in neuroblastoma patients [67]. Moreover, there are several reports of congenital neuroblastoma in patients with germline partial trisomy of chromosome 2p [68,69,70,71,72]. Javanmardi et al. investigated 365 neuroblastoma cases, showing that approximately 30% exhibited 2p-gain (arbitrarily set as more than 4× the normal copy number) or amplification (more than eight copies) in this region [66] and references therein. Furthermore, they showed that neuroblastoma cell lines express ALKAL2 protein, identifying a potential autocrine/paracrine activation of ALK in a tumor setting.
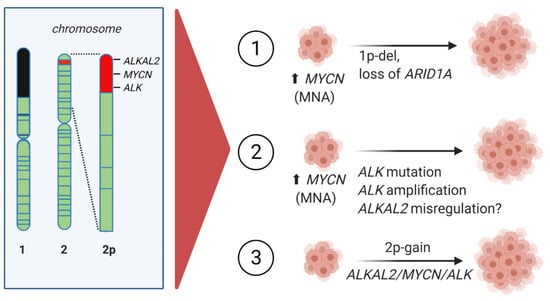
Figure 3.
Chromosome 2 chromosomal aberrations involving MYCN in high-risk NB. ALKAL2, MYCN, and ALK are localized at chromosome 2p25.3, 2p24.3, and 2p23.2, respectively. (1) MYCN amplification together with loss of 1p36, including ARID1A. (2) MYCN amplification (MNA) with ALK mutation, ALK amplification or ALKAL2 misregulation. (3) Copy number gain of chromosome 2p—2p-gain.
What happens in neuroblastoma when the troika of ALKAL2, ALK, and MYCN are aberrantly expressed, or activated, in either 2p-gain or amplification? Work from Borenäs et al. offers some insight into this question [46]. They reported that expression of the ALKAL2 ligand cooperates with MYCN to drive highly penetrant and aggressive neuroblastoma growth in mice, in the absence of ALK mutation (Figure 3). These tumors exhibited a RNA expression signature similar to that of ALK mutant/MYCN driven neuroblastoma. Importantly, they could also show that these tumors were sensitive to ALK TKI treatment [46]. This finding suggests that aberrant regulation of the ALKAL2, ALK, and MYCN troika in neuroblastoma, for example in 2p-gain, may drive ALK signaling activity and therefore could benefit from ALK TKI treatment.
4. New Candidate Tumor Suppressor Genes and Molecular Mechanisms Shed Light on 11q Deletion Neuroblastoma
An elegant cell biology experiment performed in the 1990s showed that genetic transfer of a portion of chromosome 11 (from pter to 11q22.2) consistently caused neuroblastoma cells to differentiate and blocked their ability to proliferate [73]. This dramatic finding was the first hint that this chromosomal region harbored genes important for differentiation [73]. Shortly after this, it was reported that neuroblastomas often exhibit deletion of parts of chromosome 11 [74]. Patients with 11q-deletion represent 35–45% of neuroblastoma, presenting at an older age and correlating with a more aggressive disease stage [75,76,77]. It is common that many patients with 11q-deletion (~45% of 11q-deletions) exhibit a loss of chromosome 11 in its entirety [21]. The remainder of patients with 11q-deletion (~55%) have partial loss of 11q genetic material [21], often starting at 11q13–14 and continuing out to 11qter, at the end of the chromosome. It is interesting that homozygous 11q loss has never been reported, suggesting that the tumor suppressor effect of 11q-deletion in neuroblastoma is haplosufficient. Haploinsufficiency in humans can result in a wide range of clinical outcomes, one of which is development of cancer which can occur if cell growth control or DNA repair genes are mutated [78]. While understanding haploinsufficiency, as well as potential for epigenetics to affect outcome in this context, is challenging in human cancer, a number of mouse models have reported haploinsufficiency as contributing to tumor development. These include, for example, loss of the Hint1 protein in mammary carcinogenesis, Dmp1 loss in Myc-induced lymphomas and p27Kip1 acceleration of a range of tumors [79,80,81]. In a transgenic MYCN neuroblastoma zebrafish model, tumors develop with increased penetrance when one allele of both arid1aa and arid1ab are absent, indicating a haploinsufficient effect [41]. Similarly, loss of 11q in human neuroblastoma may represent an additional example of haploinsufficiency responsible for increased tumor development, although the genes responsible have been difficult to pinpoint [23].
Over the years, several candidate genes, including CADM1, H2AFX, ATM, CHK1, MRE11, and CCND1 that lie within the chromosome 11q-deletion area, have been suggested as either driver mutations or tumor suppressor genes [23] (Figure 4). Many of these candidates fall into the categories of cellular growth control regulators or DNA repair genes that are known to contribute to tumor development in the context of haploinsufficiency [78]. In some of these cases, reduced cell proliferation, cell cycle arrest and neuronal differentiation in cell line experiments has further supported such roles [82,83,84,85]. However, no single candidate 11q gene has been confirmed to play a key role in initiation and progression of neuroblastoma, despite intensive investigation mapping of this deleted region (11q14–11qter) [23,31,74,75,86,87,88].
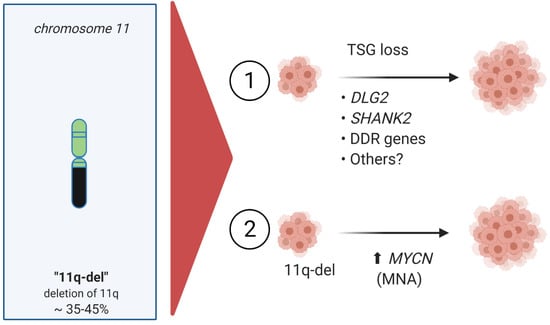
Figure 4.
High-risk neuroblastoma with 11q-deletion/non-MYCN-amplification (MNA). (1) 11q-deletion tumor suppressor gene (TSG) loss, including genes such as DLG2, Discs Large 2; SHANK2, SH3 and Multiple Ankyrin Repeat Domain 2; DDR, DNA Damage Response genes; Others, including ATM, CADM1, CHK1, MRE11, and CCND1. (2) 11q-deletion tumor suppressor gene loss combined with MYCN-amplification (MNA) comprises are rare, but very high-risk neuroblastoma group.
Three recent articles shed new light on the role of 11q-deletion in neuroblastoma from slightly different angles [21,89,90]. Two groups examined somatic structural variation using whole genome sequence and single-nucleotide polymorphism analysis of neuroblastoma samples from their own samples or from publicly available databases [89,90], while Siaw et al. employed a combination of genetics from patient neuroblastomas and developmental cell biology approaches [21]. The work of Lopez et al. analyzed a set of neuroblastoma tumors with 11q deletion that led them to propose that disruption and deregulation of the SHANK2 gene at chromosome 11q13 promotes an undifferentiated neuroblastoma state (Figure 4). SHANK2, which encodes a post-synaptic protein, is disrupted in 14% of 11q-deleted high-risk neuroblastoma. Further, overexpression of SHANK2 resulted in inhibition of neuroblastoma cell growth, as well as increased differentiation upon treatment with retinoic acid. Retinoic acid is a known differentiation factor in neuronal cells that is employed as a maintenance therapy in high-risk neuroblastoma [91,92]. Keane et al. employed public microarray data, concluding that DLG2, located on chromosome 11q14, correlated positively with better prognosis and went on to show decreased cell proliferation in 11q-deleted cell lines upon DLG2 overexpression. A third report, by Siaw et al., combined genetic data from 11q-abberant neuroblastoma tumors with computational analysis that correlated genes expressed during neural crest development together with prognosis in neuroblastoma, also identifying DLG2 as a tumor suppressor gene in neuroblastoma (Figure 4). The approach of Siaw et al., built on the developmental findings of Furlan et al. [8], hypothesizing that genes included in the ‘bridge cell signature’ would be important for the differentiation of Schwann cell precursor to adrenal chromaffin cells (Figure 1). Indeed, in the bridge signature, expression of DLG2, is upregulated in the final step of the Schwann cell precursor to adrenal chromaffin cell differentiation process [8,21]. Supporting this, Siaw et al. were able to show that overexpression of DLG2 in neuroblastoma cell lines reduced their proliferation and led to an increased transcription of adrenal specific genes, promoting differentiation to a more chromaffin-like cell type. In an interesting twist, they could also show that ALK/MAPK signaling suppressed DLG2 transcription, suggesting that oncogenic ALK/MAPK activity blocks differentiation cues through regulation of key components such as DLG2. This mechanistic explanation of a role for DLG2 as a tumor suppressor was accompanied by a comprehensive genetic analysis of 120 11q-deleted neuroblastoma cases that was able to identify DLG2 as the most proximal gene in the shortest region of genetic overlap in patient material, with DLG2 disrupted in all 11q deleted cases examined [21]. These observations led Siaw et al. to conclude that DLG2 acts as a tumor suppressor gene in 11q-deletion neuroblastoma, playing a critical role in the differentiation of neural crest lineages. Interestingly, both SHANK2 and DLG2 are post-synaptic anchor proteins that are both able to modulate transcriptional outcome in neuroblastoma cells from proliferation cues to differentiation cues. How this occurs is not presently understood, but is certainly worthy of future investigation. Together, these three articles emphasize the importance of understanding differentiation mechanisms in a normal cellular circumstances, allowing a better understanding of the perturbations in developmental events that can arise as a result of 11q-deletion events. Indeed, such events in 11q-deletion can be further impacted on by deregulated oncogenic activities, such as MYCN amplification and aberrant ALK/MAPK signaling, resulting in normal cell development veering off-track and promoting evolution of high-risk neuroblastoma (Figure 4).
5. Friends and Foes—Implications for the Future
In recent years, a better understanding of developmental processes has solidified our understanding of neuroblastoma as a cancer arising from abnormal development of neural crest derived tissues. Key findings, such as the impact of mesenchymal or adrenergic cellular identity, as well as definition of key developmental events by lineage tracing, have provided important insight [8,93]. MYCN amplification remains one of the most important prognostic biomarkers in neuroblastoma. However, there is currently much focus on copy-number-variation, whole or segmental deletions, gains, and amplification of chromosomes in neuroblastoma. Such ‘ploidy’ disturbances, arising from gain or loss of segment or individual chromosomes can lead to aberrant protein levels, resulting in increased or decreased activity output from key signaling pathways, resulting in cellular stress that can promote transformation and tumor development [94,95]. In high-risk neuroblastoma, 2p-gain and 11q-deletion cases present developing tumors with aneuploidy challenges. What implications can we draw from the recent reports discussed here?
The reports from Javanmardi et al. and Borenäs et al. consider 2p-gain and the ALKAL2 ligand, highlighting the potential importance of addressing the activation status of ALK at the protein level in neuroblastoma [46,66] (Figure 5). Future phosphor/proteomics analyses may increase our ability to identify ALK-driven neuroblastomas that may respond to ALK TKI therapy. Here, evaluation of ALKAL2 levels in neuroblastoma could provide a potential biomarker for ALK signaling activity that is not captured by genetic mutation analysis. Furthermore, regarding mutation of ALK at the genomic level, there is currently general agreement regarding ALK activating mutations, particularly in the case of well-characterized kinase domain mutations. Although rare, there has been little investigation of ALK extracellular mutations or other ALK aberrations in patient samples, although several neuroblastoma derived cell lines are known to harbor ALK extracellular domain variants that are activating [96,97,98]. Indeed, many RTKs—such as KIT, RET, and EGFR—are activated by mutation of the extracellular domain as well as the intracellular kinase domain [99]. A recent structural study has revealed how the ALKAL ligands interact with the extracellular domain of the ALK receptor [100], offering potential for therapeutic exploitation. Further mechanistic investigation taking this information into account should extend our understanding of the importance of ALKAL ligands for ALK activity and signaling in a neuroblastoma context.
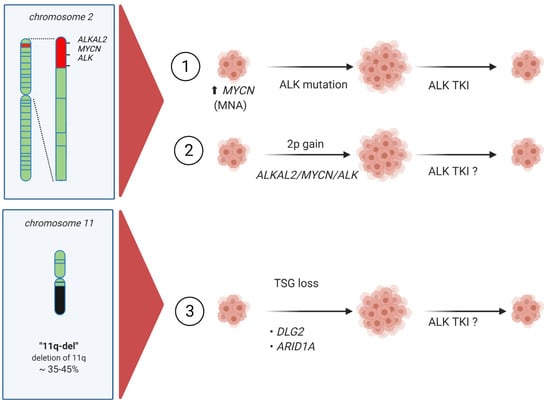
Figure 5.
New scenarios for consideration in high-risk neuroblastoma and their treatment. (1) Well established MYCN amplification with and without ALK gain-of-function mutations. (2) 2p-gain including ALKAL2, ALK and MYCN. (3) Tumor suppressor gene (TSG) loss potentially resulting in increased ALK, other RTK or MAPK signaling activity.
Together, the findings of Lopez et al., Siaw et al., and Keane et al. provide novel genetic and mechanistic insight into the complexities of 11q-deletion in neuroblastoma [21,89,90] (Figure 5). In addition to considering the loss of DDR factors, which decrease genomic integrity [101], it is clear that tumor suppressors, such as DLG2, are also influenced by oncogenic signaling activity. Hypothetically, any oncogenic activation that results in ERK pathway activity, such as ALK activation, can result in repression of DLG2 levels, tipping the balance towards mesenchymal identity and inhibiting differentiation. If this is the case in a neuroblastoma patient tumor setting, then some 11q deletions could potentially respond to ALK or ERK pathway inhibition, perhaps in combination with retinoic acid to promote differentiation (Figure 5).
Another grey area in our understanding is the interplay between copy number and epigenetic modification and resulting gene expression in neuroblastoma. Epigenetics play a critical role in the regulation of temporal and spatial gene expression, including that of oncogenes and tumor suppressor genes, and how these events integrate into the pathology of neuroblastoma is currently unclear [102]. Genetic aberrations in chromatin-modifying proteins are known to impart changes to the epigenetic landscape, promoting transcriptional programs that may be beneficial for initiation and promotion of neuroblastoma. Some well-known chromatin remodeling proteins include ARID1A/B and ATRX, which is one of the few known mutational targets in neuroblastoma [103]. The work of García-López et al. and Shi et al. implicate ARID1A, at 1p36, in neuroblastoma development, although the implications for the overall epigenetic landscape are unclear [40,41] (Figure 5). At this point, we have no epigenetic biomarker that can be employed in neuroblastoma diagnosis and prognosis. However, the work of Shi et al. suggests that high-risk neuroblastoma with deletion of 1p36 could exhibit a mesenchymal gene signature that may be exploited to identify increased resistance to chemotherapy [41]. If so, current clinical genetics analyses could be complemented by such a gene signature analysis to aid in future clinical treatment decisions.
6. Conclusions
Taken together, these recent findings shed light on the complexity of genetic imbalance that is found in neuroblastoma. How to best interpret the various chromosomal aberrations, and indeed the different combinations of them, particularly in high-risk neuroblastoma cases, is a key challenge for the field in coming years. However, increased mechanistic understanding, including identification of improper signaling and developmental events, in combination with advanced techniques and novel therapeutic options provides optimism for the future.
Author Contributions
B.H. and R.H.P. conceptualized the work with J.G., and wrote the initial draft of the manuscript. All authors contributed to the writing, review and editing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported with grants from the Swedish Childhood Cancer foundation (JG: TJ2016-0088, PR2016-0011, PR2019-0118; BH: PR2017-0110; RHP: PR2019-0078), the Swedish Cancer foundation (RHP: CAN18/729; BH: CAN18/718), the Swedish state under the LUA/ALF agreement (BH: ALFGBG-726601, RHP: ALFGBG-238561), the Swedish research council (RHP: 2019-03914; BH: 2021-01192), and the Swedish Foundation for Strategic Research (BH, RHP: RB13-0204).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Matthay, K.K.; Maris, J.M.; Schleiermacher, G.; Nakagawara, A.; Mackall, C.L.; Diller, L.; Weiss, W.A. Neuroblastoma. Nat. Rev. Dis. Prim. 2016, 2, 16078. [Google Scholar] [CrossRef]
- Maris, J.M. Recent advances in neuroblastoma. N. Engl. J. Med. 2010, 362, 2202–2211. [Google Scholar] [CrossRef] [PubMed]
- De Brouwer, S.; De Preter, K.; Kumps, C.; Zabrocki, P.; Porcu, M.; Westerhout, E.M.; Lakeman, A.; Vandesompele, J.; Hoebeeck, J.; Van Maerken, T.; et al. Meta-analysis of neuroblastomas reveals a skewed ALK mutation spectrum in tumors with MYCN amplification. Clin. Cancer Res. 2010, 16, 4353–4362. [Google Scholar] [CrossRef] [PubMed]
- Grobner, S.N.; Worst, B.C.; Weischenfeldt, J.; Buchhalter, I.; Kleinheinz, K.; Rudneva, V.A.; Johann, P.D.; Balasubramanian, G.P.; Segura-Wang, M.; Brabetz, S.; et al. The landscape of genomic alterations across childhood cancers. Nature 2018, 555, 321–327. [Google Scholar] [CrossRef]
- Pugh, T.J.; Morozova, O.; Attiyeh, E.F.; Asgharzadeh, S.; Wei, J.S.; Auclair, D.; Carter, S.L.; Cibulskis, K.; Hanna, M.; Kiezun, A.; et al. The genetic landscape of high-risk neuroblastoma. Nat. Genet. 2013, 45, 279–284. [Google Scholar] [CrossRef]
- Huber, K.; Kalcheim, C.; Unsicker, K. The development of the chromaffin cell lineage from the neural crest. Auton. Neurosci. 2009, 151, 10–16. [Google Scholar] [CrossRef]
- Saito, D.; Takase, Y.; Murai, H.; Takahashi, Y. The dorsal aorta initiates a molecular cascade that instructs sympatho-adrenal specification. Science 2012, 336, 1578–1581. [Google Scholar] [CrossRef] [PubMed]
- Furlan, A.; Dyachuk, V.; Kastriti, M.E.; Calvo-Enrique, L.; Abdo, H.; Hadjab, S.; Chontorotzea, T.; Akkuratova, N.; Usoskin, D.; Kamenev, D.; et al. Multipotent peripheral glial cells generate neuroendocrine cells of the adrenal medulla. Science 2017, 357. [Google Scholar] [CrossRef] [PubMed]
- Bedoya-Reina, O.C.; Li, W.; Arceo, M.; Plescher, M.; Bullova, P.; Pui, H.; Kaucka, M.; Kharchenko, P.; Martinsson, T.; Holmberg, J.; et al. Single-nuclei transcriptomes from human adrenal gland reveal distinct cellular identities of low and high-risk neuroblastoma tumors. Nat. Commun. 2021, 12, 5309. [Google Scholar] [CrossRef]
- Dong, R.; Yang, R.; Zhan, Y.; Lai, H.D.; Ye, C.J.; Yao, X.Y.; Luo, W.Q.; Cheng, X.M.; Miao, J.J.; Wang, J.F.; et al. Single-Cell Characterization of Malignant Phenotypes and Developmental Trajectories of Adrenal Neuroblastoma. Cancer Cell 2020, 38, 716–733.e6. [Google Scholar] [CrossRef]
- Hanemaaijer, E.S.; Margaritis, T.; Sanders, K.; Bos, F.L.; Candelli, T.; Al-Saati, H.; van Noesel, M.M.; Meyer-Wentrup, F.A.G.; van de Wetering, M.; Holstege, F.C.P.; et al. Single-cell atlas of developing murine adrenal gland reveals relation of Schwann cell precursor signature to neuroblastoma phenotype. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Ali, F.R.; Marcos, D.; Chernukhin, I.; Woods, L.M.; Parkinson, L.M.; Wylie, L.A.; Papkovskaia, T.D.; Davies, J.D.; Carroll, J.S.; Philpott, A. Dephosphorylation of the Proneural Transcription Factor ASCL1 Re-Engages a Latent Post-Mitotic Differentiation Program in Neuroblastoma. Mol. Cancer Res. 2020, 18, 1759–1766. [Google Scholar] [CrossRef]
- Zafar, A.; Wang, W.; Liu, G.; Wang, X.; Xian, W.; McKeon, F.; Foster, J.; Zhou, J.; Zhang, R. Molecular targeting therapies for neuroblastoma: Progress and challenges. Med. Res. Rev. 2021, 41, 961–1021. [Google Scholar] [CrossRef]
- Ritenour, L.E.; Randall, M.P.; Bosse, K.R.; Diskin, S.J. Genetic susceptibility to neuroblastoma: Current knowledge and future directions. Cell Tissue Res. 2018, 372, 287–307. [Google Scholar] [CrossRef] [PubMed]
- Kastriti, M.E.; Kameneva, P.; Adameyko, I. Stem cells, evolutionary aspects and pathology of the adrenal medulla: A new developmental paradigm. Mol. Cell. Endocrinol. 2020, 518, 110998. [Google Scholar] [CrossRef] [PubMed]
- Eleveld, T.F.; Oldridge, D.A.; Bernard, V.; Koster, J.; Daage, L.C.; Diskin, S.J.; Schild, L.; Bentahar, N.B.; Bellini, A.; Chicard, M.; et al. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat. Genet. 2015, 47, 864–871. [Google Scholar] [CrossRef]
- Stainczyk, S.A.; Westermann, F. Neuroblastoma-Telomere maintenance, deregulated signaling transduction and beyond. Int. J. Cancer 2021. [Google Scholar] [CrossRef] [PubMed]
- Schwab, M. MYCN Amplification in Neuroblastoma: A Paradigm for the Clinical Use of an Oncogene. Pathol. Oncol. Res. 1997, 3, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Weiss, W.A.; Aldape, K.; Mohapatra, G.; Feuerstein, B.G.; Bishop, J.M. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997, 16, 2985–2995. [Google Scholar] [CrossRef]
- Cohn, S.L.; Pearson, A.D.; London, W.B.; Monclair, T.; Ambros, P.F.; Brodeur, G.M.; Faldum, A.; Hero, B.; Iehara, T.; Machin, D.; et al. The International Neuroblastoma Risk Group (INRG) classification system: An INRG Task Force report. J. Clin. Oncol. 2009, 27, 289–297. [Google Scholar] [CrossRef]
- Siaw, J.T.; Javanmardi, N.; Van den Eynden, J.; Lind, D.E.; Fransson, S.; Martinez-Monleon, A.; Djos, A.; Sjoberg, R.M.; Ostensson, M.; Caren, H.; et al. 11q Deletion or ALK Activity Curbs DLG2 Expression to Maintain an Undifferentiated State in Neuroblastoma. Cell Rep. 2020, 32, 108171. [Google Scholar] [CrossRef]
- Maris, J.M.; Hogarty, M.D.; Bagatell, R.; Cohn, S.L. Neuroblastoma. Lancet 2007, 369, 2106–2120. [Google Scholar] [CrossRef]
- Mlakar, V.; Jurkovic Mlakar, S.; Lopez, G.; Maris, J.M.; Ansari, M.; Gumy-Pause, F. 11q deletion in neuroblastoma: A review of biological and clinical implications. Mol. Cancer 2017, 16, 114. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Weiss, W.A. Neuroblastoma and MYCN. Cold Spring Harb. Perspect. Med. 2013, 3, a014415. [Google Scholar] [CrossRef] [PubMed]
- Corvetta, D.; Chayka, O.; Gherardi, S.; D’Acunto, C.W.; Cantilena, S.; Valli, E.; Piotrowska, I.; Perini, G.; Sala, A. Physical interaction between MYCN oncogene and polycomb repressive complex 2 (PRC2) in neuroblastoma: Functional and therapeutic implications. J. Biol. Chem. 2013, 288, 8332–8341. [Google Scholar] [CrossRef]
- Higashi, M.; Sakai, K.; Fumino, S.; Aoi, S.; Furukawa, T.; Tajiri, T. The roles played by the MYCN, Trk, and ALK genes in neuroblastoma and neural development. Surg. Today 2019, 49, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Brodeur, G.M.; Sekhon, G.; Goldstein, M.N. Chromosomal aberrations in human neuroblastomas. Cancer 1977, 40, 2256–2263. [Google Scholar] [CrossRef]
- Mathew, P.; Valentine, M.B.; Bowman, L.C.; Rowe, S.T.; Nash, M.B.; Valentine, V.A.; Cohn, S.L.; Castleberry, R.P.; Brodeur, G.M.; Look, A.T. Detection of MYCN gene amplification in neuroblastoma by fluorescence in situ hybridization: A pediatric oncology group study. Neoplasia 2001, 3, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Munoz, J.; Vendrell, E.; Aiza, G.; Nistal, M.; Pestana, A.; Peinado, M.A.; Castresana, J.S. Determination of genomic damage in neuroblastic tumors by arbitrarily primed PCR: MYCN amplification as a marker for genomic instability in neuroblastomas. Neuropathology 2006, 26, 165–169. [Google Scholar] [CrossRef]
- Brodeur, G.M.; Seeger, R.C.; Schwab, M.; Varmus, H.E.; Bishop, J.M. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 1984, 224, 1121–1124. [Google Scholar] [CrossRef]
- Caren, H.; Kryh, H.; Nethander, M.; Sjoberg, R.M.; Trager, C.; Nilsson, S.; Abrahamsson, J.; Kogner, P.; Martinsson, T. High-risk neuroblastoma tumors with 11q-deletion display a poor prognostic, chromosome instability phenotype with later onset. Proc. Natl. Acad. Sci. USA 2010, 107, 4323–4328. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.; Gastier-Foster, J.M.; Mann, M.; Naranjo, A.H.; Van Ryn, C.; Bagatell, R.; Matthay, K.K.; London, W.B.; Irwin, M.S.; Shimada, H.; et al. Association of MYCN copy number with clinical features, tumor biology, and outcomes in neuroblastoma: A report from the Children’s Oncology Group. Cancer 2017, 123, 4224–4235. [Google Scholar] [CrossRef] [PubMed]
- Goto, S.; Umehara, S.; Gerbing, R.B.; Stram, D.O.; Brodeur, G.M.; Seeger, R.C.; Lukens, J.N.; Matthay, K.K.; Shimada, H. Histopathology (International Neuroblastoma Pathology Classification) and MYCN status in patients with peripheral neuroblastic tumors: A report from the Children’s Cancer Group. Cancer 2001, 92, 2699–2708. [Google Scholar] [CrossRef]
- Schwab, M.; Westermann, F.; Hero, B.; Berthold, F. Neuroblastoma: Biology and molecular and chromosomal pathology. Lancet Oncol. 2003, 4, 472–480. [Google Scholar] [CrossRef]
- Maris, J.M.; Guo, C.; Blake, D.; White, P.S.; Hogarty, M.D.; Thompson, P.M.; Rajalingam, V.; Gerbing, R.; Stram, D.O.; Matthay, K.K.; et al. Comprehensive analysis of chromosome 1p deletions in neuroblastoma. Med. Pediatric Oncol. 2001, 36, 32–36. [Google Scholar] [CrossRef]
- Schlisio, S.; Kenchappa, R.S.; Vredeveld, L.C.; George, R.E.; Stewart, R.; Greulich, H.; Shahriari, K.; Nguyen, N.V.; Pigny, P.; Dahia, P.L.; et al. The kinesin KIF1Bbeta acts downstream from EglN3 to induce apoptosis and is a potential 1p36 tumor suppressor. Genes Dev. 2008, 22, 884–893. [Google Scholar] [CrossRef]
- Munirajan, A.K.; Ando, K.; Mukai, A.; Takahashi, M.; Suenaga, Y.; Ohira, M.; Koda, T.; Hirota, T.; Ozaki, T.; Nakagawara, A. KIF1Bbeta functions as a haploinsufficient tumor suppressor gene mapped to chromosome 1p36.2 by inducing apoptotic cell death. J. Biol. Chem. 2008, 283, 24426–24434. [Google Scholar] [CrossRef]
- Hogarty, M.D.; Maris, J.M.; White, P.S.; Guo, C.; Brodeur, G.M. Analysis of genomic imprinting at 1p35-36 in neuroblastoma. Med. Pediatric Oncol. 2001, 36, 52–55. [Google Scholar] [CrossRef]
- Tolbert, V.P.; Coggins, G.E.; Maris, J.M. Genetic susceptibility to neuroblastoma. Curr. Opin. Genet. Dev. 2017, 42, 81–90. [Google Scholar] [CrossRef]
- Garcia-Lopez, J.; Wallace, K.; Otero, J.H.; Olsen, R.; Wang, Y.D.; Finkelstein, D.; Gudenas, B.L.; Rehg, J.E.; Northcott, P.; Davidoff, A.M.; et al. Large 1p36 Deletions Affecting Arid1a Locus Facilitate Mycn-Driven Oncogenesis in Neuroblastoma. Cell Rep. 2020, 30, 454–464.e5. [Google Scholar] [CrossRef]
- Shi, H.; Tao, T.; Abraham, B.J.; Durbin, A.D.; Zimmerman, M.W.; Kadoch, C.; Look, A.T. ARID1A loss in neuroblastoma promotes the adrenergic-to-mesenchymal transition by regulating enhancer-mediated gene expression. Sci. Adv. 2020, 6, eaaz3440. [Google Scholar] [CrossRef]
- Sausen, M.; Leary, R.J.; Jones, S.; Wu, J.; Reynolds, C.P.; Liu, X.; Blackford, A.; Parmigiani, G.; Diaz, L.A., Jr.; Papadopoulos, N.; et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat. Genet. 2013, 45, 12–17. [Google Scholar] [CrossRef]
- Wu, J.N.; Roberts, C.W. ARID1A mutations in cancer: Another epigenetic tumor suppressor? Cancer Discov. 2013, 3, 35–43. [Google Scholar] [CrossRef]
- Guan, J.; Umapathy, G.; Yamazaki, Y.; Wolfstetter, G.; Mendoza, P.; Pfeifer, K.; Mohammed, A.; Hugosson, F.; Zhang, H.; Hsu, A.W.; et al. FAM150A and FAM150B are activating ligands for anaplastic lymphoma kinase. elife 2015, 4, e09811. [Google Scholar] [CrossRef]
- Fadeev, A.; Mendoza-Garcia, P.; Irion, U.; Guan, J.; Pfeifer, K.; Wiessner, S.; Serluca, F.; Singh, A.P.; Nusslein-Volhard, C.; Palmer, R.H. ALKALs are in vivo ligands for ALK family receptor tyrosine kinases in the neural crest and derived cells. Proc. Natl. Acad. Sci. USA 2018, 115, E630–E638. [Google Scholar] [CrossRef]
- Borenas, M.; Umapathy, G.; Lai, W.Y.; Lind, D.E.; Witek, B.; Guan, J.; Mendoza-Garcia, P.; Masudi, T.; Claeys, A.; Chuang, T.P.; et al. ALK ligand ALKAL2 potentiates MYCN-driven neuroblastoma in the absence of ALK mutation. EMBO J. 2021, 40, e105784. [Google Scholar] [CrossRef]
- Mo, E.S.; Cheng, Q.N.; Reshetnyak, A.V.; Schlessinger, J.; Nicoli, S. Alk and Ltk ligands are essential for iridophore development in zebrafish mediated by the receptor tyrosine kinase Ltk. Proc. Natl. Acad. Sci. USA 2017, 114, 12027–12032. [Google Scholar] [CrossRef] [PubMed]
- Reshetnyak, A.V.; Murray, P.B.; Shi, X.; Mo, E.S.; Mohanty, J.; Tome, F.; Bai, H.; Gunel, M.; Lax, I.; Schlessinger, J. Augmentor alpha and beta (FAM150) are ligands of the receptor tyrosine kinases ALK and LTK: Hierarchy and specificity of ligand-receptor interactions. Proc. Natl. Acad. Sci. USA 2015, 112, 15862–15867. [Google Scholar] [CrossRef]
- George, R.E.; Sanda, T.; Hanna, M.; Frohling, S.; Luther, W., 2nd; Zhang, J.; Ahn, Y.; Zhou, W.; London, W.B.; McGrady, P.; et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature 2008, 455, 975–978. [Google Scholar] [CrossRef] [PubMed]
- Mosse, Y.P.; Laudenslager, M.; Longo, L.; Cole, K.A.; Wood, A.; Attiyeh, E.F.; Laquaglia, M.J.; Sennett, R.; Lynch, J.E.; Perri, P.; et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 2008, 455, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Caren, H.; Abel, F.; Kogner, P.; Martinsson, T. High incidence of DNA mutations and gene amplifications of the ALK gene in advanced sporadic neuroblastoma tumours. Biochem. J. 2008, 416, 153–159. [Google Scholar] [CrossRef]
- Janoueix-Lerosey, I.; Lequin, D.; Brugieres, L.; Ribeiro, A.; de Pontual, L.; Combaret, V.; Raynal, V.; Puisieux, A.; Schleiermacher, G.; Pierron, G.; et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature 2008, 455, 967–970. [Google Scholar] [CrossRef]
- Chen, Y.; Takita, J.; Choi, Y.L.; Kato, M.; Ohira, M.; Sanada, M.; Wang, L.; Soda, M.; Kikuchi, A.; Igarashi, T.; et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature 2008, 455, 971–974. [Google Scholar] [CrossRef]
- Schleiermacher, G.; Javanmardi, N.; Bernard, V.; Leroy, Q.; Cappo, J.; Rio Frio, T.; Pierron, G.; Lapouble, E.; Combaret, V.; Speleman, F.; et al. Emergence of new ALK mutations at relapse of neuroblastoma. J. Clin. Oncol. 2014, 32, 2727–2734. [Google Scholar] [CrossRef]
- Martinsson, T.; Eriksson, T.; Abrahamsson, J.; Caren, H.; Hansson, M.; Kogner, P.; Kamaraj, S.; Schonherr, C.; Weinmar, J.; Ruuth, K.; et al. Appearance of the novel activating F1174S ALK mutation in neuroblastoma correlates with aggressive tumor progression and unresponsiveness to therapy. Cancer Res. 2011, 71, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Fadeev, A.; Krauss, J.; Singh, A.P.; Nusslein-Volhard, C. Zebrafish Leucocyte tyrosine kinase controls iridophore establishment, proliferation and survival. Pigment Cell Melanoma Res. 2016, 29, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Cazes, A.; Lopez-Delisle, L.; Tsarovina, K.; Pierre-Eugene, C.; De Preter, K.; Peuchmaur, M.; Nicolas, A.; Provost, C.; Louis-Brennetot, C.; Daveau, R.; et al. Activated Alk triggers prolonged neurogenesis and Ret upregulation providing a therapeutic target in ALK-mutated neuroblastoma. Oncotarget 2014, 5, 2688–2702. [Google Scholar] [CrossRef]
- Ono, S.; Saito, T.; Terui, K.; Yoshida, H.; Enomoto, H. Generation of conditional ALK F1174L mutant mouse models for the study of neuroblastoma pathogenesis. Genesis 2019, 57, e23323. [Google Scholar] [CrossRef]
- Heukamp, L.C.; Thor, T.; Schramm, A.; De Preter, K.; Kumps, C.; De Wilde, B.; Odersky, A.; Peifer, M.; Lindner, S.; Spruessel, A.; et al. Targeted expression of mutated ALK induces neuroblastoma in transgenic mice. Sci. Transl. Med. 2012, 4, 141ra91. [Google Scholar] [CrossRef]
- Schonherr, C.; Ruuth, K.; Kamaraj, S.; Wang, C.L.; Yang, H.L.; Combaret, V.; Djos, A.; Martinsson, T.; Christensen, J.G.; Palmer, R.H.; et al. Anaplastic Lymphoma Kinase (ALK) regulates initiation of transcription of MYCN in neuroblastoma cells. Oncogene 2012, 31, 5193–5200. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.K.; Nafady, A.; Takatori, A.; Kishida, S.; Ohira, M.; Suenaga, Y.; Hossain, S.; Akter, J.; Ogura, A.; Nakamura, Y.; et al. ALK is a MYCN target gene and regulates cell migration and invasion in neuroblastoma. Sci. Rep. 2013, 3, 3450. [Google Scholar] [CrossRef]
- Berry, T.; Luther, W.; Bhatnagar, N.; Jamin, Y.; Poon, E.; Sanda, T.; Pei, D.; Sharma, B.; Vetharoy, W.R.; Hallsworth, A.; et al. The ALK(F1174L) Mutation Potentiates the Oncogenic Activity of MYCN in Neuroblastoma. Cancer Cell 2012, 22, 117–130. [Google Scholar] [CrossRef]
- Zhu, S.; Lee, J.S.; Guo, F.; Shin, J.; Perez-Atayde, A.R.; Kutok, J.L.; Rodig, S.J.; Neuberg, D.S.; Helman, D.; Feng, H.; et al. Activated ALK Collaborates with MYCN in Neuroblastoma Pathogenesis. Cancer Cell 2012, 21, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Heldin, C.H.; Lennartsson, J.; Westermark, B. Involvement of platelet-derived growth factor ligands and receptors in tumorigenesis. J. Intern. Med. 2018, 283, 16–44. [Google Scholar] [CrossRef]
- Zhang, H.; Pao, L.I.; Zhou, A.; Brace, A.D.; Halenbeck, R.; Hsu, A.W.; Bray, T.L.; Hestir, K.; Bosch, E.; Lee, E.; et al. Deorphanization of the human leukocyte tyrosine kinase (LTK) receptor by a signaling screen of the extracellular proteome. Proc. Natl. Acad. Sci. USA 2014, 111, 15741–15745. [Google Scholar] [CrossRef] [PubMed]
- Javanmardi, N.; Fransson, S.; Djos, A.; Umapathy, G.; Ostensson, M.; Milosevic, J.; Borenas, M.; Hallberg, B.; Kogner, P.; Martinsson, T.; et al. Analysis of ALK, MYCN and the ALK ligand ALKAL2 (FAM150B/AUGalpha) in neuroblastoma patient samples with chromosome arm 2p rearrangements. Genes Chromosomes Cancer 2019, 59, 50–57. [Google Scholar] [CrossRef]
- Jeison, M.; Ash, S.; Halevy-Berko, G.; Mardoukh, J.; Luria, D.; Avigad, S.; Feinberg-Gorenshtein, G.; Goshen, Y.; Hertzel, G.; Kapelushnik, J.; et al. 2p24 Gain region harboring MYCN gene compared with MYCN amplified and nonamplified neuroblastoma: Biological and clinical characteristics. Am. J. Pathol. 2010, 176, 2616–2625. [Google Scholar] [CrossRef] [PubMed]
- Say, B.; Carpenter, N.J.; Giacoia, G.; Jegathesan, S. Agenesis of the lung associated with a chromosome abnormality (46,XX,2p+). J. Med. Genet. 1980, 17, 477–478. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.S.; Pearson, J.; Willatt, L.; Andrews, T.; Beach, R.; Green, A. Germline duplication of chromosome 2p and neuroblastoma. J. Med. Genet. 1997, 34, 949–951. [Google Scholar] [CrossRef][Green Version]
- Dowa, Y.; Yamamoto, T.; Abe, Y.; Kobayashi, M.; Hoshino, R.; Tanaka, K.; Aida, N.; Take, H.; Kato, K.; Tanaka, Y.; et al. Congenital neuroblastoma in a patient with partial trisomy of 2p. J. Pediatric Hematol. Oncol. 2006, 28, 379–382. [Google Scholar] [CrossRef]
- Nagano, H.; Kano, Y.; Kobuchi, S.; Kajitani, T. A case of partial 2p trisomy with neuroblastoma. Jpn. J. Hum. Genet. 1980, 25, 39–45. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Morgenstern, D.A.; Soh, S.Y.; Stavropoulos, D.J.; Bowdin, S.; Baruchel, S.; Malkin, D.; Meyn, M.S.; Irwin, M.S. Metachronous neuroblastoma in an infant with germline translocation resulting in partial trisomy 2p: A role for ALK? J. Pediatric Hematol. Oncol. 2014, 36, e193–e196. [Google Scholar] [CrossRef]
- Bader, S.A.; Fasching, C.; Brodeur, G.M.; Stanbridge, E.J. Dissociation of suppression of tumorigenicity and differentiation in vitro effected by transfer of single human chromosomes into human neuroblastoma cells. Cell Growth Differ. 1991, 2, 245–255. [Google Scholar] [PubMed]
- Srivatsan, E.S.; Ying, K.L.; Seeger, R.C. Deletion of chromosome 11 and of 14q sequences in neuroblastoma. Genes Chromosomes Cancer 1993, 7, 32–37. [Google Scholar] [CrossRef]
- Caren, H.; Erichsen, J.; Olsson, L.; Enerback, C.; Sjoberg, R.M.; Abrahamsson, J.; Kogner, P.; Martinsson, T. High-resolution array copy number analyses for detection of deletion, gain, amplification and copy-neutral LOH in primary neuroblastoma tumors: Four cases of homozygous deletions of the CDKN2A gene. BMC Genom. 2008, 9, 353. [Google Scholar] [CrossRef] [PubMed]
- Attiyeh, E.F.; London, W.B.; Mosse, Y.P.; Wang, Q.; Winter, C.; Khazi, D.; McGrady, P.W.; Seeger, R.C.; Look, A.T.; Shimada, H.; et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N. Engl. J. Med. 2005, 353, 2243–2253. [Google Scholar] [CrossRef]
- Spitz, R.; Hero, B.; Simon, T.; Berthold, F. Loss in chromosome 11q identifies tumors with increased risk for metastatic relapses in localized and 4S neuroblastoma. Clin. Cancer Res. 2006, 12, 3368–3373. [Google Scholar] [CrossRef]
- Johnson, A.F.; Nguyen, H.T.; Veitia, R.A. Causes and effects of haploinsufficiency. Biol. Rev. 2019, 94, 1774–1785. [Google Scholar] [CrossRef]
- Fero, M.L.; Randel, E.; Gurley, K.E.; Roberts, J.M.; Kemp, C.J. The murine gene p27Kip1 is haplo-insufficient for tumour suppression. Nature 1998, 396, 177–180. [Google Scholar] [CrossRef]
- Inoue, K.; Zindy, F.; Randle, D.H.; Rehg, J.E.; Sherr, C.J. Dmp1 is haplo-insufficient for tumor suppression and modifies the frequencies of Arf and p53 mutations in Myc-induced lymphomas. Genes Dev. 2001, 15, 2934–2939. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Y.; Su, T.; Santella, R.M.; Weinstein, I.B. Hint1 is a haplo-insufficient tumor suppressor in mice. Oncogene 2006, 25, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, J.J.; Ebus, M.E.; Koster, J.; van Sluis, P.; van Noesel, C.J.; Versteeg, R.; Caron, H.N. Cyclin D1 and CDK4 activity contribute to the undifferentiated phenotype in neuroblastoma. Cancer Res. 2008, 68, 2599–2609. [Google Scholar] [CrossRef] [PubMed]
- Fagone, P.; Nicoletti, F.; Vecchio, G.M.; Parenti, R.; Magro, G. Cyclin D1 in pediatric neuroblastic tumors: A microarray analysis. Acta Histochem. 2015, 117, 820–823. [Google Scholar] [CrossRef] [PubMed]
- Russell, M.R.; Levin, K.; Rader, J.; Belcastro, L.; Li, Y.; Martinez, D.; Pawel, B.; Shumway, S.D.; Maris, J.M.; Cole, K.A. Combination therapy targeting the Chk1 and Wee1 kinases shows therapeutic efficacy in neuroblastoma. Cancer Res. 2013, 73, 776–784. [Google Scholar] [CrossRef]
- Mandriota, S.J.; Valentijn, L.J.; Lesne, L.; Betts, D.R.; Marino, D.; Boudal-Khoshbeen, M.; London, W.B.; Rougemont, A.L.; Attiyeh, E.F.; Maris, J.M.; et al. Ataxia-telangiectasia mutated (ATM) silencing promotes neuroblastoma progression through a MYCN independent mechanism. Oncotarget 2015, 6, 18558–18576. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; White, P.S.; Weiss, M.J.; Hogarty, M.D.; Thompson, P.M.; Stram, D.O.; Gerbing, R.; Matthay, K.K.; Seeger, R.C.; Brodeur, G.M.; et al. Allelic deletion at 11q23 is common in MYCN single copy neuroblastomas. Oncogene 1999, 18, 4948–4957. [Google Scholar] [CrossRef]
- Plantaz, D.; Vandesompele, J.; Van Roy, N.; Lastowska, M.; Bown, N.; Combaret, V.; Favrot, M.C.; Delattre, O.; Michon, J.; Benard, J.; et al. Comparative genomic hybridization (CGH) analysis of stage 4 neuroblastoma reveals high frequency of 11q deletion in tumors lacking MYCN amplification. Int. J. Cancer 2001, 91, 680–686. [Google Scholar] [CrossRef]
- Maris, J.M.; Guo, C.; White, P.S.; Hogarty, M.D.; Thompson, P.M.; Stram, D.O.; Gerbing, R.; Matthay, K.K.; Seeger, R.C.; Brodeur, G.M. Allelic deletion at chromosome bands 11q14-23 is common in neuroblastoma. Med. Pediatric Oncol. 2001, 36, 24–27. [Google Scholar] [CrossRef]
- Lopez, G.; Conkrite, K.L.; Doepner, M.; Rathi, K.S.; Modi, A.; Vaksman, Z.; Farra, L.M.; Hyson, E.; Noureddine, M.; Wei, J.S.; et al. Somatic structural variation targets neurodevelopmental genes and identifies SHANK2 as a tumor suppressor in neuroblastoma. Genome Res. 2020, 30, 1228–1242. [Google Scholar] [CrossRef]
- Keane, S.; Ameen, S.; Lindlof, A.; Ejeskar, K. Low DLG2 gene expression, a link between 11q-deleted and MYCN-amplified neuroblastoma, causes forced cell cycle progression, and predicts poor patient survival. Cell Commun. Signal. 2020, 18, 65. [Google Scholar] [CrossRef]
- Matthay, K.K.; Villablanca, J.G.; Seeger, R.C.; Stram, D.O.; Harris, R.E.; Ramsay, N.K.; Swift, P.; Shimada, H.; Black, C.T.; Brodeur, G.M.; et al. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. Children’s Cancer Group. N. Engl. J. Med. 1999, 341, 1165–1173. [Google Scholar] [CrossRef] [PubMed]
- Matthay, K.K.; Reynolds, C.P.; Seeger, R.C.; Shimada, H.; Adkins, E.S.; Haas-Kogan, D.; Gerbing, R.B.; London, W.B.; Villablanca, J.G. Long-term results for children with high-risk neuroblastoma treated on a randomized trial of myeloablative therapy followed by 13-cis-retinoic acid: A children’s oncology group study. J. Clin. Oncol. 2009, 27, 1007–1013. [Google Scholar] [CrossRef] [PubMed]
- van Groningen, T.; Koster, J.; Valentijn, L.J.; Zwijnenburg, D.A.; Akogul, N.; Hasselt, N.E.; Broekmans, M.; Haneveld, F.; Nowakowska, N.E.; Bras, J.; et al. Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat. Genet. 2017, 49, 1261–1266. [Google Scholar] [CrossRef]
- Torres, E.M.; Williams, B.R.; Amon, A. Aneuploidy: Cells losing their balance. Genetics 2008, 179, 737–746. [Google Scholar] [CrossRef]
- Ben-David, U.; Amon, A. Context is everything: Aneuploidy in cancer. Nat. Rev. Genet. 2020, 21, 44–62. [Google Scholar] [CrossRef]
- Fransson, S.; Hansson, M.; Ruuth, K.; Djos, A.; Berbegall, A.; Javanmardi, N.; Abrahamsson, J.; Palmer, R.H.; Noguera, R.; Hallberg, B.; et al. Intragenic anaplastic lymphoma kinase (ALK) rearrangements: Translocations as a novel mechanism of ALK activation in neuroblastoma tumors. Genes Chromosomes Cancer 2015, 54, 99–109. [Google Scholar] [CrossRef]
- Cazes, A.; Louis-Brennetot, C.; Mazot, P.; Dingli, F.; Lombard, B.; Boeva, V.; Daveau, R.; Cappo, J.; Combaret, V.; Schleiermacher, G.; et al. Characterization of Rearrangements Involving the ALK Gene Reveals a Novel Truncated Form Associated with Tumor Aggressiveness in Neuroblastoma. Cancer Res. 2013, 73, 195–204. [Google Scholar] [CrossRef]
- Okubo, J.; Takita, J.; Chen, Y.; Oki, K.; Nishimura, R.; Kato, M.; Sanada, M.; Hiwatari, M.; Hayashi, Y.; Igarashi, T.; et al. Aberrant activation of ALK kinase by a novel truncated form ALK protein in neuroblastoma. Oncogene 2012, 31, 4667–4676. [Google Scholar] [CrossRef]
- Lemmon, M.A.; Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef]
- De Munck, S.; Provost, M.; Kurikawa, M.; Omori, I.; Mukohyama, J.; Felix, J.; Bloch, Y.; Abdel-Wahab, O.; Bazan, J.F.; Yoshimi, A.; et al. Structural basis of cytokine-mediated activation of ALK family receptors. Nature 2021. [Google Scholar] [CrossRef] [PubMed]
- Liptay, M.; Barbosa, J.S.; Rottenberg, S. Replication Fork Remodeling and Therapy Escape in DNA Damage Response-Deficient Cancers. Front. Oncol. 2020, 10, 670. [Google Scholar] [CrossRef] [PubMed]
- Fetahu, I.S.; Taschner-Mandl, S. Neuroblastoma and the epigenome. Cancer Metastasis Rev. 2021, 40, 173–189. [Google Scholar] [CrossRef] [PubMed]
- Zeineldin, M.; Federico, S.; Chen, X.; Fan, Y.; Xu, B.; Stewart, E.; Zhou, X.; Jeon, J.; Griffiths, L.; Nguyen, R.; et al. MYCN amplification and ATRX mutations are incompatible in neuroblastoma. Nat. Commun 2020, 11, 913. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).