Knockdown of microRNA-214-3p Promotes Tumor Growth and Epithelial-Mesenchymal Transition in Prostate Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
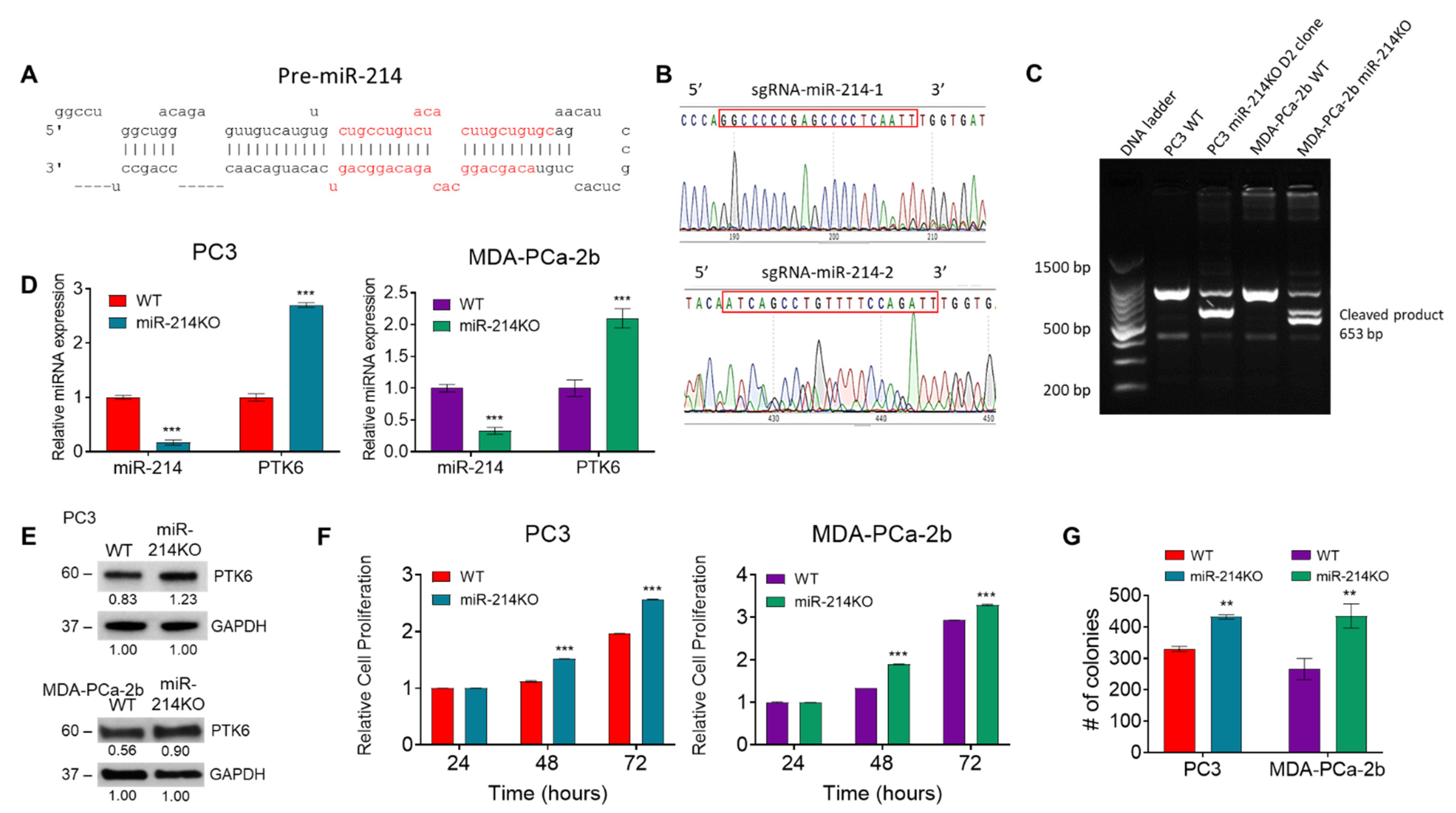
2.2. Establishment of miR-214 Knockout Cells
2.3. miR-214 and Negative Control (NC) Mimic Transfection
2.4. MTT Assay
2.5. Clonogenic Assay
2.6. Anoikis Assay
2.7. RNA Isolation and Quantification
2.8. Wound Healing Migration Assay
2.9. Migration and Invasion Assay
2.10. Western Blot Analysis
2.11. Immunofluorescence and Phalloidin Staining
2.12. RNA Isolation, Library Preparation, and RNA-Seq
2.13. In Vivo Tumorigenesis Assay
2.14. Immunohistochemical (IHC) Staining
2.15. Statistical Analysis
3. Results
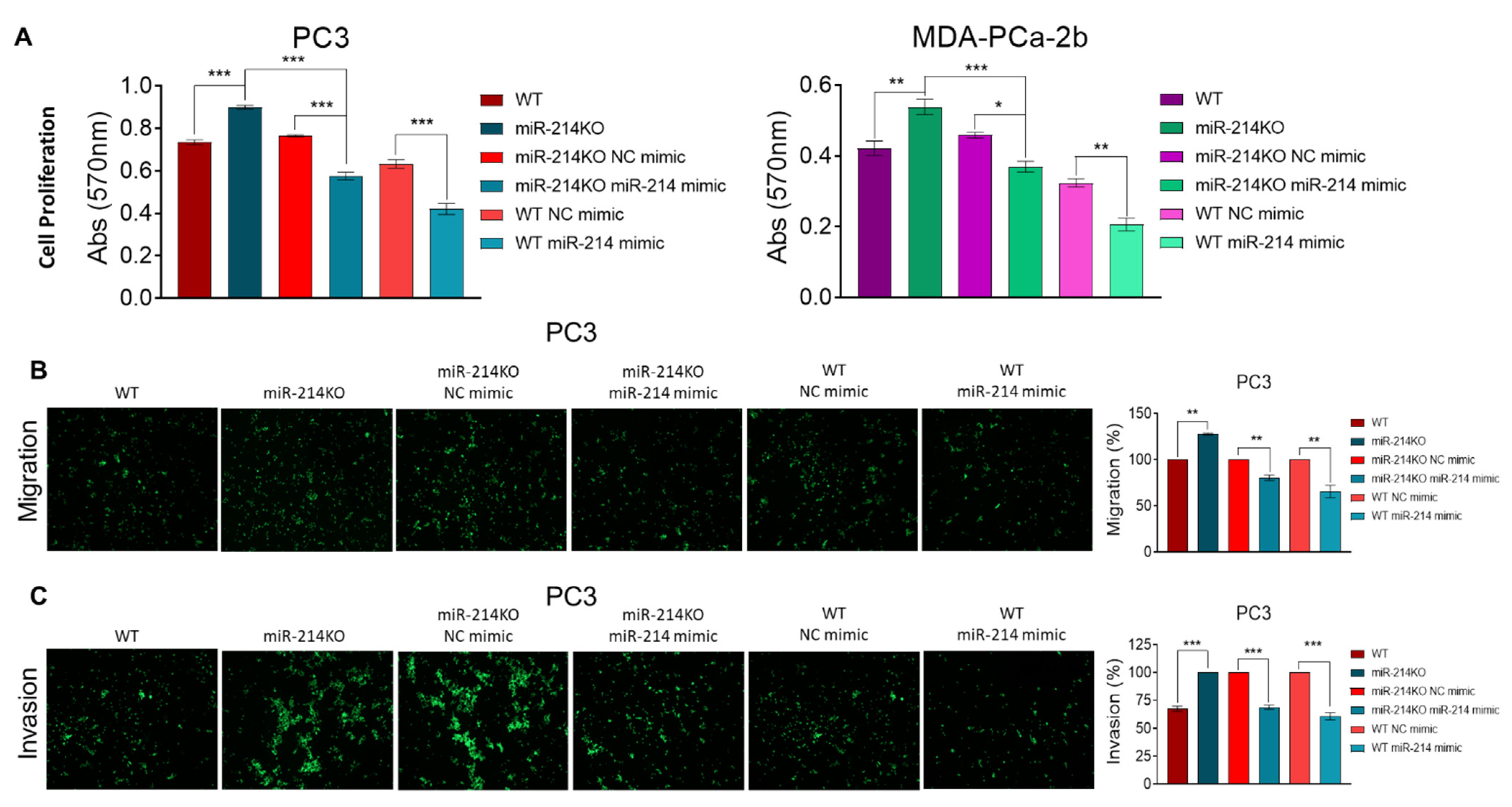
3.1. Effects of miR-214 Gene Knockdown on Cell Proliferation and Colony Formation in Prostate Cancer Cells
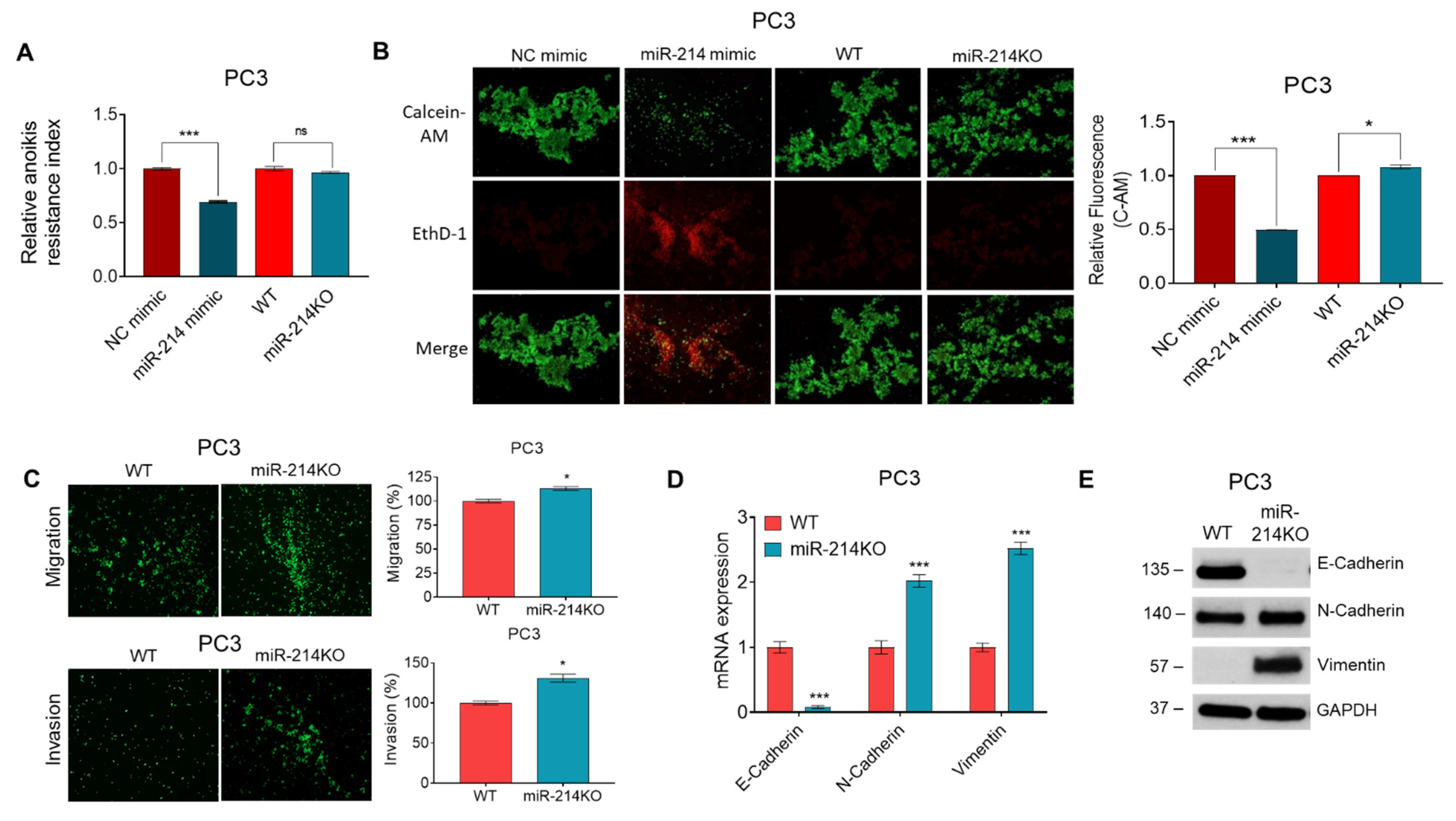
3.2. miR-214 Knockdown Increases Anoikis Resistance, Invasiveness, and EMT in PCa Cells
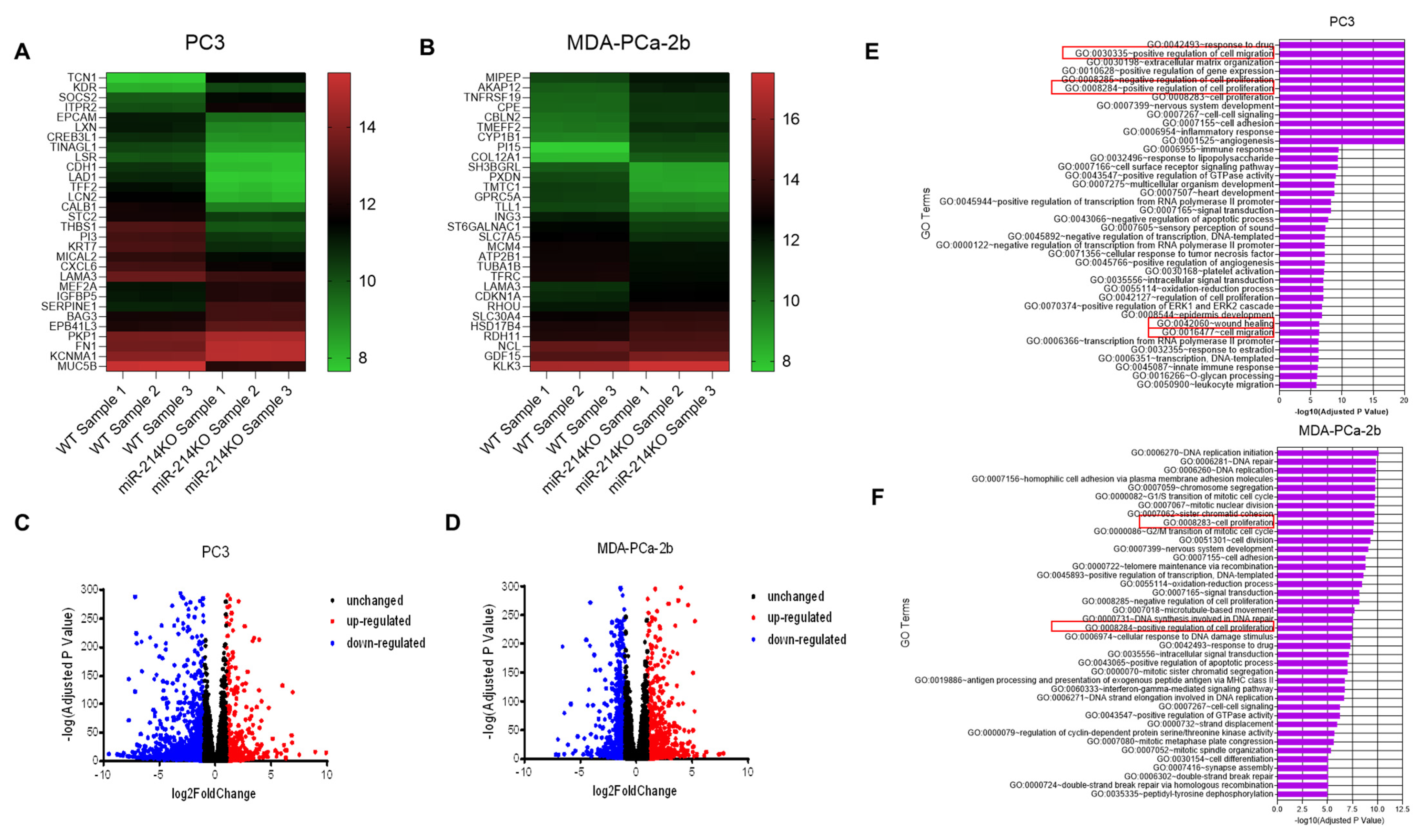
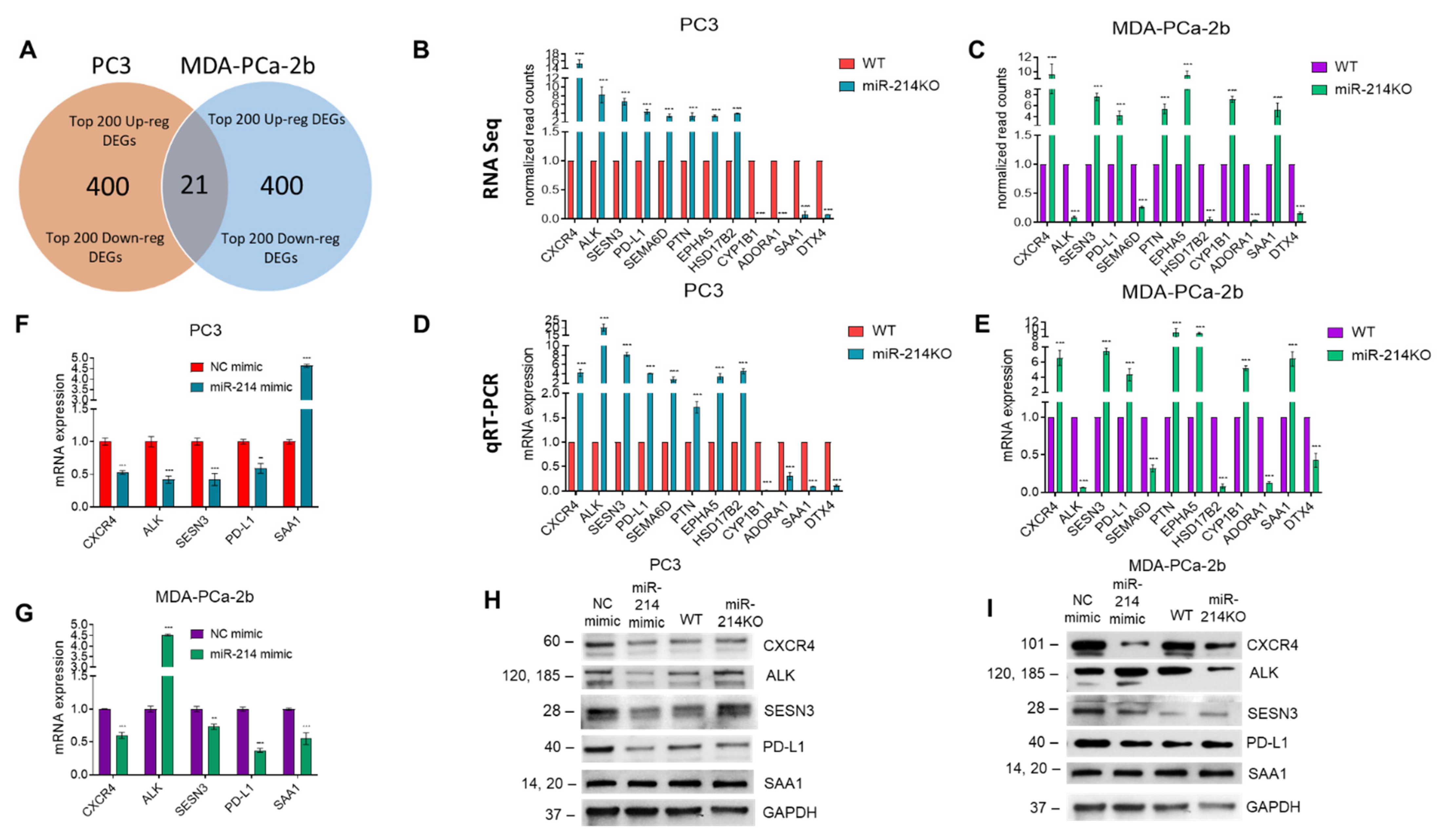
3.3. Differential Expression of mRNAs in PCa Cells with miR-214 Knockdown
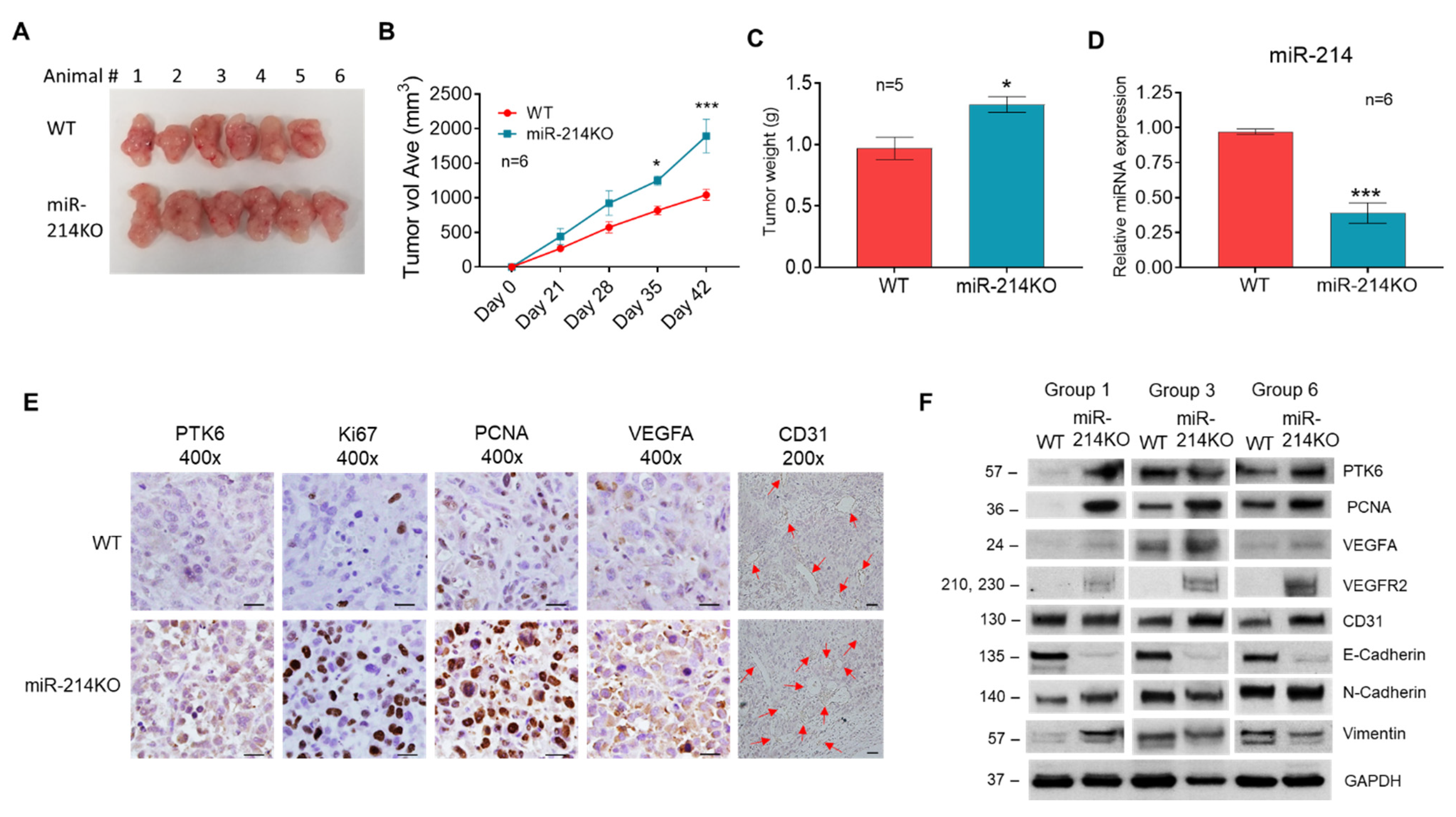
3.4. Knockdown of miR-214 Enhanced PCa Tumor Growth, Angiogenesis, and EMT in Tumor Xenografts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Howlader, N.; Noone, A.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.; et al. SEER Cancer Statistics Review, 1975–2017; National Cancer Institute Bethesda: Maryland, MD, USA, 2020.
- Lin, X.; Kapoor, A.; Gu, Y.; Chow, M.J.; Xu, H.; Major, P.; Tang, D. Assessment of biochemical recurrence of prostate cancer. Int. J. Oncol. 2019, 55, 1194–1212. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Kang, D.H.; Chung, D.Y.; Kwon, J.K.; Lee, H.; Cho, N.H.; Choi, Y.D.; Hong, S.J.; Cho, K.S. Meta-Analysis of the Relationship between CXCR4 Expression and Metastasis in Prostate Cancer. World J. Men’s Health 2014, 32, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Livas, T.; Kyprianou, N. Anoikis and EMT: Lethal “Liaisons” during Cancer Progression. Crit. Rev. Oncog. 2016, 21, 155–168. [Google Scholar] [CrossRef]
- Ardekani, A.M.; Naeini, M.M. The Role of MicroRNAs in Human Diseases. Avicenna J. Med. Biotechnol. 2010, 2, 161–179. [Google Scholar]
- Mendell, J.T.; Olson, E.N. MicroRNAs in Stress Signaling and Human Disease. Cell 2012, 148, 1172–1187. [Google Scholar] [CrossRef]
- Chen, B.; Li, H.; Zeng, X.; Yang, P.; Liu, X.; Zhao, X.; Liang, S. Roles of microRNA on cancer cell metabolism. J. Transl. Med. 2012, 10, 228. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Ding, X.-M. MicroRNAs: Regulators of cancer metastasis and epithelial-mesenchymal transition (EMT). Chin. J. Cancer 2014, 33, 140–147. [Google Scholar] [CrossRef]
- Tang, J.; Li, Y.; Wang, J.; Wen, Z.; Lai, M.; Zhang, H. Molecular mechanisms of microRNAs in regulating epithelial-mesenchymal transitions in human cancers. Cancer Lett. 2016, 371, 301–313. [Google Scholar] [CrossRef]
- Penna, E.; Orso, F.; Taverna, D. miR-214 as a Key Hub that Controls Cancer Networks: Small Player, Multiple Functions. J. Investig. Dermatol. 2015, 135, 960–969. [Google Scholar] [CrossRef] [PubMed]
- Sharma, T.; Hamilton, R.; Mandal, C.C. miR-214: A potential biomarker and therapeutic for different cancers. Future Oncol. 2015, 11, 349–363. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Lou, K.; Zhao, X.; Zhang, J.; Chen, W.; Qian, Y.; Zhao, Y.; Zhu, Y.; Zhang, Y. miR-214 regulates papillary thyroid carcinoma cell proliferation and metastasis by targeting PSMD10. Int. J. Mol. Med. 2018, 42, 3027–3036. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Sun, P.; Wang, Y.-L.; Yu, X.-F.; Tong, J.-J. MiR-214 promotes proliferation and inhibits apoptosis of oral cancer cells through MAPK/ERK signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 3710–3716. [Google Scholar] [CrossRef] [PubMed]
- Cagle, P.; Niture, S.; Srivastava, A.; Ramalinga, M.; Aqeel, R.; Rios-Colon, L.; Chimeh, U.; Suy, S.; Collins, S.P.; Dahiya, R.; et al. MicroRNA-214 targets PTK6 to inhibit tumorigenic potential and increase drug sensitivity of prostate cancer cells. Sci. Rep. 2019, 9, 9776. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Peng, J.; Chenhui, Q.; Xuefeng, L.; Lei, W.; Chunhong, W. miR-214 sensitizes human colorectal cancer cells to doxorubicin by p53 targeting. Iran. Red. Crescent Med. J. 2020, 22, 1–6. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, L.; Lu, L.; Wang, Y. miR-214-3p Regulates Multi-Drug Resistance and Apoptosis in Retinoblastoma Cells by Targeting ABCB1 and XIAP. OncoTargets Ther. 2020, 13, 803–811. [Google Scholar] [CrossRef]
- Yang, Y.; Bao, Y.; Yang, G.-K.; Wan, J.; Du, L.-J.; Ma, Z.-H. MiR-214 sensitizes human colon cancer cells to 5-FU by targeting Hsp27. Cell. Mol. Biol. Lett. 2019, 24, 22. [Google Scholar] [CrossRef]
- Yu, X.; Luo, A.; Liu, Y.; Wang, S.; Li, Y.; Shi, W.; Liu, Z.; Qu, X. MiR-214 increases the sensitivity of breast cancer cells to tamoxifen and fulvestrant through inhibition of autophagy. Mol. Cancer 2015, 14, 208. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.L.; Wang, Z.Q.; Zeng, Z.L.; Wu, W.J.; Zhang, D.S.; Luo, H.Y.; Wang, F.; Qiu, M.Z.; Wang, D.S.; Ren, C.; et al. Identification of MicroRNA-214 as a negative regulator of colorectal cancer liver metastasis by way of regulation of fibroblast growth factor receptor 1 expression. Hepatology 2014, 60, 598–609. [Google Scholar] [CrossRef]
- Chandrasekaran, K.S.; Sathyanarayanan, A.; Karunagaran, D. MicroRNA-214 suppresses growth, migration and invasion through a novel target, high mobility group AT-hook 1, in human cervical and colorectal cancer cells. Br. J. Cancer 2016, 115, 741–751. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, H.; Ma, L.; Hou, Y.; Pan, J.; Sun, C.; Yang, Y.; Zhang, J. MiR-214 suppressed ovarian cancer and negatively regulated semaphorin 4D. Tumor Biol. 2016, 37, 8239–8248. [Google Scholar] [CrossRef]
- Lu, Q.; Xu, L.; Li, C.; Yuan, Y.; Huang, S.; Chen, H. miR-214 inhibits invasion and migration via downregulating GALNT7 in esophageal squamous cell cancer. Tumor Biol. 2016, 37, 14605–14614. [Google Scholar] [CrossRef]
- Phatak, P.; Byrnes, K.A.; Mansour, D.; Liu, L.; Cao, S.; Li, R.; Rao, J.N.; Turner, D.J.; Wang, J.Y.; Donahue, J.M. Overexpression of miR-214-3p in esophageal squamous cancer cells enhances sensitivity to cisplatin by targeting survivin directly and indirectly through CUG-BP1. Oncogene 2016, 35, 2087–2097. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.; Men, J.; Ma, R.; Wang, Q.; Wang, Y.; Sun, Y.; Ren, J. miR-214 down-regulates ARL2 and suppresses growth and invasion of cervical cancer cells. Biochem. Biophys. Res. Commun. 2017, 484, 623–630. [Google Scholar] [CrossRef]
- Qiang, R.; Wang, F.; Shi, L.-Y.; Liu, M.; Chen, S.; Wan, H.-Y.; Li, Y.-X.; Li, X.; Gao, S.-Y.; Sun, B.-C.; et al. Plexin-B1 is a target of miR-214 in cervical cancer and promotes the growth and invasion of HeLa cells. Int. J. Biochem. Cell Biol. 2011, 43, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.; Cheng, X.; Zhang, Y.; Lu, X.; Hu, Z. miR-214 down-regulates MKK3 and suppresses malignant phenotypes of cervical cancer cells. Gene 2020, 724, 144146. [Google Scholar] [CrossRef]
- Zheng, C.; Guo, K.; Chen, B.; Wen, Y.; Xu, Y. miR-214-5p inhibits human prostate cancer proliferation and migration through regulating CRMP5. Cancer Biomark. 2019, 26, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Goldberger, H.; Dimtchev, A.; Ramalinga, M.; Chijioke, J.; Marian, C.; Oermann, E.K.; Uhm, S.; Kim, J.S.; Chen, L.N.; et al. MicroRNA Profiling in Prostate Cancer—The Diagnostic Potential of Urinary miR-205 and miR-214. PLoS ONE 2013, 8, e76994. [Google Scholar] [CrossRef]
- Shi, M.; Ren, S.; Chen, H.; Li, J.; Huang, C.; Li, Y.; Han, Y.; Li, Y.; Sun, Z.; Chen, X.; et al. Alcohol drinking inhibits NOTCH–PAX9 signaling in esophageal squamous epithelial cells. J. Pathol. 2021, 253, 384–395. [Google Scholar] [CrossRef]
- Paoli, P.; Giannoni, E.; Chiarugi, P. Anoikis molecular pathways and its role in cancer progression. Biochim. Biophys. Acta Mol. Cell Res. 2013, 1833, 3481–3498. [Google Scholar] [CrossRef] [PubMed]
- Strilic, B.; Offermanns, S. Intravascular Survival and Extravasation of Tumor Cells. Cancer Cell 2017, 32, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Abdollah, N.A.; Kumitaa, T.D.; Narazah, M.Y.; Abdul Razak, S.R. Sequence-specific inhibition of microRNA-130a gene by CRISPR/Cas9 system in breast cancer cell line. J. Phys.Conf. Ser. 2017, 851, 012037. [Google Scholar] [CrossRef]
- Zhao, Y.; Dai, Z.; Liang, Y.; Yin, M.; Ma, K.; He, M.; Ouyang, H.; Teng, C.-B. Sequence-specific inhibition of microRNA via CRISPR/CRISPRi system. Sci. Rep. 2014, 4, 3943. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819. [Google Scholar] [CrossRef]
- Michael, I.P.; Saghafinia, S.; Hanahan, D. A set of microRNAs coordinately controls tumorigenesis, invasion, and metastasis. Proc. Natl. Acad. Sci. USA 2019, 116, 24184–24195. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Ooi, L.L.P.J.; Hui, K.M. MiR-214 Targets β-Catenin Pathway to Suppress Invasion, Stem-Like Traits and Recurrence of Human Hepatocellular Carcinoma. PLoS ONE 2012, 7, e44206. [Google Scholar] [CrossRef]
- Ostrander, J.H.; Daniel, A.R.; Lange, C.A. Brk/PTK6 signaling in normal and cancer cell models. Curr. Opin. Pharmacol. 2010, 10, 662–669. [Google Scholar] [CrossRef]
- Ono, H.; Basson, M.D.; Ito, H. PTK6 promotes cancer migration and invasion in pancreatic cancer cells dependent on ERK signaling. PLoS ONE 2014, 9, e96060. [Google Scholar] [CrossRef][Green Version]
- Alwanian, W.M.; Tyner, A.L. Protein tyrosine kinase 6 signaling in prostate cancer. Am. J. Clin. Exp. Urol. 2020, 8, 1–8. [Google Scholar]
- Zheng, Y.; Wang, Z.; Bie, W.; Brauer, P.M.; White, B.E.P.; Li, J.; Nogueira, V.; Raychaudhuri, P.; Hay, N.; Tonetti, D.A.; et al. PTK6 activation at the membrane regulates epithelial-mesenchymal transition in prostate cancer. Cancer Res. 2013, 73, 5426–5437. [Google Scholar] [CrossRef]
- Zheng, Y.; Gierut, J.; Wang, Z.; Miao, J.; Asara, J.M.; Tyner, A.L. Protein tyrosine kinase 6 protects cells from anoikis by directly phosphorylating focal adhesion kinase and activating AKT. Oncogene 2013, 32, 4304–4312. [Google Scholar] [CrossRef]
- Ito, K.; Park, S.H.; Nayak, A.; Byerly, J.H.; Irie, H.Y. PTK6 Inhibition Suppresses Metastases of Triple-Negative Breast Cancer via SNAIL-Dependent E-Cadherin Regulation. Cancer Res. 2016, 76, 4406. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wan, Y.; Su, Z.; Li, J.; Han, M.; Zhou, C. SRF Potentiates Colon Cancer Metastasis and Progression in a microRNA-214/PTK6-Dependent Manner. Cancer Manag. Res. 2020, 12, 6477–6491. [Google Scholar] [CrossRef] [PubMed]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Montanari, M.; Rossetti, S.; Cavaliere, C.; D’Aniello, C.; Malzone, M.G.; Vanacore, D.; Di Franco, R.; La Mantia, E.; Iovane, G.; Piscitelli, R.; et al. Epithelial-mesenchymal transition in prostate cancer: An overview. Oncotarget 2017, 8, 35376–35389. [Google Scholar] [CrossRef]
- Wade, C.A.; Kyprianou, N. Profiling Prostate Cancer Therapeutic Resistance. Int. J. Mol. Sci. 2018, 19, 904. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, S.; Kyprianou, N. Targeting anoikis resistance in prostate cancer metastasis. Mol. Aspects Med. 2010, 31, 205–214. [Google Scholar] [CrossRef]
- Tang, Y.; Pan, J.; Huang, S.; Peng, X.; Zou, X.; Luo, Y.; Ren, D.; Zhang, X.; Li, R.; He, P.; et al. Downregulation of miR-133a-3p promotes prostate cancer bone metastasis via activating PI3K/AKT signaling. J. Exp. Clin. Cancer Res. 2018, 37, 160. [Google Scholar] [CrossRef]
- Ishteiwy, R.A.; Ward, T.M.; Dykxhoorn, D.M.; Burnstein, K.L. The microRNA -23b/-27b cluster suppresses the metastatic phenotype of castration-resistant prostate cancer cells. PLoS ONE 2012, 7, e52106. [Google Scholar] [CrossRef] [PubMed]
- Rice, M.A.; Ishteiwy, R.A.; Magani, F.; Udayakumar, T.; Reiner, T.; Yates, T.J.; Miller, P.; Perez-Stable, C.; Rai, P.; Verdun, R.; et al. The microRNA-23b/-27b cluster suppresses prostate cancer metastasis via Huntingtin-interacting protein 1-related. Oncogene 2016, 35, 4752–4761. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, H.; Yonemori, M.; Miyamoto, K.; Tatarano, S.; Kofuji, S.; Nohata, N.; Nakagawa, M.; Enokida, H. microRNA-210-3p depletion by CRISPR/Cas9 promoted tumorigenesis through revival of TWIST1 in renal cell carcinoma. Oncotarget 2017, 8, 20881–20894. [Google Scholar] [CrossRef]
- Li, Y.; Chen, P.; Zu, L.; Liu, B.; Wang, M.; Zhou, Q. MicroRNA-338-3p suppresses metastasis of lung cancer cells by targeting the EMT regulator Sox4. Am. J. Cancer Res. 2016, 6, 127–140. [Google Scholar] [PubMed]
- Liu, P.; Long, P.; Huang, Y.; Sun, F.; Wang, Z. CXCL12/CXCR4 axis induces proliferation and invasion in human endometrial cancer. Am. J. Transl. Res. 2016, 8, 1719–1729. [Google Scholar] [PubMed]
- Zlotnik, A. Chemokines and cancer. Int. J. Cancer 2006, 119, 2026–2029. [Google Scholar] [CrossRef] [PubMed]
- Long, P.; Sun, F.; Ma, Y.; Huang, Y. Inhibition of CXCR4 and CXCR7 for reduction of cell proliferation and invasion in human endometrial cancer. Tumor Biol. 2016, 37, 7473–7480. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Singh, U.P.; Grizzle, W.E.; Lillard, J.W. CXCL12–CXCR4 interactions modulate prostate cancer cell migration, metalloproteinase expression and invasion. Lab. Investig. 2004, 84, 1666–1676. [Google Scholar] [CrossRef]
- Adekoya, T.O.; Richardson, R.M. Cytokines and Chemokines as Mediators of Prostate Cancer Metastasis. Int. J. Mol. Sci. 2020, 21, 4449. [Google Scholar] [CrossRef]
- Hallberg, B.; Palmer, R.H. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat. Rev. Cancer 2013, 13, 685–700. [Google Scholar] [CrossRef]
- Holla, V.R.; Elamin, Y.Y.; Bailey, A.M.; Johnson, A.M.; Litzenburger, B.C.; Khotskaya, Y.B.; Sanchez, N.S.; Zeng, J.; Shufean, M.A.; Shaw, K.R.; et al. ALK: A tyrosine kinase target for cancer therapy. Cold Spring Harb. Mol. Case Stud. 2017, 3, a001115. [Google Scholar] [CrossRef]
- Kong, X.; Pan, P.; Sun, H.; Xia, H.; Wang, X.; Li, Y.; Hou, T. Drug Discovery Targeting Anaplastic Lymphoma Kinase (ALK). J. Med. Chem. 2019, 62, 10927–10954. [Google Scholar] [CrossRef] [PubMed]
- Drake, J.M.; Graham, N.A.; Lee, J.K.; Stoyanova, T.; Faltermeier, C.M.; Sud, S.; Titz, B.; Huang, J.; Pienta, K.J.; Graeber, T.G.; et al. Metastatic castration-resistant prostate cancer reveals intrapatient similarity and interpatient heterogeneity of therapeutic kinase targets. Proc. Natl. Acad. Sci. USA 2013, 110, E4762–E4769. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, B.A.; Pamarthy, S.; Shah, A.N.; Sagar, V.; Unno, K.; Han, H.; Yang, X.J.; Costa, R.B.; Nagy, R.J.; Lanman, R.B.; et al. Anaplastic Lymphoma Kinase Mutation (ALK F1174C) in Small Cell Carcinoma of the Prostate and Molecular Response to Alectinib. Clin. Cancer Res. 2018, 24, 2732–2739. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Kim, H.G.; Dong, E.; Dong, C.; Huang, M.; Liu, Y.; Liangpunsakul, S.; Dong, X.C. Sesn3 deficiency promotes carcinogen-induced hepatocellular carcinoma via regulation of the hedgehog pathway. Biochim. Biophys. Acta Mol. Basis. Dis. 2019, 1865, 2685–2693. [Google Scholar] [CrossRef]
- Kosaka, T.; Hongo, H.; Miyazaki, Y.; Nishimoto, K.; Miyajima, A.; Oya, M. Reactive oxygen species induction by cabazitaxel through inhibiting Sestrin-3 in castration resistant prostate cancer. Oncotarget 2017, 8, 87675–87683. [Google Scholar] [CrossRef] [PubMed]
- Alsuliman, A.; Colak, D.; Al-Harazi, O.; Fitwi, H.; Tulbah, A.; Al-Tweigeri, T.; Al-Alwan, M.; Ghebeh, H. Bidirectional crosstalk between PD-L1 expression and epithelial to mesenchymal transition: Significance in claudin-low breast cancer cells. Mol. Cancer 2015, 14, 149. [Google Scholar] [CrossRef]
- Sun, L.; Ye, R.D. Serum amyloid A1: Structure, function and gene polymorphism. Gene 2016, 583, 48–57. [Google Scholar] [CrossRef]
- Takehara, M.; Sato, Y.; Kimura, T.; Noda, K.; Miyamoto, H.; Fujino, Y.; Miyoshi, J.; Nakamura, F.; Wada, H.; Bando, Y.; et al. Cancer-associated adipocytes promote pancreatic cancer progression through SAA1 expression. Cancer Sci. 2020, 111, 2883–2894. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cagle, P.; Smith, N.; Adekoya, T.O.; Li, Y.; Kim, S.; Rios-Colon, L.; Deep, G.; Niture, S.; Albanese, C.; Suy, S.; et al. Knockdown of microRNA-214-3p Promotes Tumor Growth and Epithelial-Mesenchymal Transition in Prostate Cancer. Cancers 2021, 13, 5875. https://doi.org/10.3390/cancers13235875
Cagle P, Smith N, Adekoya TO, Li Y, Kim S, Rios-Colon L, Deep G, Niture S, Albanese C, Suy S, et al. Knockdown of microRNA-214-3p Promotes Tumor Growth and Epithelial-Mesenchymal Transition in Prostate Cancer. Cancers. 2021; 13(23):5875. https://doi.org/10.3390/cancers13235875
Chicago/Turabian StyleCagle, Patrice, Nikia Smith, Timothy O. Adekoya, Yahui Li, Susy Kim, Leslimar Rios-Colon, Gagan Deep, Suryakant Niture, Christopher Albanese, Simeng Suy, and et al. 2021. "Knockdown of microRNA-214-3p Promotes Tumor Growth and Epithelial-Mesenchymal Transition in Prostate Cancer" Cancers 13, no. 23: 5875. https://doi.org/10.3390/cancers13235875
APA StyleCagle, P., Smith, N., Adekoya, T. O., Li, Y., Kim, S., Rios-Colon, L., Deep, G., Niture, S., Albanese, C., Suy, S., Collins, S. P., & Kumar, D. (2021). Knockdown of microRNA-214-3p Promotes Tumor Growth and Epithelial-Mesenchymal Transition in Prostate Cancer. Cancers, 13(23), 5875. https://doi.org/10.3390/cancers13235875









