Identification of an Immunogenic Medulloblastoma-Specific Fusion Involving EPC2 and GULP1
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Material
2.2. Nucleic Acid Extraction
2.3. RNA-Seq
2.4. RT-PCR and qRT-PCR
2.5. Determination of the Full Sequence
2.6. In Silico Epitope Prediction
2.7. In Vitro Assay for Potential Peptide Antigens
2.8. IFN-γ ELISpot
2.9. De Novo Structure Prediction
2.10. Immunohistochemistry
3. Results
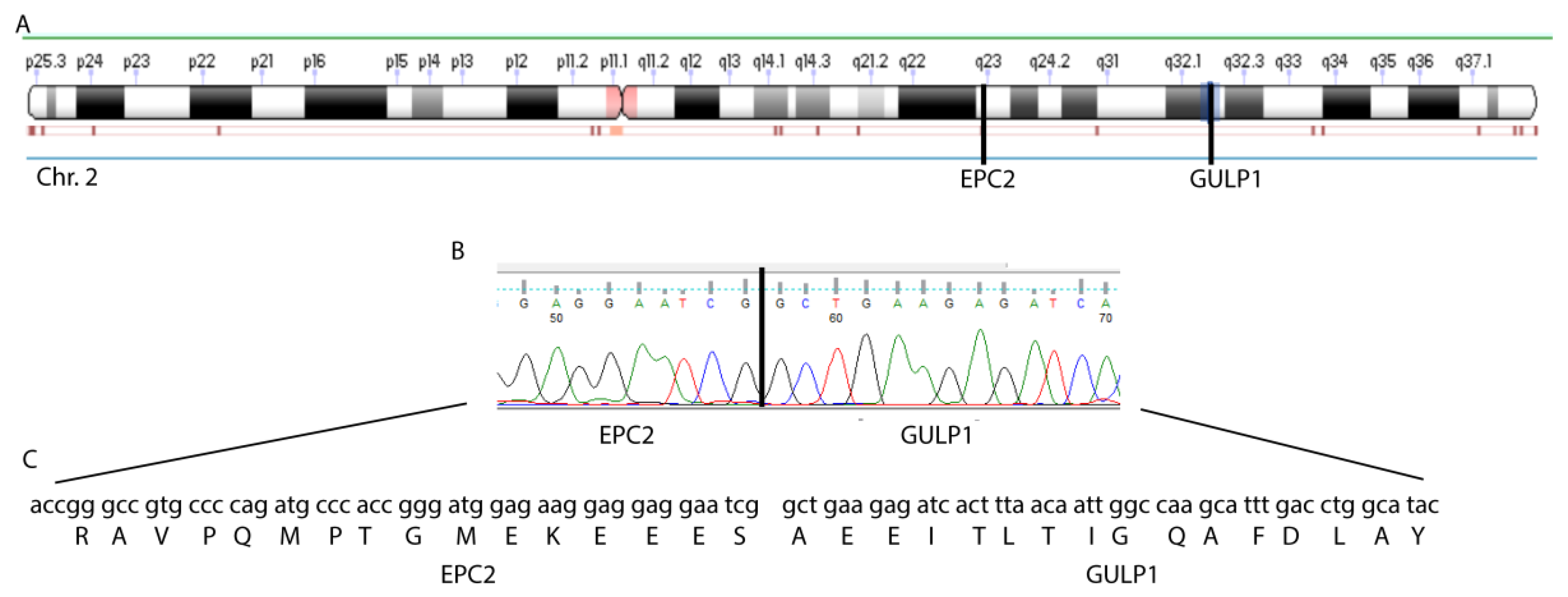
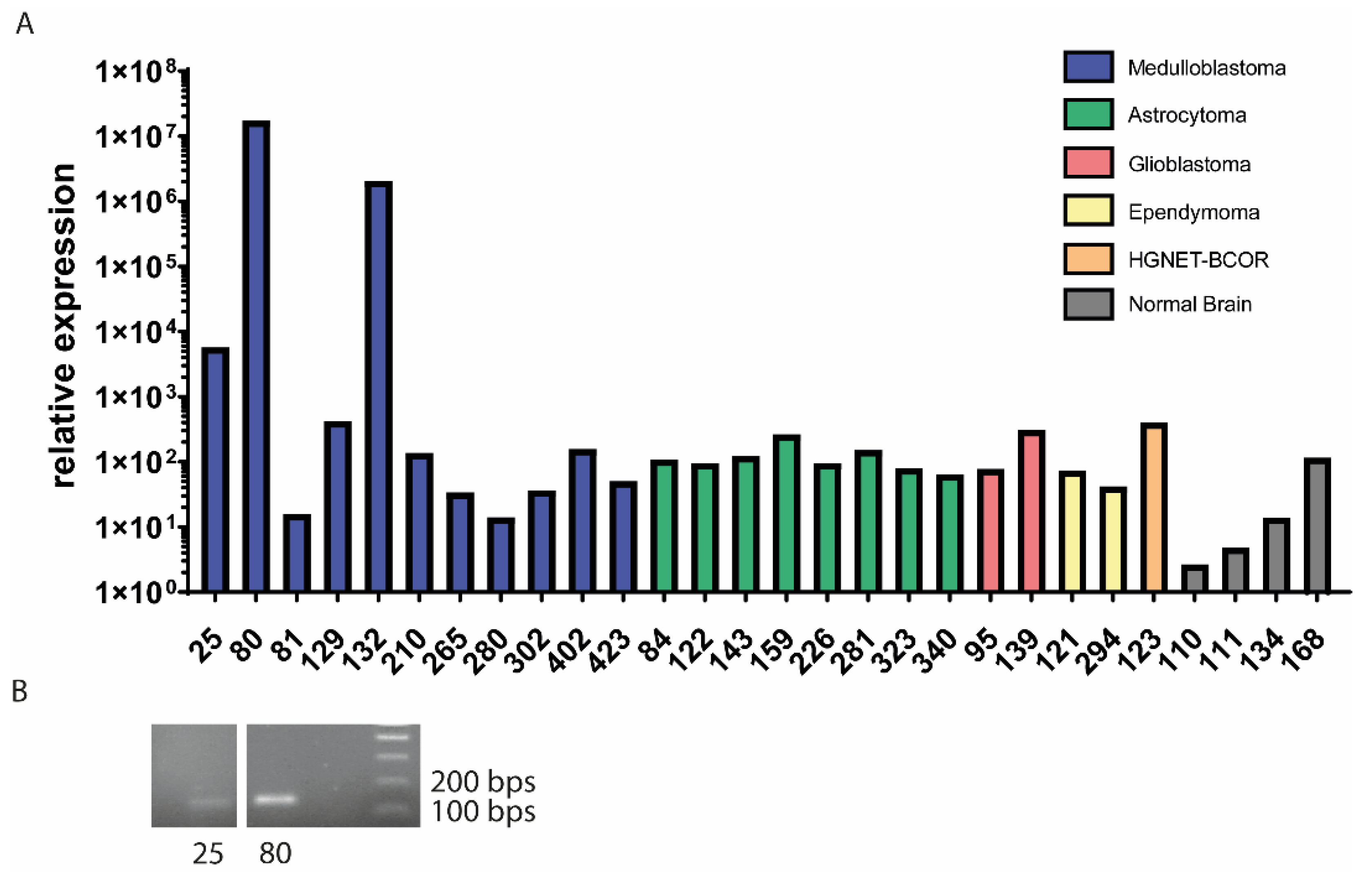
3.1. EPC2–GULP1 Is a Medulloblastoma Specific Fusion
3.2. The Structure of the EPC2–GULP1 Fusion
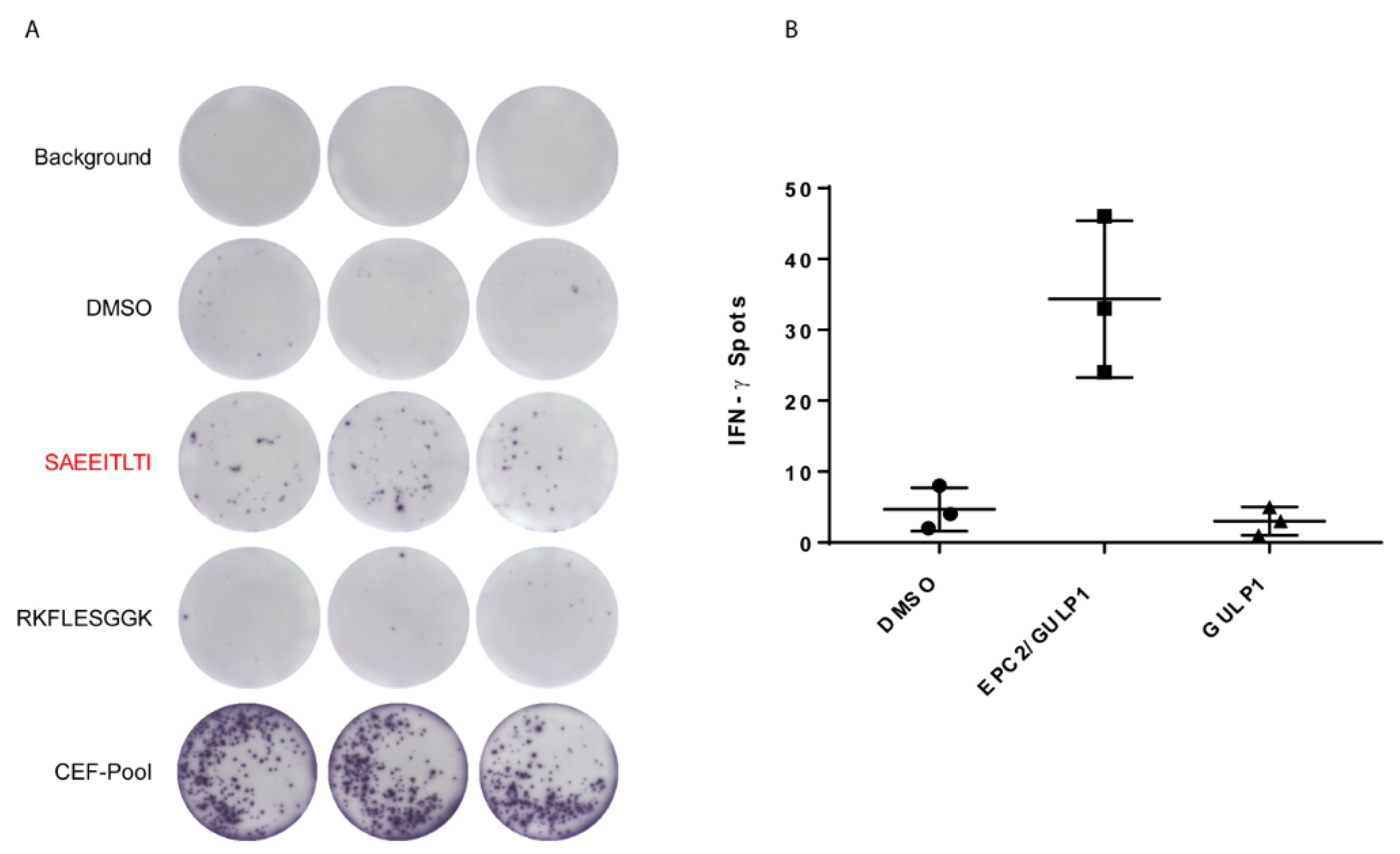
3.3. EPC2–GULP1 Fusion Peptide Has Immunogenic Potential to Activate CD8+ T Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| EPC2 | Enhancer of polycomb homolog 2 |
| GULP1 | GULP PTB domain containing engulfment adaptor 1 |
| Chr | Chromosome |
| PcG | Polycomb group of genes |
| EPcA | Enhancer of polycomb-like domain A |
| PTB | Phosphotyrosine-binding |
| LRP1 | Lipoprotein receptor-related protein 1 |
| LICD | Liposaccharide cholinephosphotransferase |
References
- Dhall, G. Medulloblastoma. J. Child Neurol. 2009, 24, 1418–1430. [Google Scholar] [CrossRef]
- Northcott, P.A.; Buchhalter, I.; Morrissy, A.S.; Hovestadt, V.; Weischenfeldt, J.; Ehrenberger, T.; Grobner, S.; Segura-Wang, M.; Zichner, T.; Rudneva, V.A.; et al. The whole-genome landscape of medulloblastoma subtypes. Nature 2017, 547, 311–317. [Google Scholar] [CrossRef] [Green Version]
- Kijima, N.; Kanemura, Y. Molecular Classification of Medulloblastoma. Neurol. Med.-Chir. 2016, 56, 687–697. [Google Scholar] [CrossRef] [Green Version]
- Okada, H. Brain tumor immunotherapy with type-1 polarizing strategies. Ann. N. Y. Acad. Sci. 2009, 1174, 18–23. [Google Scholar] [CrossRef]
- Tomaszewski, W.; Sanchez-Perez, L.; Gajewski, T.F.; Sampson, J.H. Brain Tumor Microenvironment and Host State: Implications for Immunotherapy. Clin. Cancer Res. 2019, 25, 4202–4210. [Google Scholar] [CrossRef] [Green Version]
- Haen, S.P.; Loffler, M.W.; Rammensee, H.G.; Brossart, P. Towards new horizons: Characterization, classification and implications of the tumour antigenic repertoire. Nat. Rev. Clin. Oncol. 2020, 17, 595–610. [Google Scholar] [CrossRef] [PubMed]
- Rowley, J.D. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature 1973, 243, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Wang, Q.; Torres-Garcia, W.; Zheng, S.; Vegesna, R.; Kim, H.; Verhaak, R.G. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene 2015, 34, 4845–4854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitelman, F.; Johansson, B.; Mertens, F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer 2007, 7, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Kadash-Edmondson, K.E.; Wang, R.; Phillips, J.; Liu, S.; Ribas, A.; Aplenc, R.; Witte, O.N.; Xing, Y. RNA Dysregulation: An Expanding Source of Cancer Immunotherapy Targets. Trends Pharmacol. Sci. 2021, 42, 268–282. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Zhou, C.; Zhang, Z.; Guan, M.; Zhang, C.; Liu, Z.; Liu, Q. The Landscape of Tumor Fusion Neoantigens: A Pan-Cancer Analysis. iScience 2019, 21, 249–260. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Shi, T.; Song, X.; Liu, B.; Wei, J. Gene fusion neoantigens: Emerging targets for cancer immunotherapy. Cancer Lett. 2021, 506, 45–54. [Google Scholar] [CrossRef]
- Northcott, P.A.; Shih, D.J.; Peacock, J.; Garzia, L.; Morrissy, A.S.; Zichner, T.; Stutz, A.M.; Korshunov, A.; Reimand, J.; Schumacher, S.E.; et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 2012, 488, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Dong, X.; Yu, J.; Xia, Y.; Berry, K.P.; Rao, R.; Xu, L.; Xue, P.; Chen, T.; Lin, Y.; et al. Genomic and Transcriptomic Analyses Reveals ZNF124 as a Critical Regulator in Highly Aggressive Medulloblastomas. Front. Cell Dev. Biol. 2021, 9, 634056. [Google Scholar] [CrossRef]
- Jones, D.T.; Jager, N.; Kool, M.; Zichner, T.; Hutter, B.; Sultan, M.; Cho, Y.J.; Pugh, T.J.; Hovestadt, V.; Stutz, A.M.; et al. Dissecting the genomic complexity underlying medulloblastoma. Nature 2012, 488, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Skowron, P.; Farooq, H.; Cavalli, F.M.G.; Morrissy, A.S.; Ly, M.; Hendrikse, L.D.; Wang, E.Y.; Djambazian, H.; Zhu, H.; Mungall, K.L.; et al. The transcriptional landscape of Shh medulloblastoma. Nat. Commun. 2021, 12, 1749. [Google Scholar] [CrossRef] [PubMed]
- Rammensee, H.; Bachmann, J.; Emmerich, N.P.; Bachor, O.A.; Stevanovic, S. SYFPEITHI: Database for MHC ligands and peptide motifs. Immunogenetics 1999, 50, 213–219. [Google Scholar] [CrossRef]
- Schubert, B.; Kohlbacher, O. Designing string-of-beads vaccines with optimal spacers. Genome Med. 2016, 8, 9. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Anishchenko, I.; Park, H.; Peng, Z.; Ovchinnikov, S.; Baker, D. Improved protein structure prediction using predicted interresidue orientations. Proc. Natl. Acad. Sci. USA 2020, 117, 1496–1503. [Google Scholar] [CrossRef]
- Reynolds, C.R.; Islam, S.A.; Sternberg, M.J.E. EzMol: A Web Server Wizard for the Rapid Visualization and Image Production of Protein and Nucleic Acid Structures. J. Mol. Biol. 2018, 430, 2244–2248. [Google Scholar] [CrossRef]
- Selleck, W.; Fortin, I.; Sermwittayawong, D.; Cote, J.; Tan, S. The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the Enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes. Mol. Cell. Biol. 2005, 25, 5535–5542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Lee, K.W.; Srivastava, R.M.; Kuo, F.; Krishna, C.; Chowell, D.; Makarov, V.; Hoen, D.; Dalin, M.G.; Wexler, L.; et al. Immunogenic neoantigens derived from gene fusions stimulate T cell responses. Nat. Med. 2019, 25, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.R.; Ramkissoon, S.H.; Ross, J.; Weintraub, L. Tumor mutational burden and driver mutations: Characterizing the genomic landscape of pediatric brain tumors. Pediatr. Blood Cancer 2020, 67, e28338. [Google Scholar] [CrossRef] [PubMed]
- Stankunas, K.; Berger, J.; Ruse, C.; Sinclair, D.A.; Randazzo, F.; Brock, H.W. The enhancer of polycomb gene of Drosophila encodes a chromatin protein conserved in yeast and mammals. Development 1998, 125, 4055–4066. [Google Scholar] [CrossRef]
- Shimono, Y.; Murakami, H.; Hasegawa, Y.; Takahashi, M. RET finger protein is a transcriptional repressor and interacts with enhancer of polycomb that has dual transcriptional functions. J. Biol. Chem. 2000, 275, 39411–39419. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Tan, S. Piccolo NuA4-catalyzed acetylation of nucleosomal histones: Critical roles of an Esa1 Tudor/chromo barrel loop and an Epl1 enhancer of polycomb A (EPcA) basic region. Mol. Cell. Biol. 2013, 33, 159–169. [Google Scholar] [CrossRef] [Green Version]
- Nakahata, S.; Saito, Y.; Hamasaki, M.; Hidaka, T.; Arai, Y.; Taki, T.; Taniwaki, M.; Morishita, K. Alteration of enhancer of polycomb 1 at 10p11.2 is one of the genetic events leading to development of adult T-cell leukemia/lymphoma. Genes Chromosomes Cancer 2009, 48, 768–776. [Google Scholar] [CrossRef]
- Brunetti, M.; Gorunova, L.; Davidson, B.; Heim, S.; Panagopoulos, I.; Micci, F. Identification of an EPC2-PHF1 fusion transcript in low-grade endometrial stromal sarcoma. Oncotarget 2018, 9, 19203–19208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyon, Y.; Selleck, W.; Lane, W.S.; Tan, S.; Cote, J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 2004, 24, 1884–1896. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Spencer, G.J.; Lynch, J.T.; Ciceri, F.; Somerville, T.D.; Somervaille, T.C. Enhancers of Polycomb EPC1 and EPC2 sustain the oncogenic potential of MLL leukemia stem cells. Leukemia 2014, 28, 1081–1091. [Google Scholar] [CrossRef] [Green Version]
- Gotoh, M.; Ichikawa, H.; Arai, E.; Chiku, S.; Sakamoto, H.; Fujimoto, H.; Hiramoto, M.; Nammo, T.; Yasuda, K.; Yoshida, T.; et al. Comprehensive exploration of novel chimeric transcripts in clear cell renal cell carcinomas using whole transcriptome analysis. Genes Chromosomes Cancer 2014, 53, 1018–1032. [Google Scholar] [CrossRef] [Green Version]
- Kiss, R.S.; Ma, Z.; Nakada-Tsukui, K.; Brugnera, E.; Vassiliou, G.; McBride, H.M.; Ravichandran, K.S.; Marcel, Y.L. The lipoprotein receptor-related protein-1 (LRP) adapter protein GULP mediates trafficking of the LRP ligand prosaposin, leading to sphingolipid and free cholesterol accumulation in late endosomes and impaired efflux. J. Biol. Chem. 2006, 281, 12081–12092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wahler, A.; Beyer, A.S.; Keller, I.E.; Schnack, C.; von Einem, B.; Propper, C.; Boeckers, T.M.; Peltan, I.D.; Strickland, D.K.; Hyman, B.T.; et al. Engulfment adaptor phosphotyrosine-binding-domain-containing 1 (GULP1) is a nucleocytoplasmic shuttling protein and is transactivationally active together with low-density lipoprotein receptor-related protein 1 (LRP1). Biochem. J. 2013, 450, 333–343. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.I.; Martin, C.; Ma, Z.; Hafiane, A.; Dai, M.; Lebrun, J.J.; Kiss, R.S. Engulfment protein GULP is regulator of transforming growth factor-beta response in ovarian cells. J. Biol. Chem. 2012, 287, 20636–20651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maldonado, L.; Brait, M.; Begum, S.; Chatterjee, A.; Loyo, M.; Barbosa, A.; Poeta, M.L.; Fazio, V.M.; Roberto, A.; Tarquini, E.; et al. GULP1, a potential tumor suppressor gene in ovarian tumors and its utility as a biomarker. Cell. Mol. Biol. 2010, 70. [Google Scholar] [CrossRef]
- Hayashi, M.; Hoque, M.O. Engulfment gene GULP1 is a functional tumor suppressor through influencing TGF-β pathway and is silenced by promoter methylation in urothelial carcinoma. Mol. Cell. Biol. 2015, 75, 4943. [Google Scholar] [CrossRef]
- Su, H.P.; Brugnera, E.; Van Criekinge, W.; Smits, E.; Hengartner, M.; Bogaert, T.; Ravichandran, K.S. Identification and characterization of a dimerization domain in CED-6, an adapter protein involved in engulfment of apoptotic cells. J. Biol. Chem. 2000, 275, 9542–9549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalina, J.L.; Neilson, D.S.; Lin, Y.Y.; Hamilton, P.T.; Comber, A.P.; Loy, E.M.H.; Sahinalp, S.C.; Collins, C.C.; Hach, F.; Lum, J.J. Mutational Analysis of Gene Fusions Predicts Novel MHC Class I-Restricted T-Cell Epitopes and Immune Signatures in a Subset of Prostate Cancer. Clin. Cancer Res. 2017, 23, 7596–7607. [Google Scholar] [CrossRef] [Green Version]
- Cathcart, K.; Pinilla-Ibarz, J.; Korontsvit, T.; Schwartz, J.; Zakhaleva, V.; Papadopoulos, E.B.; Scheinberg, D.A. A multivalent bcr-abl fusion peptide vaccination trial in patients with chronic myeloid leukemia. Blood 2004, 103, 1037–1042. [Google Scholar] [CrossRef]
- Bocchia, M.; Gentili, S.; Abruzzese, E.; Fanelli, A.; Iuliano, F.; Tabilio, A.; Amabile, M.; Forconi, F.; Gozzetti, A.; Raspadori, D.; et al. Effect of a p210 multipeptide vaccine associated with imatinib or interferon in patients with chronic myeloid leukaemia and persistent residual disease: A multicentre observational trial. Lancet 2005, 365, 657–662. [Google Scholar] [CrossRef]
- Chang, T.C.; Carter, R.A.; Li, Y.; Li, Y.; Wang, H.; Edmonson, M.N.; Chen, X.; Arnold, P.; Geiger, T.L.; Wu, G.; et al. The neoepitope landscape in pediatric cancers. Genome Med. 2017, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.A.; Bemis, J.G.T.; Bagby, S.M.; Capasso, A.; Yacob, B.W.; Chimed, T.S.; Van Gulick, R.; Lee, H.; Tobin, R.; Tentler, J.J.; et al. BRAF fusions identified in melanomas have variable treatment responses and phenotypes. Oncogene 2019, 38, 1296–1308. [Google Scholar] [CrossRef] [PubMed]
- Biernacki, M.A.; Foster, K.A.; Woodward, K.B.; Coon, M.E.; Cummings, C.; Cunningham, T.M.; Dossa, R.G.; Brault, M.; Stokke, J.; Olsen, T.M.; et al. CBFB-MYH11 fusion neoantigen enables T cell recognition and killing of acute myeloid leukemia. J. Clin. Investig. 2020, 130, 5127–5141. [Google Scholar] [CrossRef] [PubMed]
- Kabir, T.F.; Kunos, C.A.; Villano, J.L.; Chauhan, A. Immunotherapy for Medulloblastoma: Current Perspectives. Immunotargets Ther. 2020, 9, 57–77. [Google Scholar] [CrossRef] [Green Version]
- Vermeulen, J.F.; Van Hecke, W.; Adriaansen, E.J.M.; Jansen, M.K.; Bouma, R.G.; Villacorta Hidalgo, J.; Fisch, P.; Broekhuizen, R.; Spliet, W.G.M.; Kool, M.; et al. Prognostic relevance of tumor-infiltrating lymphocytes and immune checkpoints in pediatric medulloblastoma. Oncoimmunology 2018, 7, e1398877. [Google Scholar] [CrossRef]
- Miller, L.D.; Chou, J.A.; Black, M.A.; Print, C.; Chifman, J.; Alistar, A.; Putti, T.; Zhou, X.; Bedognetti, D.; Hendrickx, W.; et al. Immunogenic Subtypes of Breast Cancer Delineated by Gene Classifiers of Immune Responsiveness. Cancer Immunol. Res. 2016, 4, 600–610. [Google Scholar] [CrossRef] [Green Version]
- Gao, G.; Yu, Z.; Zhao, X.; Fu, X.; Liu, S.; Liang, S.; Deng, A. Immune classification and identification of prognostic genes for uveal melanoma based on six immune cell signatures. Sci. Rep. 2021, 11, 22244. [Google Scholar] [CrossRef]
- Galon, J.; Pages, F.; Marincola, F.M.; Angell, H.K.; Thurin, M.; Lugli, A.; Zlobec, I.; Berger, A.; Bifulco, C.; Botti, G.; et al. Cancer classification using the Immunoscore: A worldwide task force. J. Transl. Med. 2012, 10, 205. [Google Scholar] [CrossRef]
- Jividen, K.; Li, H. Chimeric RNAs generated by intergenic splicing in normal and cancer cells. Genes Chromosomes Cancer 2014, 53, 963–971. [Google Scholar] [CrossRef]

| Transcript 1 | Chr | Position | Exon | Transcript 2 | Chr | Position | Exon | In-Frame |
|---|---|---|---|---|---|---|---|---|
| KPNA3 | chr13 | 50321083–50321128 | exon 16 | TNFSF13B | chr13 | 108955599–108955712 | exon 4 | no |
| KPNA3 | chr13 | 50321083–50321128 | exon 16 | TNFSF13B | chr13 | 108939207–108955598 | intron 3 | no |
| EPC2 | chr2 | 149402558–149402738 | exon 1 | GULP1 | chr2 | 189433964–189434081 | exon 8 | yes |
| EPC2 | chr2 | 149402739–149447780 | intron 1 | GULP1 | chr2 | 189433964–189434081 | exon 8 | no |
| NFIB | chr9 | 14306986–14307518 | exon 8 | ENSG00000237137 | chr9 | 14531912–14532039 | exon 1 | no |
| Sample | Age at Diagnosis | Genetic Subtype | Histologic Subtype | EPC2–GULP1 Fusion |
|---|---|---|---|---|
| 25 | 16 | WNT | desmoplastic medulloblastoma | yes |
| 80 | 6 | SHH | large cell/anaplastic medulloblastoma | yes |
| 81 | 5 | Group 3/4 | classic medulloblastoma | no |
| 129 | 8 | WNT | classic medulloblastoma | no |
| 132 * | 7 | SHH | large cell/anaplastic medulloblastoma | yes |
| 210 | 5 | Group 3/4 | classic medulloblastoma | no |
| 265 | 11 | Group 3/4 | large cell/anaplastic medulloblastoma | no |
| 280 | 6 | Group 3 | classic medulloblastoma | no |
| 302 | 1 | SHH | desmoplastic medulloblastoma | no |
| 402 | 1 | SHH | desmoplastic nodular medulloblastoma | no |
| 423 | 6 | Group 3/4 | classic medulloblastoma | no |
| Healthy Donor | IFN-γ Secretion of CD8+ T Cells |
|---|---|
| 1 | 1144% |
| 2 | 131% |
| 3 | 354% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paret, C.; Lehmann, N.; Bender, H.; Sprang, M.; Sommer, C.J.; Cana, D.; Seidmann, L.; Wingerter, A.; Neu, M.A.; El Malki, K.; et al. Identification of an Immunogenic Medulloblastoma-Specific Fusion Involving EPC2 and GULP1. Cancers 2021, 13, 5838. https://doi.org/10.3390/cancers13225838
Paret C, Lehmann N, Bender H, Sprang M, Sommer CJ, Cana D, Seidmann L, Wingerter A, Neu MA, El Malki K, et al. Identification of an Immunogenic Medulloblastoma-Specific Fusion Involving EPC2 and GULP1. Cancers. 2021; 13(22):5838. https://doi.org/10.3390/cancers13225838
Chicago/Turabian StyleParet, Claudia, Nadine Lehmann, Hannah Bender, Maximilian Sprang, Clemens J. Sommer, Denis Cana, Larissa Seidmann, Arthur Wingerter, Marie A. Neu, Khalifa El Malki, and et al. 2021. "Identification of an Immunogenic Medulloblastoma-Specific Fusion Involving EPC2 and GULP1" Cancers 13, no. 22: 5838. https://doi.org/10.3390/cancers13225838
APA StyleParet, C., Lehmann, N., Bender, H., Sprang, M., Sommer, C. J., Cana, D., Seidmann, L., Wingerter, A., Neu, M. A., El Malki, K., Alt, F., Roth, L., Marini, F., Ottenhausen, M., Glaser, M., Knuf, M., Russo, A., & Faber, J. (2021). Identification of an Immunogenic Medulloblastoma-Specific Fusion Involving EPC2 and GULP1. Cancers, 13(22), 5838. https://doi.org/10.3390/cancers13225838







