hENT1 Predicts Benefit from Gemcitabine in Pancreatic Cancer but Only with Low CDA mRNA
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Tissue Microarray Manufacture
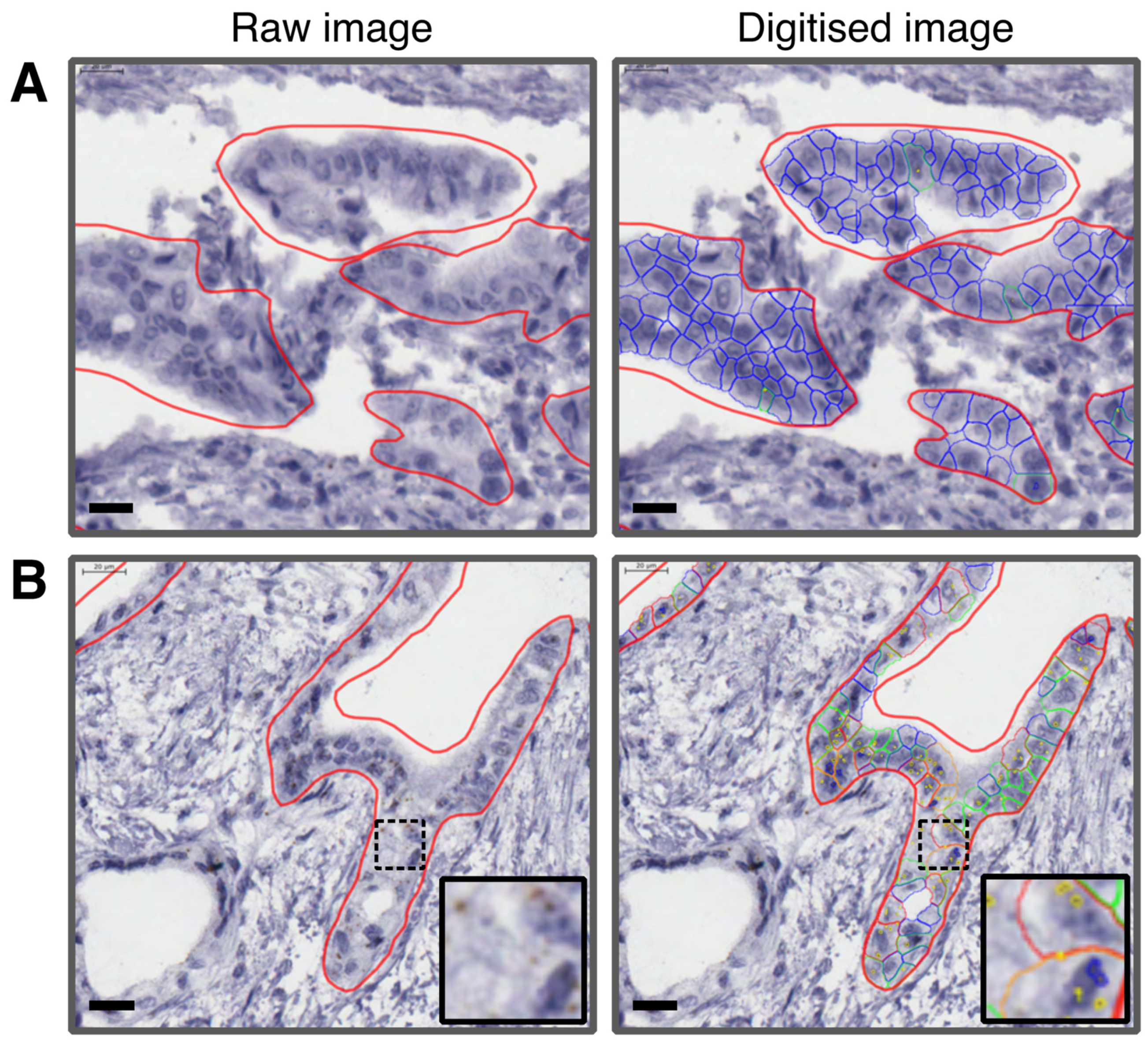
2.3. RNAscope® In Situ Hybridization (ISH)
2.4. Quantification Using RNAscope SpotStudio® Software
2.5. Immunohistochemistry
2.6. Statistical Analysis
3. Results
3.1. Determining Expression Levels of hENT1 and CDA in PDAC
3.2. Univariate and Multivariable Analyses of Clinical and Pathological Characteristics
3.3. Overall Survival Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Kleeff, J.; Korc, M.; Apte, M.; La Vecchia, C.; Johnson, C.D.; Biankin, A.V.; Neale, R.E.; Tempero, M.; Tuveson, D.A.; Hruban, R.H.; et al. Pancreatic cancer. Nat. Rev. Dis. Primers 2016, 2, 16022. [Google Scholar] [CrossRef]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef]
- Neoptolemos, J.; Stocken, D.D.; Friess, H.; Bassi, C.; Dunn, J.; Hickey, H.; Beger, H.; Fernandez-Cruz, L.; Dervenis, C.; Lacaine, F.; et al. A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer. N. Engl. J. Med. 2004, 350, 1200–1210. [Google Scholar] [CrossRef]
- Neoptolemos, J.P.; Stocken, D.D.; Bassi, C.; Ghaneh, P.; Cunningham, D.; Goldstein, D.; Padbury, R.; Moore, M.J.; Gallinger, S.; Mariette, C.; et al. Adjuvant chemotherapy with fluorouracil plus folinic acid vs gemcitabine following pancreatic cancer resection: A randomized controlled trial. JAMA 2010, 304, 1073–1081. [Google Scholar] [CrossRef]
- Oettle, H.; Neuhaus, P.; Hochhaus, A.; Hartmann, J.T.; Gellert, K.; Ridwelski, K.; Niedergethmann, M.; Zulke, C.; Fahlke, J.; Arning, M.B.; et al. Adjuvant chemotherapy with gemcitabine and long-term outcomes among patients with resected pancreatic cancer: The CONKO-001 randomized trial. JAMA 2013, 310, 1473–1481. [Google Scholar] [CrossRef]
- Oettle, H.; Post, S.; Neuhaus, P.; Gellert, K.; Langrehr, J.; Ridwelski, K.; Schramm, H.; Fahlke, J.; Zuelke, C.; Burkart, C.; et al. Adjuvant chemotherapy with gemcitabine vs observation in patients undergoing curative-intent resection of pancreatic cancer-A Randomized controlled trial. JAMA 2007, 297, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Valle, J.; Palmer, D.; Jackson, R.; Cox, T.; Neoptolemos, J.; Ghaneh, P.; Rawcliffe, C.L.; Bassi, C.; Stocken, D.D.; Cunningham, D.; et al. Optimal Duration and Timing of Adjuvant Chemotherapy After Definitive Surgery for Ductal Adenocarcinoma of the Pancreas: Ongoing Lessons From the ESPAC-3 Study. J. Clin. Oncol. 2014, 32, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Neoptolemos, J.P.; Dunn, J.A.; Stocken, D.D.; Almond, J.; Link, K.; Beger, H.; Bassi, C.; Falconi, M.; Pederzoli, P.; Dervenis, C.; et al. ESPAC-1: A European, randomized controlled study of adjuvant chemoradiotherapy and chemotherapy in resectable pancreatic cancer. Lancet 2001, 358, 1576–1585. [Google Scholar] [CrossRef]
- Neoptolemos, J.P.; Stocken, D.D.; Smith, C.T.; Bassi, C.; Ghaneh, P.; Owen, E.; Moore, M.; Padbury, R.; Doi, R.; Büchler, M.W. Adjuvant 5-fluorouracil and folinic acid vs observation for pancreatic cancer: Composite data from the ESPAC-1 and -3(v1) trials. Br. J. Cancer 2009, 100, 246–250. [Google Scholar] [CrossRef]
- Neoptolemos, J.P.; Palmer, D.H.; Ghaneh, P.; Psarelli, E.E.; Valle, J.W.; Halloran, C.M.; Faluyi, O.; O’Reilly, D.A.; Cunningham, D.; Wadsley, J.; et al. Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): A multicentre, open-label, randomised, phase 3 trial. Lancet 2017, 389, 1011–1024. [Google Scholar] [CrossRef]
- Conroy, T.; Hammel, P.; Hebbar, M.; Ben Abdelghani, M.; Wei, A.C.; Raoul, J.-L.; Choné, L.; Francois, E.; Artru, P.; Biagi, J.J.; et al. FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic cancer. N. Engl. J. Med. 2018, 379, 2395–2406. [Google Scholar] [CrossRef] [PubMed]
- Massard, C.; Michiels, S.; Ferté, C.; Le Deley, M.-C.; Lacroix, L.; Hollebecque, A.; Verlingue, L.; Ileana, E.; Rosellini, S.; Ammari, S.; et al. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: Results of the MOSCATO 01 trial. Cancer Discov. 2017, 7, 586–595. [Google Scholar] [CrossRef] [PubMed]
- Biankin, A.V.; Waddell, N.; Kassahn, K.S.; Gingras, M.-C.; Muthuswamy, L.B.; Johns, A.L.; Miller, D.K.; Wilson, P.J.; Patch, A.-M.; Wu, J.; et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012, 491, 399–405. [Google Scholar] [CrossRef]
- Waddell, N.; Pajic, M.; Patch, A.-M.; Chang, D.K.; Kassahn, K.S.; Bailey, P.; Johns, A.L.; Miller, D.; Nones, K.; Quek, K.; et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015, 518, 495–501. [Google Scholar] [CrossRef]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.-M.; Gingras, M.-C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.C.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef]
- Lowery, M.; Jordan, E.J.; Basturk, O.; Ptashkin, R.N.; Zehir, A.; Berger, M.F.; Leach, T.; Herbst, B.; Askan, G.; Maynard, H.; et al. Real-Time Genomic Profiling of Pancreatic Ductal Adenocarcinoma: Potential Actionability and Correlation with Clinical Phenotype. Clin. Cancer Res. 2017, 23, 6094–6100. [Google Scholar] [CrossRef]
- Aguirre, A.J.; Nowak, J.A.; Camarda, N.; Moffitt, R.A.; Ghazani, A.A.; Hazar-Rethinam, M.; Raghavan, S.; Kim, J.; Brais, L.K.; Ragon, D.; et al. Real-time genomic characterization of advanced pancreatic cancer to enable precision medicine. Cancer Discov. 2018, 8, 1096–1111. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Electronic address aadhe, Cancer Genome Atlas Research N. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 2017, 32, e113. [Google Scholar]
- Moffitt, R.A.; Marayati, R.; Flate, E.L.; Volmar, K.E.; Loeza, S.G.H.; Hoadley, K.A.; Rashid, N.U.; Williams, L.A.; Eaton, S.C.; Chung, A.H.; et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015, 47, 1168–1178. [Google Scholar] [CrossRef] [PubMed]
- Costello, E.; Greenhalf, W.; Neoptolemos, J.P. New biomarkers and targets in pancreatic cancer and their application to treatment. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Young, J.D.; Yao, S.Y.M.; Sun, L.; Cass, C.E.; Baldwin, S.A. Human equilibrative nucleoside transporter (ENT) family of nucleoside and nucleobase transporter proteins. Xenobiotica 2008, 38, 995–1021. [Google Scholar] [CrossRef]
- Greenhalf, W.; Ghaneh, P.; Neoptolemos, J.; Palmer, D.H.; Cox, T.; Lamb, R.F.; Garner, E.; Campbell, F.; Mackey, J.R.; Costello, E.; et al. Pancreatic cancer hENT1 expression and survival from gemcitabine in patients from the ESPAC-3 trial. J. Natl. Cancer Inst. 2014, 106, djt347. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.L.; Sherali, A.; Mo, Z.-P.; Tse, C.-M. Kinetic and pharmacological properties of cloned human equilibrative nucleoside transporters, ENT1 and ENT2, stably expressed in nucleoside transporter-deficient PK15 cells. ENT2 exhibits a low affinity for guanosine and cytidine but a high affinity for inosine. J. Biol. Chem. 2000, 275, 8375–8381. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.Y.; Ng, A.M.; Cass, C.E.; Baldwin, S.A.; Young, J.D. Nucleobase transport by human equilibrative nucleoside transporter 1 (hENT1). J. Biol. Chem. 2011, 286, 32552–32562. [Google Scholar] [CrossRef]
- Derissen, E.J.B.; Huitema, A.D.R.; Rosing, H.; Schellens, J.H.M.; Beijnen, J.H. Intracellular pharmacokinetics of gemcitabine, its deaminated metabolite 2′,2′-difluorodeoxyuridine and their nucleotides. Br. J. Clin. Pharmacol. 2018, 84, 1279–1289. [Google Scholar] [CrossRef]
- Somasekaram, A.; Jarmuz, A.; How, A.; Scott, J.; Navaratnam, N. Intracellular localization of human cytidine deaminase. Identification of a functional nuclear localization signal. J. Biol. Chem. 1999, 274, 28405–28412. [Google Scholar] [CrossRef]
- Ge, Y.; Jensen, T.L.; Stout, M.L.; Flatley, R.M.; Grohar, P.J.; Ravindranath, Y.; Matherly, L.H.; Taub, J.W. The role of cytidine deaminase and GATA1 mutations in the increased cytosine arabinoside sensitivity of Down syndrome myeloblasts and leukemia cell lines. Cancer Res. 2004, 64, 728–735. [Google Scholar] [CrossRef]
- Bjånes, T.K.; Jordheim, L.P.; Schjøtt, J.; Kamceva, T.; Cros-Perrial, E.; Langer, A.; de Garibay, G.R.; Kotopoulis, S.; Mc Cormack, E.; Riedel, B. Intracellular cytidine deaminase regulates gemcitabine metabolism in pancreatic cancer cell lines. Drug Metab. Dispos. 2020, 48, 153–158. [Google Scholar] [CrossRef]
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156–1160. [Google Scholar] [CrossRef]
- Giovannetti, E.; Peters, G.J. Molecular targets of gemcitabine action: Rationale for development of novel drugs and drug combinations. Curr. Pharm. Des. 2012, 18, 2811–2829. [Google Scholar] [CrossRef]
- Honeywell, R.J.; Ruiz van Haperen, V.W.; Veerman, G.; Smid, K.; Peters, G.J. Inhibition of thymidylate synthase by 2′,2′-difluoro-2′-deoxycytidine (Gemcitabine) and its metabolite 2′,2′-difluoro-2′-deoxyuridine. Int. J. Biochem. Cell Biol. 2015, 60C, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Alvarellos, M.L.; Lamba, J.; Sangkuhl, K.; Thorn, C.F.; Wang, L.; Klein, D.J.; Altman, R.B.; Klein, T.E. PharmGKB summary: Gemcitabine pathway. Pharm. Genom. 2014, 24, 564–574. [Google Scholar] [CrossRef]
- Tibaldi, C.; Giovannetti, E.; Vasile, E.; Mey, V.; Laan, A.C.; Nannizzi, S.; Di Marsico, R.; Antonuzzo, A.; Orlandini, C.; Ricciardi, S.; et al. Correlation of CDA, ERCC1, and XPD polymorphisms with response and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin. Cancer Res. 2008, 14, 1797–1803. [Google Scholar] [CrossRef] [PubMed]
- Weizman, N.; Krelin, Y.; Shabtayorbach, A.; Amit, M.; Binenbaum, Y.; Wong, R.J.; Gil, Z. Macrophages mediate gemcitabine resistance of pancreatic adenocarcinoma by upregulating cytidine deaminase. Oncogene 2013, 33, 3812–3819. [Google Scholar] [CrossRef]
- Binenbaum, Y.; Fridman, E.; Yaari, Z.; Milman, N.; Schroeder, A.; Ben David, G.; Shlomi, T.; Gil, Z. Transfer of miRNA in macrophage-derived exosomes induces drug resistance in pancreatic adenocarcinoma. Cancer Res. 2018, 78, 5287–5299. [Google Scholar] [CrossRef] [PubMed]
- McShane, L.M.; Altman, D.G.; Sauerbrei, W.; Taube, S.E.; Gion, M.; Clark, G.M. Statistics Subcommittee of the NCIEWGoCD. REporting recommendations for tumour MARKer prognostic studies (REMARK). Br. J. Cancer 2005, 93, 387–391. [Google Scholar] [CrossRef]
- Kaplan, E.; Meier, P. Non-parametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958, 53, 457–481. [Google Scholar] [CrossRef]
- Zhong, Y.; Macgregor-Das, A.; Saunders, T.; Whittle, M.C.; Makohon-Moore, A.; Kohutek, Z.A.; Poling, J.; Herbst, B.T.; Javier, B.M.; Cope, L.; et al. Mutant p53 together with TGFbeta signaling influence organ-specific hematogenous colonization patterns of pancreatic cancer. Clin. Cancer Res. 2017, 23, 1607–1620. [Google Scholar] [CrossRef]
- Farrell, J.J.; Elsaleh, H.; Garcia, M.; Lai, R.; Ammar, A.; Regine, W.F.; Abrams, R.; Benson, A.B.; Macdonald, J.; Cass, C.E.; et al. Human equilibrative nucleoside transporter 1 levels predict response to gemcitabine in patients with pancreatic cancer. Gastroenterology 2009, 136, 187–195. [Google Scholar] [CrossRef]
- Giovannetti, E.; Del Tacca, M.; Mey, V.; Funel, N.; Nannizzi, S.; Ricci, S.; Orlandini, C.; Boggi, U.; Campani, D.; Del Chiaro, M.; et al. Transcription analysis of human equilibrative nucleoside transporter-1 predicts survival in pancreas cancer patients treated with gemcitabine. Cancer Res. 2006, 66, 3928–3935. [Google Scholar] [CrossRef] [PubMed]
- Tavano, F.; Fontana, A.; Pellegrini, F.; Burbaci, F.P.; Rappa, F.; Cappello, F.; Copetti, M.; Maiello, E.; Lombardi, L.; Graziano, P.; et al. Modeling interactions between Human Equilibrative Nucleoside Transporter-1 and other factors involved in the response to gemcitabine treatment to predict clinical outcomes in pancreatic ductal adenocarcinoma patients. J. Transl. Med. 2014, 12, 248. [Google Scholar] [CrossRef] [PubMed]
- Funamizu, N.; Okamoto, A.; Kamata, Y.; Misawa, T.; Uwagawa, T.; Gocho, T.; Yanaga, K.; Manome, Y. Is the resistance of gemcitabine for pancreatic cancer settled only by overexpression of deoxycytidine kinase? Oncol. Rep. 2009, 23, 471–475. [Google Scholar] [CrossRef]
- Nio, Y.; Toga, T.; Maruyama, R.; Fukushima, M. Expression of orotate phosphoribosyl transferase in human pancreatic cancer: Implication for the efficacy of uracil and tegafur-based adjuvant chemotherapy. Oncol. Rep. 2007, 18, 59–64. [Google Scholar] [CrossRef][Green Version]
- Traut, T.W.; Jones, M.E. Uracil metabolism—UMP synthesis from orotic acid or uridine and conversion of uracil to beta-alanine: Enzymes and cDNAs. Prog. Nucleic Acid Res. Mol. Biol. 1996, 53, 1–78. [Google Scholar]
- Raffenne, J.; Nicolle, R.; Puleo, F.; Le Corre, D.; Boyez, C.; Marechal, R.; Emile, J.F.; Demetter, P.; Bardier, A.; Laurent-Puig, P.; et al. hENT1 Testing in Pancreatic Ductal Adenocarcinoma: Are We Ready? A Multimodal Evaluation of hENT1 Status. Cancers 2019, 11, 1808. [Google Scholar] [CrossRef]
| Summary Statistics | Hazard Ratio (95% Confidence Interval) | |||||
|---|---|---|---|---|---|---|
| Characteristic | Level | 5-Fluorouracil | Gemcitabine | 5-Fluorouracil | Gemcitabine | Total |
| Resection Margin | n = 132 | n = 145 | n = 277 | |||
| Negative | 70 (53%) | 86 (59%) | 1 | 1 | 1 | |
| Positive | 62 (47%) | 59 (41%) | 1.80 (1.23–2.63) | 1.35 (0.94–1.94) | 1.56 (1.20–2.03) | |
| Wald χ2 = 9.05, p = 0.003 * | Wald χ2 = 2.62 p = 0.106 | Wald χ2 = 11.22, p = 0.001 | ||||
| WHO | n = 132 | n = 145 | n = 277 | |||
| 0 | 52 (39%) | 49 (34%) | 1 | 1 | 1 | |
| 1 | 67 (51%) | 80 (55%) | 1.26 (0.86–1.96) | 1.40 (0.94–2.07) | 1.33 (1.01–1.76) | |
| 2 | 13 (10%) | 16 (11%) | 0.68 (0.29–1.59) | 1.24 (0.68–2.25) | 0.92 (0.56–1.53) | |
| Wald χ2 = 2.89, p = 0.236 | Wald χ2 = 2.75 p = 0.253 | Wald χ2 = 5.24, p = 0.073 | ||||
| Lymph Node Status | n = 132 | n = 145 | n = 277 | |||
| Negative | 29 (22%) | 29 (20%) | 1 | 1 | 1 | |
| Positive | 103 (78%) | 116 (80%) | 2.40 (1.47–3.90) | 1.56 (0.99–2.46) | 1.94 (1.39–2.71) | |
| Wald χ2 = 12.30, p = 0.001 | Wald χ2 = 3.63, p = 0.057 | Wald χ2 = 15.18 p < 0.001 | ||||
| Tumor Stage | n = 131 | n = 144 | n = 275 | |||
| 01/02 | 38 (29%) | 46 (32%) | 1 | 1 | 1 | |
| 03/04 | 93 (70%) | 98 (68%) | 1.69 (1.10–2.59) | 1.39 (0.95–2.01) | 1.51 (1.15–2.00) | |
| Wald χ2= 5.81, p = 0.016 | Wald χ2 = 2.95, p = 0.086 | Wald χ2 = 8.47, p = 0.004 | ||||
| Tumor Grade | N = 129 | n = 142 | n = 271 | |||
| Well | 7 (5%) | 10 (7%) | 1 | 1 | 1 | |
| Moderately | 89 (67%) | 98 (68%) | 0.60 (0.37–0.96) | 0.83 (0.44–1.58) | 0.72 (0.47–1.01) | |
| Poorly | 33 (25%) | 34 (23%) | 0.67 (0.35–1.28) | 1.19 (0.58–2.44) | 0.91 (0.55–1.51) | |
| Wald χ2 = 4.54, p = 0.103 | Wald χ2 = 2.75, p = 0.753 | Wald χ2 = 3.60 p = 0.165 | ||||
| Local Invasion | n = 132 | n = 142 | n = 274 | |||
| No | 73 (55%) | 72 (50%) | 1 | 1 | 1 | |
| Yes | 59 (45%) | 70 (48%) | 1.25 (0.85–1.84) | 1.10 (0.77–1.58) | 1.17 (0.90–1.52) | |
| Wald χ2 = 1.31, p = 0.252 | Wald χ2 = 0.26, p = 0.607 | Wald χ2 = 1.37 p = 0.242 | ||||
| Maximum Tumor diameter | n = 128 | n = 139 | n = 267 | |||
| <30 mm | 69 (52%) | 58 (40%) | 1 | 1 | 1 | |
| ≥30 mm | 59 (45%) | 81 (56%) | 1.31 (0.89–1.95) | 1.64 (1.13–2.39) | 1.47 (1.12–1.92) | |
| Wald χ2 = 1.82, p = 0.177 | Wald χ2 = 6.73, p = 0.010 | Wald χ2 = 7.84 p = 0.005 | ||||
| Diabetes mellitus | n = 129 | n = 141 | n = 270 | |||
| No | 102 (77%) | 106 (73%) | 1 | 1 | 1 | |
| Yes | 27 (20%) | 35 (24%) | 1.06 (0.65–1.75) | 0.99 (0.64–1.53) | 1.02 (0.74–1.41) | |
| Wald χ2 = 0.07, p = 0.797 | Wald χ2 = 0.00, p = 0.951 | Wald χ2 = 0.01, p = 0.905 | ||||
| Gender | n = 132 | n = 145 | n = 277 | |||
| Male | 75 (57%) | 88 (61%) | 1 | 1 | 1 | |
| Female | 57 (43%) | 57 (39%) | 0.88 (0.59–1.32) | 1.25 (0.86–1.81) | 1.06 (0.81–1.39) | |
| Wald χ2= 0.38, p = 0.537 | Wald χ2 = 1.42, p = 0.234 | Wald χ2 = 0.16, p = 0.686 | ||||
| Age, years | n = 132 | n = 145 | n = 277 | |||
| ≥64 | 65 (49%) | 80 (55%) | 1 | 1 | 1 | |
| <64 | 67 (51%) | 65 (45%) | 1.37 (0.93–2.02) | 0.84 (0.58–1.21) | 1.07 (0.83–1.40) | |
| Wald χ2= 2.55, p = 0.110 | Wald χ2 = 0.90, p = 0.342 | Wald χ2 = 0.28, p = 0.598 | ||||
| Smoking | n = 125 | n = 128 | n = 253 | |||
| Never | 52 (39%) | 58 (40%) | 1 | 1 | 1 | |
| Ex | 51 (39%) | 52 (36%) | 0.93 (0.60–1.43) | 1.08 (0.70–1.66) | 1.00 (0.74–1.35) | |
| Current | 22 (17%) | 18 (12%) | 0.91 (0.53–1.57) | 1.76 (1.02–3.05) | 1.20 (0.81–1.78) | |
| Wald χ2 = 0.17, p = 0.917 | Wald χ2 = 4.16, p = 0.125 | Wald χ2 = 0.95, p = 0.623 | ||||
| 5FU | Gemcitabine | ||||||
|---|---|---|---|---|---|---|---|
| Characteristic | est (se) | HR (95% CI) | Pval | est (se) | HR (95% CI) | p-Value | |
| Single biomarker: CDA or hENT1 combined with stage, resection margin and lymph node involvement | |||||||
| CDA mRNA Expression | Per unit increase in MSPC | 1.47 (0.685) | 4.35 (1.14, 16.62) | 0.032 | 1.15 (0.623) | 3.15 (0.93, 10.68) | 0.066 |
| CDA mRNA Expression (Low vs. High) | Low | 1 (Reference) | 1 (Reference) | ||||
| High | 0.34 (0.221) | 1.41 (0.91, 2.17) | 0.120 | 0.49 (0.192) | 1.62 (1.12, 2.39) | 0.011 * | |
| hENT1 protein expression | Per unit increase in (log) H-score | −0.03 (0.159) | 0.97 (0.71, 1.33) | 0.861 | −0.26 (0.129) | 0.77 (0.60, 0.99) | 0.047 |
| hENT1 protein expression (Low vs. High) | Low | 1 (Reference) | 1 (Reference) | ||||
| High | 0.18 (0.207) | 1.21 (0.80, 1.81) | 0.374 | −0.43 (0.192) | 0.65 (0.45, 0.95) | 0.025 | |
| Multiple biomarker: CDA and hENT1 combined with stage, resection margin and lymph node involvement | |||||||
| CDA mRNA Expression (Low vs. High) | Low | 1 (Reference) | 1 (Reference) | ||||
| High | 0.39 (0.226) | 1.48 (0.95, 2.31) | 0.082 | 0.50 (0.193) | 1.65 (1.13, 2.41) | 0.009 | |
| hENT1 protein expression (Low vs. High) | Low | 1 (Reference) | 1 (Reference) | ||||
| High | 0.28 (0.212) | 1.32 (0.87, 1.99) | 0.193 | −0.41 (0.191) | 0.66 (0.46, 0.96) | 0.030 | |
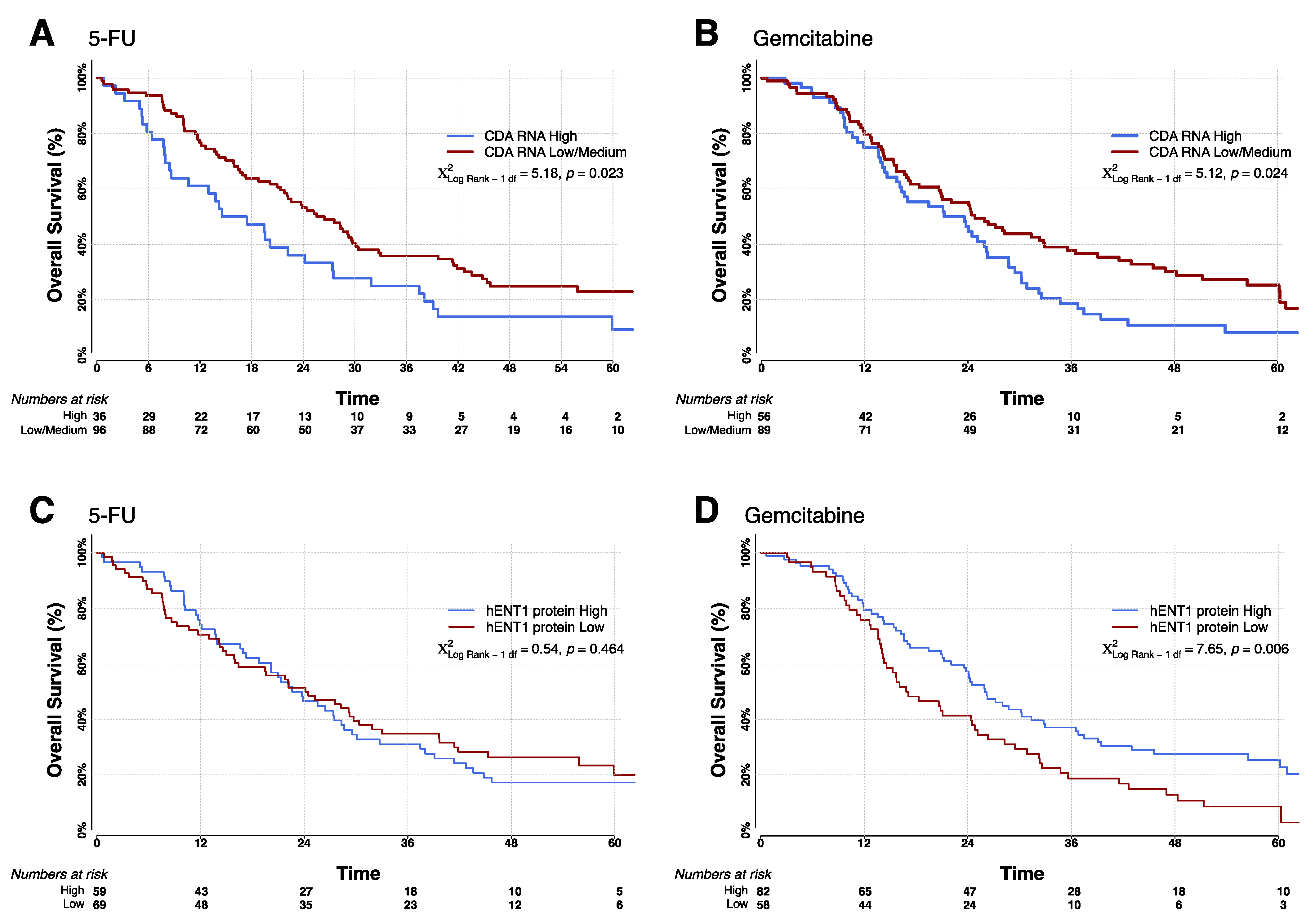
| Arm | Biomarker Expression (High or Low) | Number | Median OS | 95% Confidence Interval | Log Rank | p-Value |
|---|---|---|---|---|---|---|
| 5-FU/FA | CDA High | 36 | 14.6 | 8.4–24.1 | 5.17 | 0.0229 |
| CDA Low | 96 | 26.4 | 21.4–29.7 | |||
| GEM | CDA High | 56 | 21.2 | 15.7–26.2 | 5.14 | 0.0234 |
| CDA Low | 89 | 24.8 | 18.3–33.0 | |||
| 5-FU/FA | hENT1 High | 59 | 22.6 | 17.3–28.6 | 0.53 | 0.4658 |
| hENT1 Low | 69 | 24.1 | 15.9–30.4 | |||
| GEM | hENT1 High | 82 | 26.0 | 21.2–32.8 | 7.58 | 0.0059 |
| hENT1 Low | 58 | 16.8 | 14.1–24.8 | |||
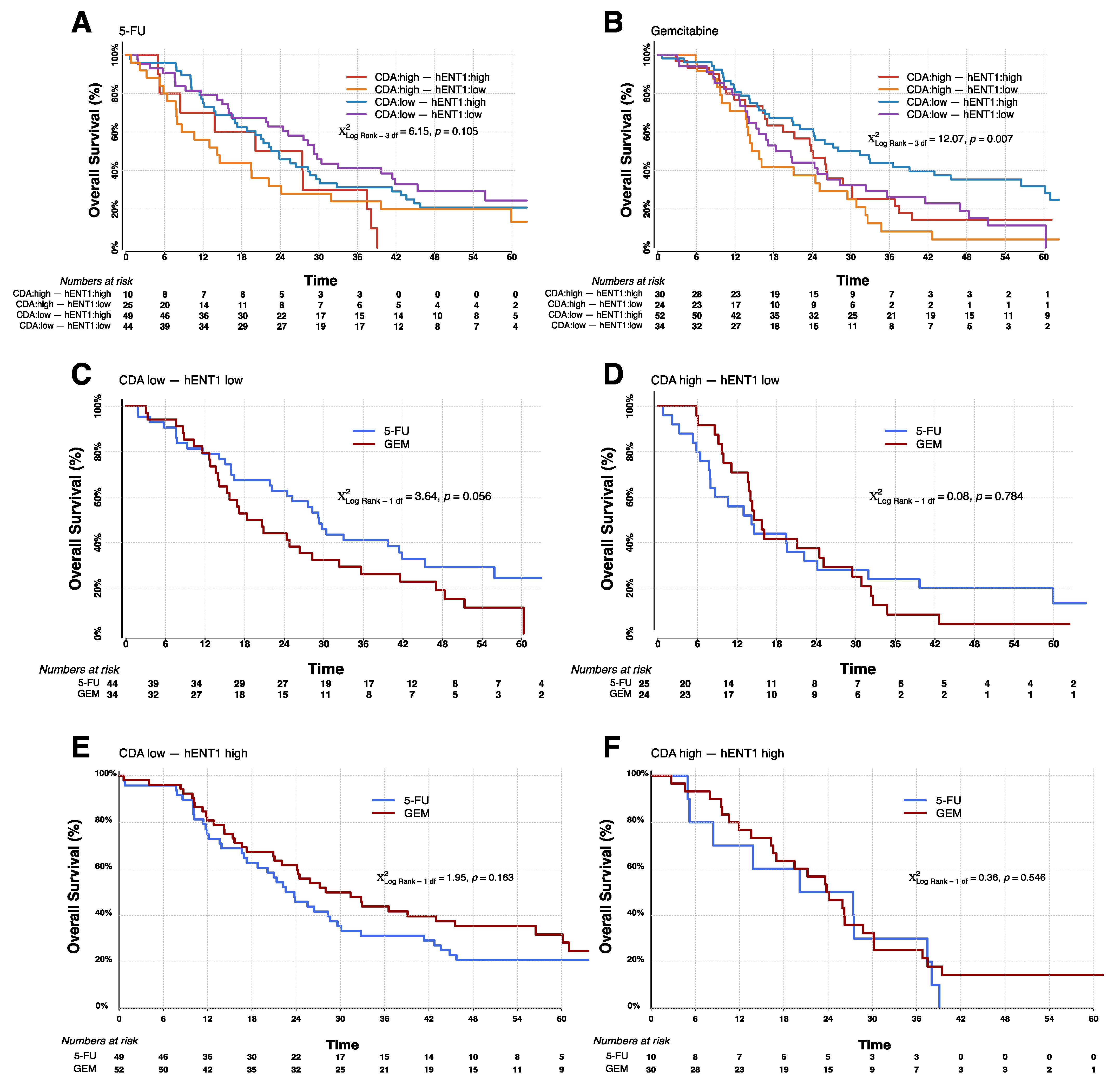
| 5-FU | CDA Low, hENT1 Low | 44 | 29.3 | 21.9–41.9 | 6.14 | 0.1050 |
| CDA High, hENT1 Low | 25 | 14.2 | 7.9–24.1 | |||
| CDA Low, hENT1 High | 49 | 22.6 | 16.9–29.6 | |||
| CDA High, hENT1 High | 10 | 20.1 | 5.0–37.5 | |||
| GEM | CDA Low, hENT1 Low | 34 | 18.3 | 13.9–28.3 | 12.0 | 0.0073 |
| CDA High, hENT1 Low | 24 | 14.6 | 11.1–25.1 | |||
| CDA Low, hENT1 High | 52 | 28.0 | 21.1–45.5 | |||
| CDA High, hENT1 High | 30 | 23.8 | 16.6–28.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aughton, K.; Elander, N.O.; Evans, A.; Jackson, R.; Campbell, F.; Costello, E.; Halloran, C.M.; Mackey, J.R.; Scarfe, A.G.; Valle, J.W.; et al. hENT1 Predicts Benefit from Gemcitabine in Pancreatic Cancer but Only with Low CDA mRNA. Cancers 2021, 13, 5758. https://doi.org/10.3390/cancers13225758
Aughton K, Elander NO, Evans A, Jackson R, Campbell F, Costello E, Halloran CM, Mackey JR, Scarfe AG, Valle JW, et al. hENT1 Predicts Benefit from Gemcitabine in Pancreatic Cancer but Only with Low CDA mRNA. Cancers. 2021; 13(22):5758. https://doi.org/10.3390/cancers13225758
Chicago/Turabian StyleAughton, Karen, Nils O. Elander, Anthony Evans, Richard Jackson, Fiona Campbell, Eithne Costello, Christopher M. Halloran, John R. Mackey, Andrew G. Scarfe, Juan W. Valle, and et al. 2021. "hENT1 Predicts Benefit from Gemcitabine in Pancreatic Cancer but Only with Low CDA mRNA" Cancers 13, no. 22: 5758. https://doi.org/10.3390/cancers13225758
APA StyleAughton, K., Elander, N. O., Evans, A., Jackson, R., Campbell, F., Costello, E., Halloran, C. M., Mackey, J. R., Scarfe, A. G., Valle, J. W., Carter, R., Cunningham, D., Tebbutt, N. C., Goldstein, D., Shannon, J., Glimelius, B., Hackert, T., Charnley, R. M., Anthoney, A., ... Greenhalf, W. (2021). hENT1 Predicts Benefit from Gemcitabine in Pancreatic Cancer but Only with Low CDA mRNA. Cancers, 13(22), 5758. https://doi.org/10.3390/cancers13225758









