Topological Analysis of γH2AX and MRE11 Clusters Detected by Localization Microscopy during X-ray-Induced DNA Double-Strand Break Repair
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Preparation, Irradiation, and SMLM Acquisition of the Data Set
2.2. Cluster Evaluation
2.3. Persistence Homology, Barcode Calculation, and Topological Similarities
2.4. Topological Similarities
2.5. Heatmaps
2.6. Confocal Microscopy
3. Results
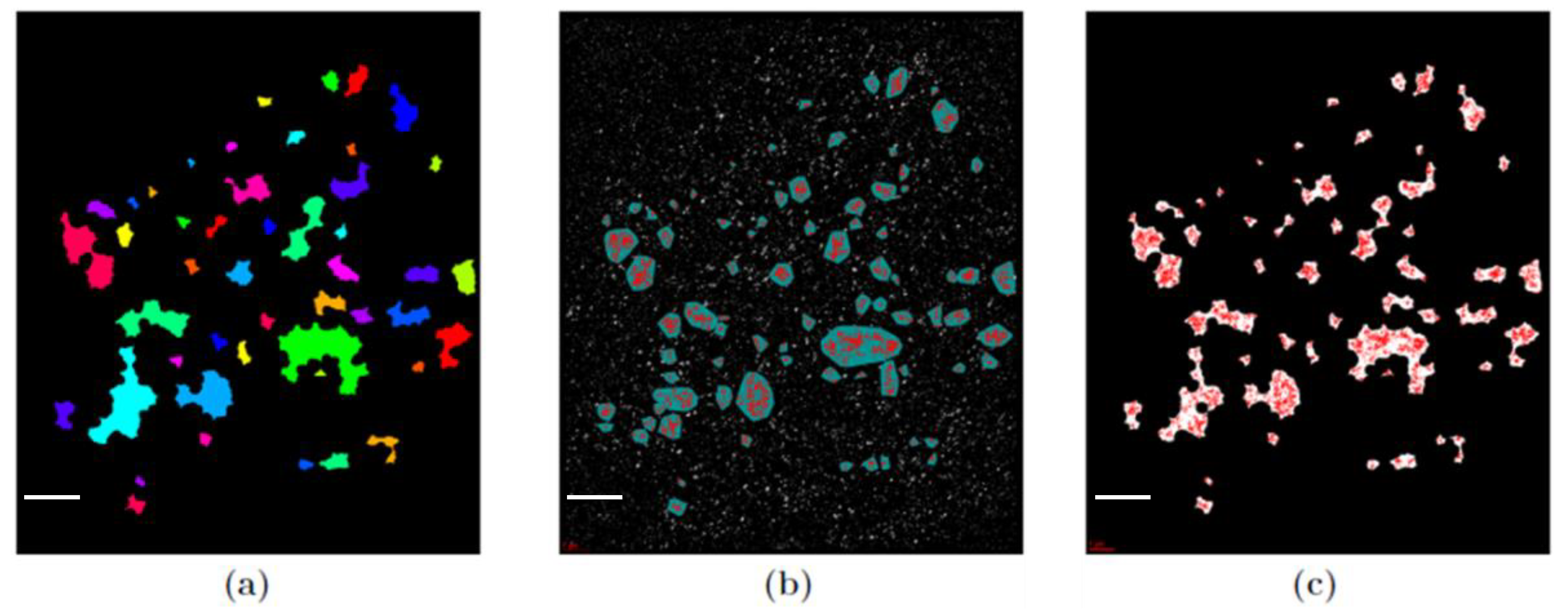
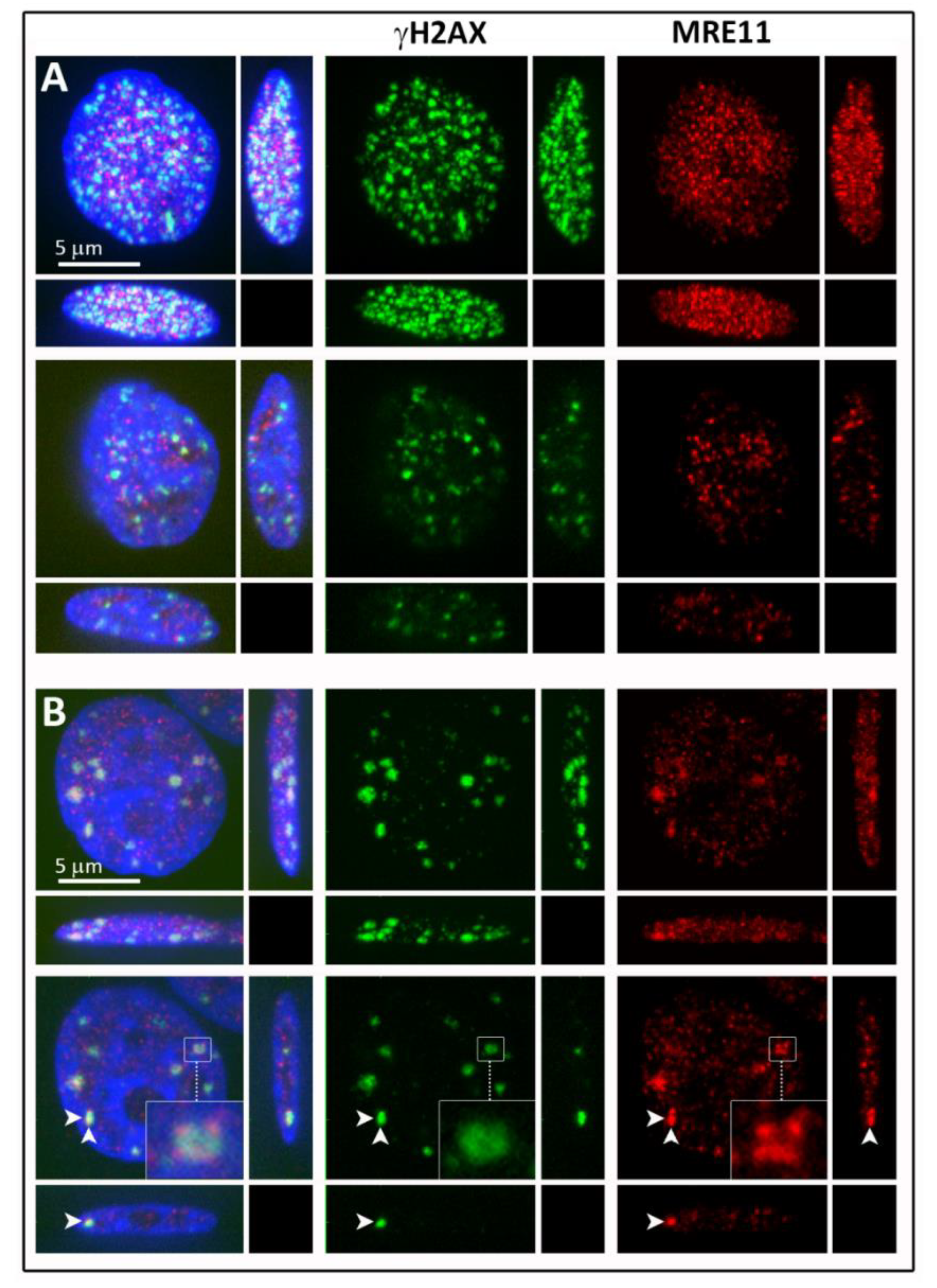
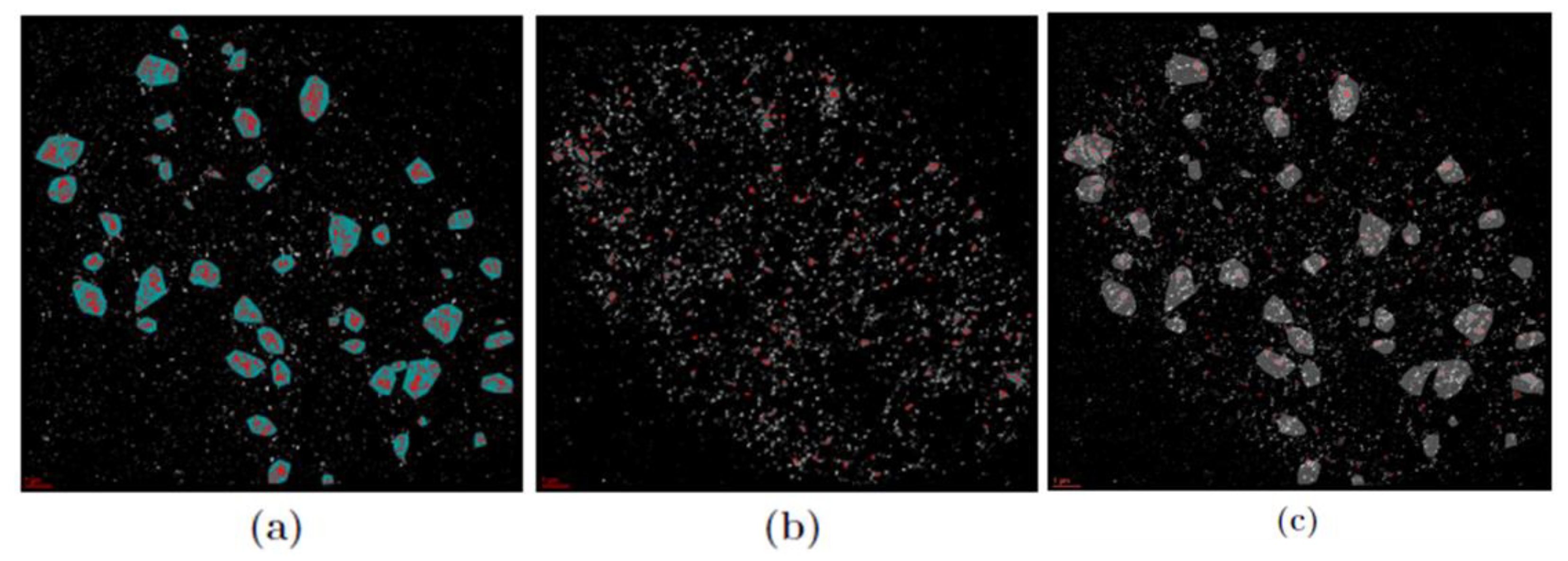
3.1. Cluster Formation
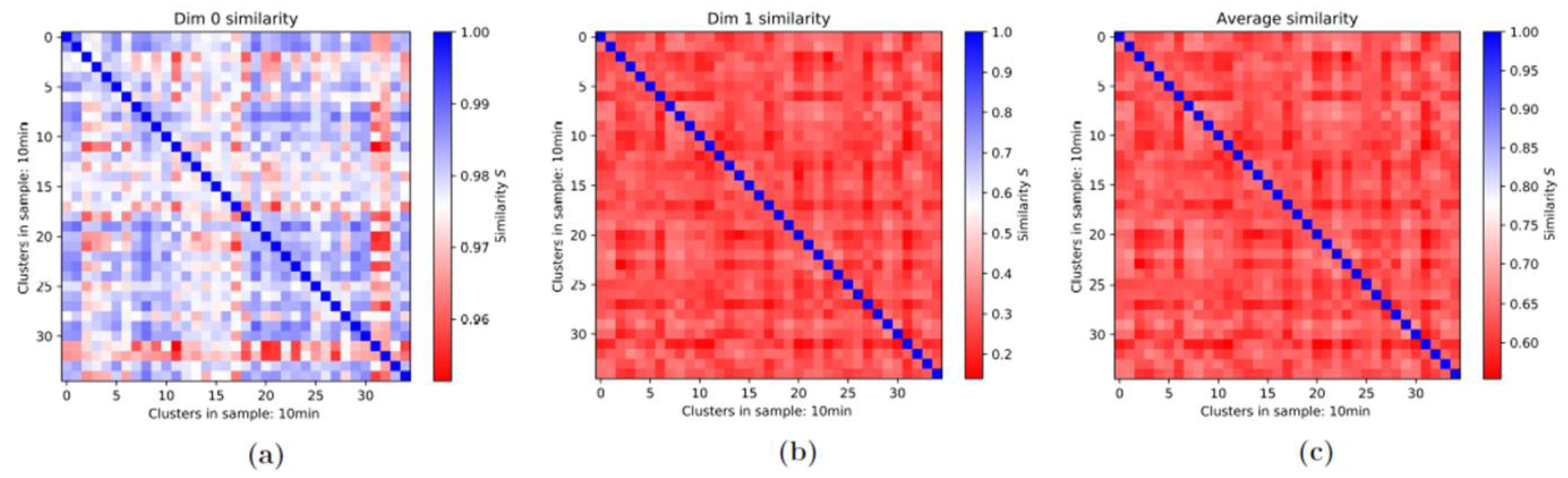
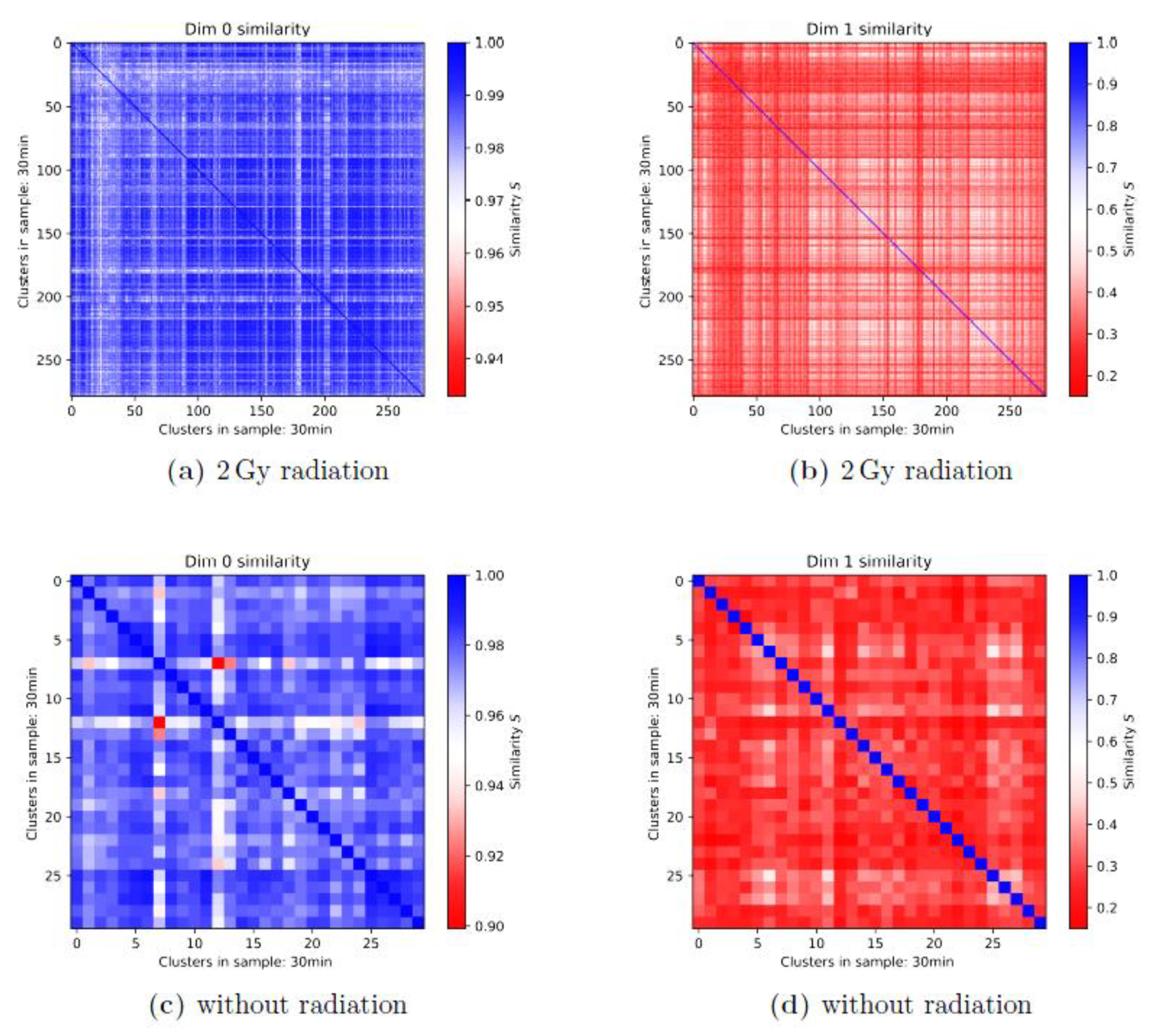
3.2. Persistence Homology and Similarities: 1st-Generation Heatmaps
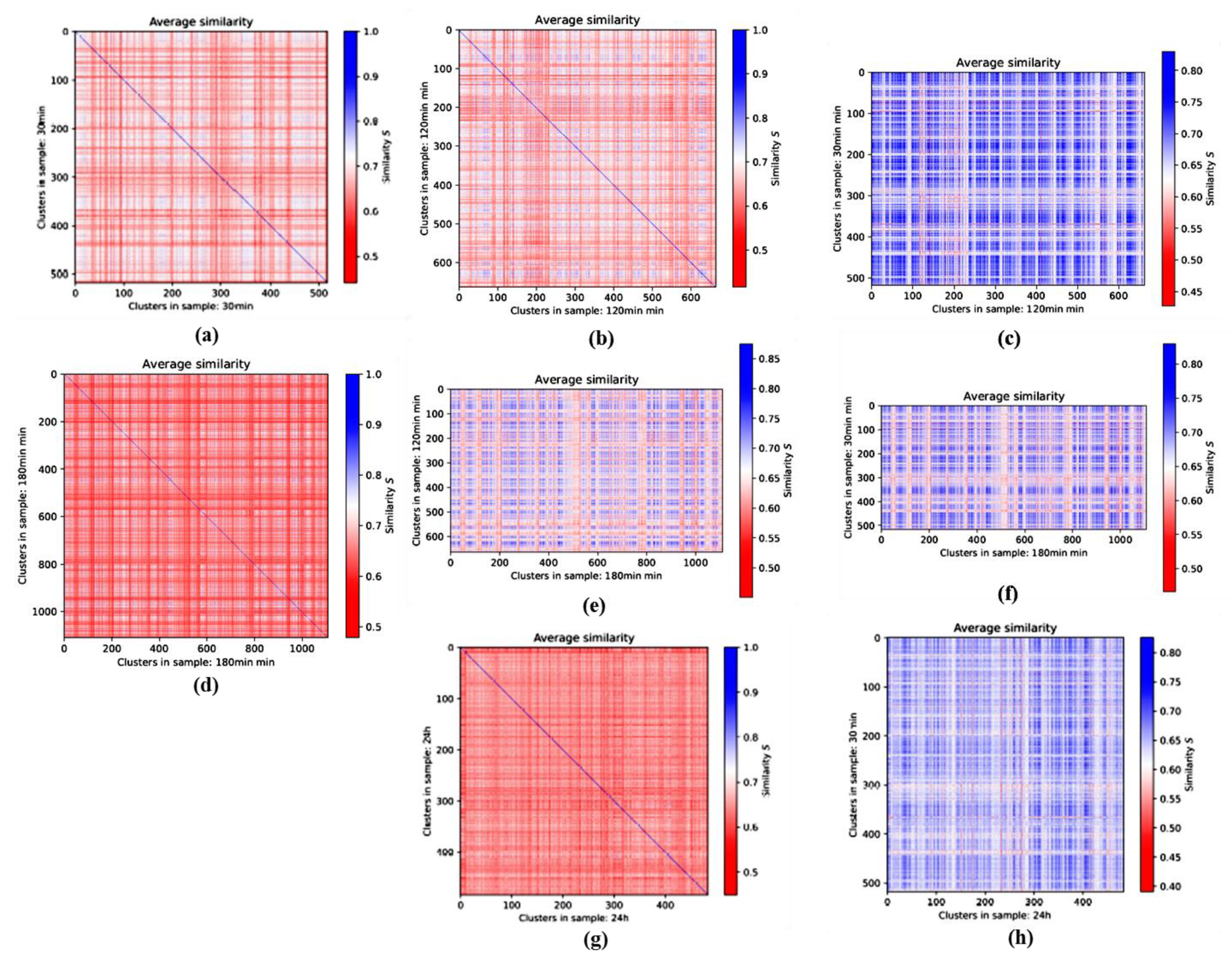
3.3. Averaged Similarities along the Repair Period: 2nd-Generation Heatmaps
3.3.1. 2nd-Generation Heatmaps of γH2AX
3.3.2. 2nd-Generation Heatmaps of MRE11
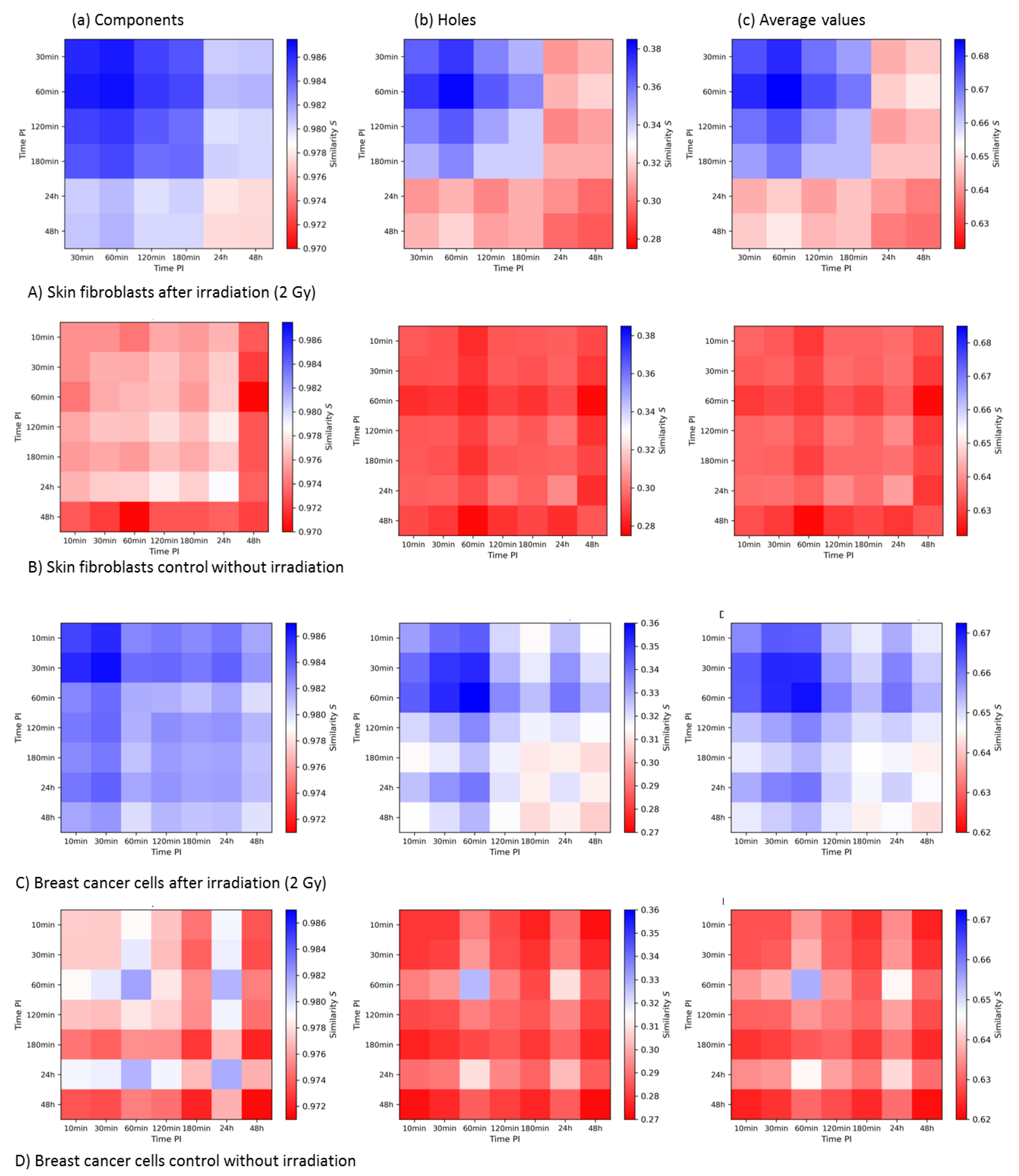
3.3.3. Summary and Comparison of 2nd-Generation Heatmaps of γH2AX and MRE11
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Falk, M.; Hausmann, M. A paradigm revolution or just better resolution—Will newly emerging superresolution techniques identify chromatin architecture as a key factor in radiation-induced DNA damage and repair regulation? Cancers 2021, 13, 18. [Google Scholar] [CrossRef]
- Lee, J.-H.; Hausmann, M. Super-resolution radiation biology: From bio-dosimetry towards nano-studies of DNA repair mechanisms. In DNA-Repair; Behzadi, P., Ed.; Intech-Open: Rijeka, Hrvatska, 2021; ISBN 978-1-83881-094-8. [Google Scholar] [CrossRef]
- Jezkova, L.; Zadneprianetc, M.; Kulikova, E.; Smirnova, E.; Bulanova, T.; Depes, D.; Falkova, I.; Boreyko, A.; Krasavin, E.; Davidkova, M.; et al. Particles with similar LET values generate DNA breaks of different complexity and reparability: A high-resolution microscopy analysis of γH2AX/53BP1 foci. Nanoscale 2018, 10, 1162–1179. [Google Scholar] [CrossRef] [Green Version]
- Depes, D.; Lee, J.-H.; Bobkova, E.; Jezkova, L.; Falkova, I.; Bestvater, F.; Pagacova, E.; Kopecna, O.; Zadneprianetc, M.; Bacikova, A.; et al. Single-molecule localization microscopy as a promising tool for γH2AX/53BP1 foci exploration. Eur. Phys. J. 2018, 72, 158. [Google Scholar] [CrossRef]
- Falk, M.; Hausmann, M.; Lukášová, E.; Biswas, A.; Hildenbrand, G.; Davídková, M.; Krasavin, E.; Kleibl, Z.; Falková, I.; Ježková, L.; et al. Determining OMICS spatiotemporal dimensions using exciting new nanoscopy techniques to asses complex cell responses to DNA damage—PART A (Radiomics). Crit. Rev. Eukaryot. Gene Express. 2014, 24, 205–223. [Google Scholar] [CrossRef] [PubMed]
- Falk, M.; Hausmann, M.; Lukášová, E.; Biswas, A.; Hildenbrand, G.; Davídková, M.; Krasavin, E.; Kleibl, Z.; Falková, I.; Ježková, L.; et al. Determining OMICS spatiotemporal dimensions using exciting new nanoscopy techniques to asses complex cell responses to DNA damage—PART B (Structuromics). Crit. Rev. Eukaryot. Gene Express. 2014, 24, 225–247. [Google Scholar] [CrossRef] [PubMed]
- Roobol, S.J.; van den Bent, I.; van Cappellen, W.A.; Abraham, T.E.; Paul, M.W.; Kanaar, R.; Houtsmuller, A.B.; van Gent, D.C.; Essers, J. Comparison of high- and low-LET radiation-induced DNA double-strand break processing in living cells. Int. J. Mol. Sci. 2020, 21, 6602. [Google Scholar] [CrossRef]
- Schipler, A.; Iliakis, G. DNA double-strand–break complexity levels and their possible contributions to the probability for error-prone processing and repair pathway choice. Nucleic Acids Res. 2013, 41, 7589–7605. [Google Scholar] [CrossRef] [Green Version]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA double-starnd breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redon, C.E.; Dickey, J.S.; Bonner, W.M.; Sedelnikova, O.A. γ-H2AX as a biomarker of DNA damage induced by ionizing radiation in human peripheral blood lymphocytes and artificial skin. Adv. Space Res. 2009, 43, 1171–1178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eberlein, U.; Peper, M.; Fernandez, M.; Lassmann, M.; Scherthan, H. Calibration of the γ-H2AX DNA double strand break focus assay for internal radiation exposure of blood lymphocytes. PLoS ONE 2015, 10, e0123174. [Google Scholar] [CrossRef]
- Hausmann, M.; Wagner, E.; Lee, J.-H.; Schrock, G.; Schaufler, W.; Krufczik, M.; Papenfuß, F.; Port, M.; Bestvater, F.; Scherthan, H. Super-resolution localization microscopy of radiation-induced histone H2AX-phosphorylation in relation to H3K9-trimethylation in HeLa cells. Nanoscale 2018, 10, 4320–4331. [Google Scholar] [CrossRef]
- Belyaev, I.Y. Radiation-induced DNA repair foci: Spatio-temporal aspects of formation, application for assessment of radiosensitivity and biological dosimetry. Mutat. Res. Rev. Mutat. Res. 2010, 704, 132–141. [Google Scholar] [CrossRef]
- Moquet, J.; Rothkamm, K.; Barnard, S.; Ainsbury, E. Radiation Biomarkers in Large Scale Human Health Effects Studies. J. Pers. Med. 2020, 10, 155. [Google Scholar] [CrossRef] [PubMed]
- Cremer, C.; Kaufmann, R.; Gunkel, M.; Pres, S.; Weiland, Y.; Müller, P.; Ruckelshausen, T.; Lemmer, P.; Geiger, F.; Degenhard, M.; et al. Superresolution imaging of biological nanostructures by Spectral Precision Distance Microscopy (SPDM). Biotechnol. J. 2011, 6, 1037–1051. [Google Scholar] [CrossRef]
- Cremer, C.; Masters, B.R. Resolution Enhancement Techniques in Microscopy. Eur. Phys. J. H 2013, 38, 281–344. [Google Scholar] [CrossRef] [Green Version]
- Miné-Hattab, J.; Heltberg, M.; Villemeur, M.; Guedj, C.; Mora, T.; Walczak, A.M.; Dahan, M.; Taddei, A. Single molecule microscopy reveals key physical features of repair foci in living cells. eLife 2021, 10, e60577. [Google Scholar] [CrossRef]
- Natale, F.; Rapp, A.; Yu, W.; Maiser, A.; Harz, H.; Scholl, A.; Grulich, S.; Anton, T.; Hörl, D.; Chen, W.; et al. Identification of the elementary structural units of the DNA damage response. Nat. Commun. 2017, 8, 15760. [Google Scholar] [CrossRef]
- Hausmann, M.; Falk, M.; Neitzel, C.; Hofmann, A.; Biswas, A.; Gier, T.; Falkova, I.; Heermann, D.W.; Hildenbrand, G. Elucidation of the clustered nano-architecture of radiation-induced DNA damage sites and surrounding chromatin in cancer cells: A Single Molecule Localization Microscopy approach. Int. J. Mol. Sci. 2021, 22, 3636. [Google Scholar] [CrossRef]
- Varga, D.; Majoros, H.; Ujfaludi, Z.; Erdélyi, M.; Pankotai, T. Quantification of DNA damage induced repair focus formation via super-resolution dSTORM localization microscopy. Nanoscale 2019, 11, 14226–14236. [Google Scholar] [CrossRef] [Green Version]
- Eryilmaz, M.; Schmitt, E.; Krufczik, M.; Theda, F.; Lee, J.-H.; Cremer, C.; Bestvater, F.; Schaufler, W.; Hausmann, M.; Hildenbrand, G. Localization microscopy analyses of MRE11 clusters in 3D-conserved cell nuclei of different cell lines. Cancers 2018, 10, 25. [Google Scholar] [CrossRef] [Green Version]
- Máté, G.; Hofmann, A.; Wenzel, N.; Heermann, D.W. A topological similarity measure for proteins. Biochim. Biophys. Acta 2014, 1838, 1180–1190. [Google Scholar] [CrossRef] [Green Version]
- Ghrist, R. Barcodes: The persistent topology of data. Bull. Am. Math. Soc. 2007, 45, 61–76. [Google Scholar] [CrossRef] [Green Version]
- Hofmann, A.; Krufczik, M.; Heermann, D.W.; Hausmann, M. Using Persistent Homology as a New Approach for Super-Resolution Localization Microscopy Data Analysis and Classification of γH2AX Foci/Clusters. Int. J. Mol. Sci. 2018, 19, 2263. [Google Scholar] [CrossRef] [Green Version]
- Hausmann, M.; Neitzel, C.; Bobkova, E.; Nagel, D.; Hofmann, A.; Chramko, T.; Smirnova, E.; Kopečná, O.; Pagáčová, E.; Boreyko, A.; et al. Single Molecule Localization Microscopy Analyses of DNA-Repair Foci and Clusters Detected along Particle Damage Tracks. Front. Phys. Sect. Med. Phys. Imaging 2020, 8, 473. [Google Scholar] [CrossRef]
- Jakob, B.; Splinter, J.; Conrad, S.; Voss, K.-O.; Zink, D.; Durante, M.; Löbrich, M.; Taucher-Scholz, G. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res. 2011, 39, 6489–6499. [Google Scholar] [CrossRef]
- Jakob, B.; Splinter, J.; Durante, M.; Taucher-Scholz, G. Live cell microscopy analysis of radiation-induced DNA double-strand break motion. Proc. Natl. Acad. Sci. USA 2009, 106, 3172–3177. [Google Scholar] [CrossRef] [Green Version]
- Falk, M.; Lukasova, E.; Gabrielova, B.; Ondrej, V.; Kozubek, S. Chromatin dynamics during DSB repair. Biochim. Biophys. Acta 2007, 1773, 1534–1545. [Google Scholar] [CrossRef] [Green Version]
- Falk, M.; Lukasova, E.; Kozubek, S. Higher-order chromatin structure in DSB induction, repair and misrepair. Mutat. Res. 2010, 704, 88–100. [Google Scholar] [CrossRef]
- Rothkamm, K.; Krüger, I.; Thompson, L.H.; Löbrich, M. Pathways of DNA doublestrand break repair during the mammalian cell cycle. Mol. Cell Biol. 2003, 23, 5706–5715. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell. Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Iliakis, G.; Murmann, T.; Soni, A. Alternative end-joining repair pathways are the ultimate backup for abrogated classical non-homologous end-joining and homologous recombination repair: Implications for the formation of chromosome translocations. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015, 793, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Jasin, M.; Rothstein, R. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 2013, 5, a012740. [Google Scholar] [CrossRef] [PubMed]
- Iliakis, G. Backup pathways of NHEJ in cells of higher eukaryotes: Cell cycle dependence. Radiother. Oncol. 2009, 92, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Shibata, A.; Jeggo, P.A. Canonical DNA non-homologous end-joining; capacity versus fidelity. Br. J. Radiol. 2020, 23, 20190966. [Google Scholar] [CrossRef]
- Löbrich, M.; Jeggo, P.A. Process of resection-dependent nonhomologous end joining involving the Goddess Artemis. Trends Biochem. Sci. 2017, 42, 690–701. [Google Scholar] [CrossRef] [Green Version]
- Ceccaldi, R.; Rondinelli, B.; Andrea, A.D.D. Repair pathway choices and consequences at the double-strand break. Trends Biol. 2016, 26, 52–64. [Google Scholar] [CrossRef] [Green Version]
- Shrivastav, M.; De Haro, L.P.; Nickoloff, J.A. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008, 18, 134–147. [Google Scholar] [CrossRef] [Green Version]
- Brandsma, I.; Gent, D.C. Pathway choice in DNA double strand break repair: Observations of a balancing act. Genome Integr. 2012, 3, 9. [Google Scholar] [CrossRef] [Green Version]
- Aparicio, T.; Baer, R.; Gautier, J. DNA double-strand break repair pathway choice and cancer. DNA Repair 2014, 19, 169–175. [Google Scholar] [CrossRef] [Green Version]
- Kakarougkas, A.; Jeggo, P.A. DNA DSB repair pathway choice: An orchestrated handover mechanism. Br. J. Radiol. 2014, 87, 20130685. [Google Scholar] [CrossRef]
- Clouaire, T.; Legube, G. DNA double strand break repair pathway choice: A chromatin based decision? Nucleus 2015, 6, 107–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maier, P.; Hartmann, L.; Wenz, F.; Herskind, C. Cellular pathways in response to ionizing radiation and their targetability for tumor radiosensitization. Int. J. Mol. Sci. 2016, 17, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Xu, X. DNA double-strand break repair: A tale of pathway choices. Acta Biochim. Biophys. Sin. 2016, 48, 641–646. [Google Scholar] [CrossRef] [Green Version]
- Cremer, T.; Cremer, M.; Hübner, B.; Strickfaden, H.; Smeets, D.; Popken, J.; Sterr, M.; Markaki, Y.; Rippe, K.; Cremer, C. The 4D nucleome: Evidence for a dynamic nuclear landscape based on co-aligned active and inactive nuclear compartments. FEBS Lett. 2015, 589, 2931–2943. [Google Scholar] [CrossRef] [Green Version]
- Cremer, T.; Cremer, M.; Hübner, B.; Silahtaroglu, A.; Hendzel, M.; Lanctôt, C.H.; Strickfaden, H.; Cremer, C. The interchromatin compartment participates in the structural and functional organization of the cell nucleus. BioEssays 2020, 42, 1900132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibata, A.; Jeggo, P.; Löbrich, M. The pendulum of the Ku-Ku clock. DNA Repair 2018, 71, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Bakkenist, C.J.; Kastan, M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003, 421, 499–506. [Google Scholar] [CrossRef]
- Du, F.; Zhang, M.; Li, X.; Yang, C.; Meng, H.; Wang, D.; Chang, S.; Xu, Y.; Price, B.; Sun, Y. Dimer monomer transition and dimer re-formation play important role for ATM cellular function during DNA repair. Biochem. Biophys. Res. Commun. 2014, 452, 1034–1039. [Google Scholar] [CrossRef] [Green Version]
- Myler, L.R.; Gallardo, I.F.; Soniat, M.M.; Deshpande, R.A.; Gonzalez, X.B.; Kim, Y.; Paull, T.T.; Finkelstein, I.J. Single-Molecule Imaging Reveals How Mre11-Rad50-Nbs1 Initiates DNA Break Repair. Mol. Cell 2017, 67, 891–898. [Google Scholar] [CrossRef] [Green Version]
- Lisby, M.; Barlow, J.H.; Burgess, R.C.; Rothstein, R. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell 2004, 118, 699–713. [Google Scholar] [CrossRef] [Green Version]
- Scherthan, H.; Lee, J.-H.; Maus, E.; Schumann, S.; Muhtadi, R.; Chojowski, R.; Port, M.; Lassmann, M.; Bestvater, F.; Hausmann, M. Nanostructure of Clustered DNA Damage in Leukocytes after In-Solution Irradiation with the Alpha Emitter Ra-223. Cancers 2019, 11, 1877. [Google Scholar] [CrossRef] [Green Version]
- Hausmann, M.; Ilić, N.; Pilarczyk, G.; Lee, J.-H.; Logeswaran, A.; Borroni, A.P.; Krufczik, M.; Theda, F.; Waltrich, N.; Bestvater, F.; et al. Challenges for super-resolution localization microscopy and biomolecular fluorescent nano-probing in cancer research. Int. J. Mol. Sci. 2017, 18, 2066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaccard, P. Etude Comparative de La Distribution Florale Dans Une Portion des Alpes et Des Jura. Bull. Soc. Vaud. Sci. Nat. 1901, 37, 547–579. [Google Scholar]
- Falk, M.; Falková, I.; Kopečná, O.; Bačíková, A.; Pagáčová, E.; Šimek, D.; Golan, M.; Kozubek, S.; Pekarová, M.; Follett, S.E.; et al. Chromatin architecture changes and DNA replication fork collapse are critical features in cryopreserved cells that are differentially controlled by cryoprotectants. Sci. Rep. 2018, 8, 14694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, J.; Chen, J. Mre11-rad50-nbs1 complex dictates DNA repair independent of H2AX. J. Biol. Chem. 2010, 285, 1097–1104. [Google Scholar] [CrossRef] [Green Version]
- Haas, K.T.; Lee, M.Y.; Esposito, A.; Venkitaraman, A.R. Single-molecule localization microscopy reveals molecular transactions during RAD51 filament assembly at cellular DNA damage sites. Nucl. Acids Res. 2018, 46, 2398–2416. [Google Scholar] [CrossRef] [Green Version]
- Whelan, D.R.; Rothenberg, E. Super-resolution mapping of cellular double-strand break resection complexes during homologous recombination. Proc. Nat. Acad. Sci. USA 2021, 118, e2021963118. [Google Scholar] [CrossRef] [PubMed]
- Murmann-Konda, T.; Soni, A.; Stuschke, M.; Iliakis, G. Analysis of chromatid-break-repair detects a homologous recombination to non-homologous end-joining switch with increasing load of DNA double-strand breaks. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2021, 867, 503372. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hahn, H.; Neitzel, C.; Kopečná, O.; Heermann, D.W.; Falk, M.; Hausmann, M. Topological Analysis of γH2AX and MRE11 Clusters Detected by Localization Microscopy during X-ray-Induced DNA Double-Strand Break Repair. Cancers 2021, 13, 5561. https://doi.org/10.3390/cancers13215561
Hahn H, Neitzel C, Kopečná O, Heermann DW, Falk M, Hausmann M. Topological Analysis of γH2AX and MRE11 Clusters Detected by Localization Microscopy during X-ray-Induced DNA Double-Strand Break Repair. Cancers. 2021; 13(21):5561. https://doi.org/10.3390/cancers13215561
Chicago/Turabian StyleHahn, Hannes, Charlotte Neitzel, Olga Kopečná, Dieter W. Heermann, Martin Falk, and Michael Hausmann. 2021. "Topological Analysis of γH2AX and MRE11 Clusters Detected by Localization Microscopy during X-ray-Induced DNA Double-Strand Break Repair" Cancers 13, no. 21: 5561. https://doi.org/10.3390/cancers13215561
APA StyleHahn, H., Neitzel, C., Kopečná, O., Heermann, D. W., Falk, M., & Hausmann, M. (2021). Topological Analysis of γH2AX and MRE11 Clusters Detected by Localization Microscopy during X-ray-Induced DNA Double-Strand Break Repair. Cancers, 13(21), 5561. https://doi.org/10.3390/cancers13215561







