Simple Summary
TP53 is the most frequently mutated genes in cancer, and mutations of TP53 are observed in 5–10% of patients in myelodysplastic syndrome (MDS). In patients with MDS, TP53 mutations are associated with adverse outcomes; however, there is still significant heterogeneity in these disease courses. We performed retrospective review of 107 patients with untreated TP53-mutated MDS, and identified that the functional impact of TP53 mutations, represented by phenotypic annotation of TP53 mutations (PHANTM) combined phenotype score is associated with prognosis. In patients with TP53-mutated MDS, we found that a higher PHANTM combined phenotype score is associated with poorer clinical outcome, and this has independent influence on prognosis accounting for IPSS-R and other risk variables. Our findings suggest that TP53-mutated MDS is heterogeneous and not all TP53 mutations harbor the same impact on prognosis. The PHANTM combined score adds to prognostic precision in MDS beyond previously reported TP53 allelic state.
Abstract
Mutations of TP53 are observed in 5–10% of patients in myelodysplastic syndrome (MDS) and are associated with adverse outcomes. Previous studies indicate that the TP53 allelic state and variant allele frequency of TP53 mutation impact patient outcomes, but there is significant heterogeneity within this MDS subgroup. We performed retrospective review of clinicopathologic and genomic information of 107 patients with TP53-mutated MDS. We assessed each mutation according to the phenotypic annotation of TP53 mutations (PHANTM) and analyzed the associations between predicted TP53 mutant function, represented by the PHANTM combined phenotype score, and overall survival (OS) using the log rank test and Cox regression. Our results indicated that patients with PHANTM combined phenotype score above the median (>1) had significantly shorter OS compared to those with scores below the median (median OS: 10.59 and 16.51 months, respectively, p = 0.025). This relationship remained significant in multivariable analysis (HR (95%CI): 1.62 (1.01–2.58), p = 0.044) and identified to have an independent prognostic influence, accounting for known risk such as IPSS-R and other standard risk variables. Our results suggest that the functional information of TP53 mutations, represented by PHANTM combined phenotype score, are associated with the clinical outcome of patients with TP53-mutated MDS.
1. Introduction
Myelodysplastic syndrome (MDS) is a heterogeneous group of myeloid disorders characterized by ineffective hematopoiesis leading to cytopenias and risk of transformation to acute myeloid leukemia (AML) [1]. TP53, a tumor suppressor gene, is the most frequently mutated gene in cancer, and mutations of TP53 are observed in 5–10% of patients with MDS [1,2,3,4]. In patients with MDS, TP53 mutations are associated with high-risk disease, rapid transformation to AML, resistance to conventional therapies, and shorter survival [5,6,7,8,9,10].
While TP53 mutations are known to be associated with complex karyotypes and generally predict poor prognosis in myeloid neoplasms, associations between outcome and the predicted functional deficit of specific TP53 mutations have not been well characterized [5]. A tumor suppressor gene is a gene that regulates cell division and replication, and a loss or reduction in its function caused by gene deletion, truncation, or alteration in promoter lesions results in uncontrollable cell growth and leads to oncogenesis [11,12]. However, unlike most tumor suppressor genes, the most common genetic alterations in TP53 are missense mutations which can happen throughout the gene [3,13]. It has previously been argued that loss of p53 function is the critical determinant in cancer, yet the preponderance of missense mutations relative to truncation mutations also argues that full-length mutant p53 actively promotes tumor development [14,15]. Mutant p53 protein is often abundantly expressed in cancers and specific allelic variants exhibit dominant-negative or gain-of-function activities in experimental models [16,17,18,19,20,21]. Experimental models also show that over 80% of full-length TP53 DNA-binding domain missense mutants that displays loss-of-function also display dominant-negative activity, suggesting that the ability of mutant p53 to interfere with wild type p53 is critical during tumorigenesis [15]. To determine the function of each missense or nonsense TP53 mutations, Giacomelli et al. created a comprehensive library of p53 mutants and evaluated the function of these alleles in the presence or absence of endogenous p53 [15]. From these data, they developed a classifier which provides functional classification for any missense and nonsense variant of TP53 (phenotypic annotation of TP53 mutations (PHANTM); http://mutantp53.broadinstitute.org, accessed on 4 February 2020) [15].
In this study, we estimated p53 mutant function, represented by the PHANTM combined phenotype score, and determined the impact of this score on the overall survival (OS) of patients with TP53-mutated MDS.
2. Materials and Methods
2.1. Study Group
We searched the medical records of Memorial Sloan Kettering Cancer Center and Massachusetts General Hospital, Harvard Medical School, for cases diagnosed with MDS with TP53 mutation between January 2007 and December 2019. Search was focused on identifying the patients with no prior disease-modifying therapy for MDS, i.e., no prior cytotoxic therapy for MDS with or without a history of transfusion and growth factor administration. The diagnosis of MDS was based on criteria specified in the World Health Organization classification [1]. We performed a retrospective review of the medical records. As part of the chart review, we collected demographic information, history of any prior cytotoxic therapies, the clinical presentation, findings on physical examination, underlying diseases, laboratory data, therapy, and clinical follow-up. Prognostic risk was calculated using the revised International Prognostic Scoring System (IPSS-R) [22]. These data were collected according to protocols approved by the institutional review boards of all institutions in accordance with the Declaration of Helsinki.
2.2. Conventional Cytogenetics and Fluorescence In Situ Hybridization Analysis
Conventional karyotyping was performed on bone marrow aspirate and reported by using the International System for Human Cytogenetic Nomenclature (ISCN) 2013 [23]. Fluorescence in situ hybridization (FISH) analysis was performed with BM aspirate on a subset of cases to assess for loss of 17p using LSI TP53/CEP17 FISH Prove Kit (Abbott Molecular/Vysis, Des Plaines, IL, USA). A total of 300 interphases were analyzed. The cutoff established in our laboratory was 3% for TP53 (17p13.1) deletion.
2.3. Next-Generation Sequencing
Next-generation sequencing was performed using different platforms that evaluated similarly compiled 30 myeloid gene panels in addition to TP53. Many of these cases were evaluated by a next-generation sequencing-based custom-designed assay using the Illumina MiSeq platform. TP53 mutations were characterized by variant type and exon location, and variant allele frequency (VAF). The number of other co-mutated genes was also recorded. The limit of detection for variant calling was 2%.
2.4. TP53 Mutation Phenotype Score (PHANTM Combined Phenotype Score)
We applied phenotypic annotation of TP53 mutations (PHANTM); http://mutantp53.broadinstitute.org (accessed on 4 February 2020) for detected missense and nonsense TP53 mutations, and PHANTM combined phenotype score was calculated for each patient. This supports functional classification for missense and nonsense variants in the gene TP53; therefore, for the cases with frameshift mutation, in-frame deletion, or splice site mutation, we applied the score of 1 based on literatures that shows pathogenic features of these gene alterations [24,25].
2.5. Statistical Analysis
The primary endpoint of this study was overall survival (OS) time, defined as the time from the date of MDS diagnosis until the date of death or last follow-up. Only patients with TP53 mutations were included in this study; therefore, we account for the interval from diagnosis of MDS until TP53 sequencing, when patients enter the risk set, using left-truncation [26]. Median OS was estimated using the Kaplan–Meier method and compared between groups using the log rank test. Continuous variables were assessed using Cox regression. We explored combined phenotype score as both a categorical variable dichotomized at the median and as a continuous variable. Combined phenotype score was evaluated for independent association with OS in multivariable Cox regression models controlling for clinically and statistically significant factors. Combined phenotype score was also evaluated for association with important clinical factors using Pearson’s Chi-squared test or the Wilcoxon rank sum test. AML transformation and transplant were analyzed for association with OS as time-dependent covariates in Cox regression models. A p value of <0.05 was considered statistically significant. All statistical analyses were performed in R version 3.6.1 (R Foundation for Statistical Computing, Vienna, Austria).
3. Results
3.1. Patient Characteristics
We identified 107 patients with TP53-mutated MDS with no prior disease modifying therapy. Patient characteristics are summarized in Table 1.

Table 1.
Clinical characteristics of patients (n = 107) with MDS with TP53 mutation.
3.2. TP53 Mutations, Other Co-Mutations, and Cytogenetic Abnormalities
3.2.1. TP53 Mutations
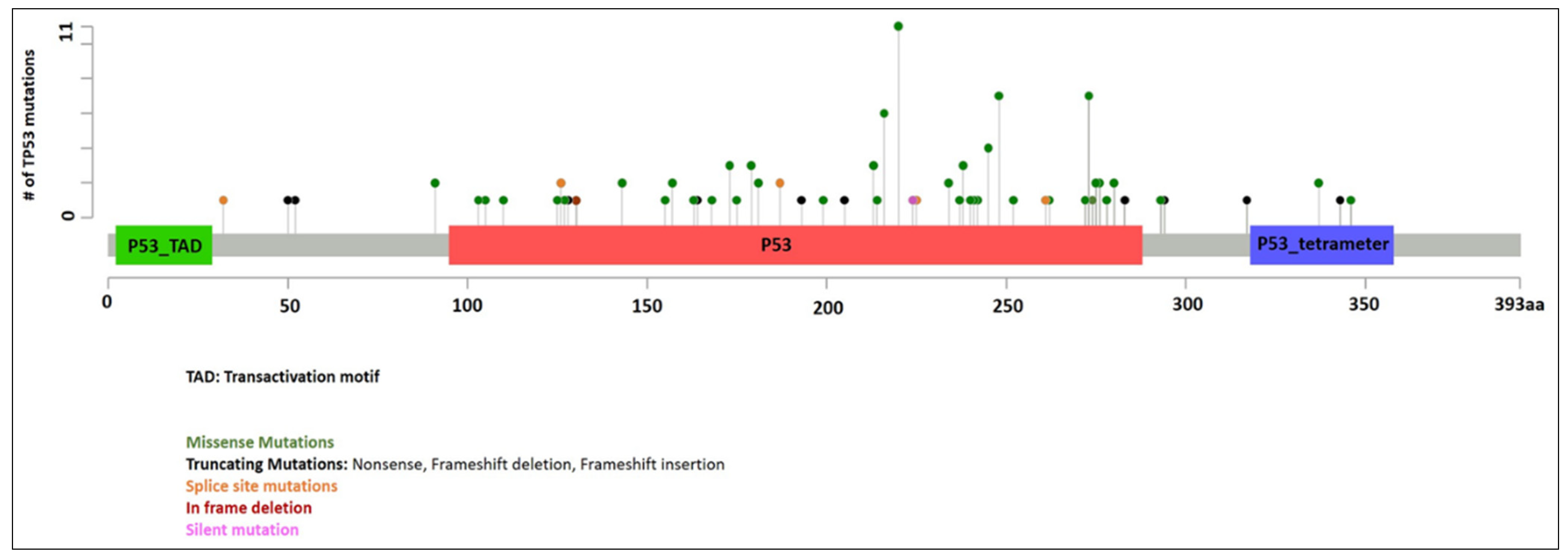
Information of TP53 mutations is summarized in Figure 1. A total of 128 mutations were identified in 107 patients with median VAF of 0.29 (range 0.02 to 1.00). Twenty-one patients had two TP53 mutations. VAF was not available in three patients (all with one TP53 mutation). In patients with two mutations, the TP53 mutations with the higher VAF were designated as the primary TP53 mutations in this study for evaluation purpose. The median VAF of the primary TP53 mutations were 0.34 (range 0.02 to 1.00, n = 104). Missense mutations were the most common (86 patients, 80%), followed by splice site mutations (seven patients, 6.5%), frameshift mutations (six patients, 5.6%), nonsense mutations (five patients, 4.7%), and in-frame deletions (three patients, 2.8%). The median VAF of secondary TP53 mutations were 0.22 (range 0.04 to 0.44, n = 21). Missense mutations were again the most common (12 patients, 57%), followed by nonsense mutations (four patients, 19%), splice site mutations (four patients, 19%), and frameshift mutations (one patient, 4.8%).
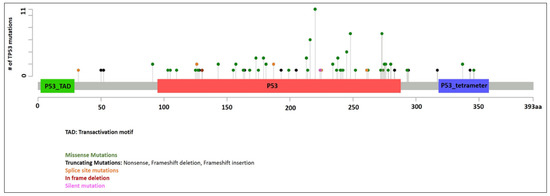
Figure 1.
TP53 mutations identified in our cohort.
3.2.2. Other Co-Mutations
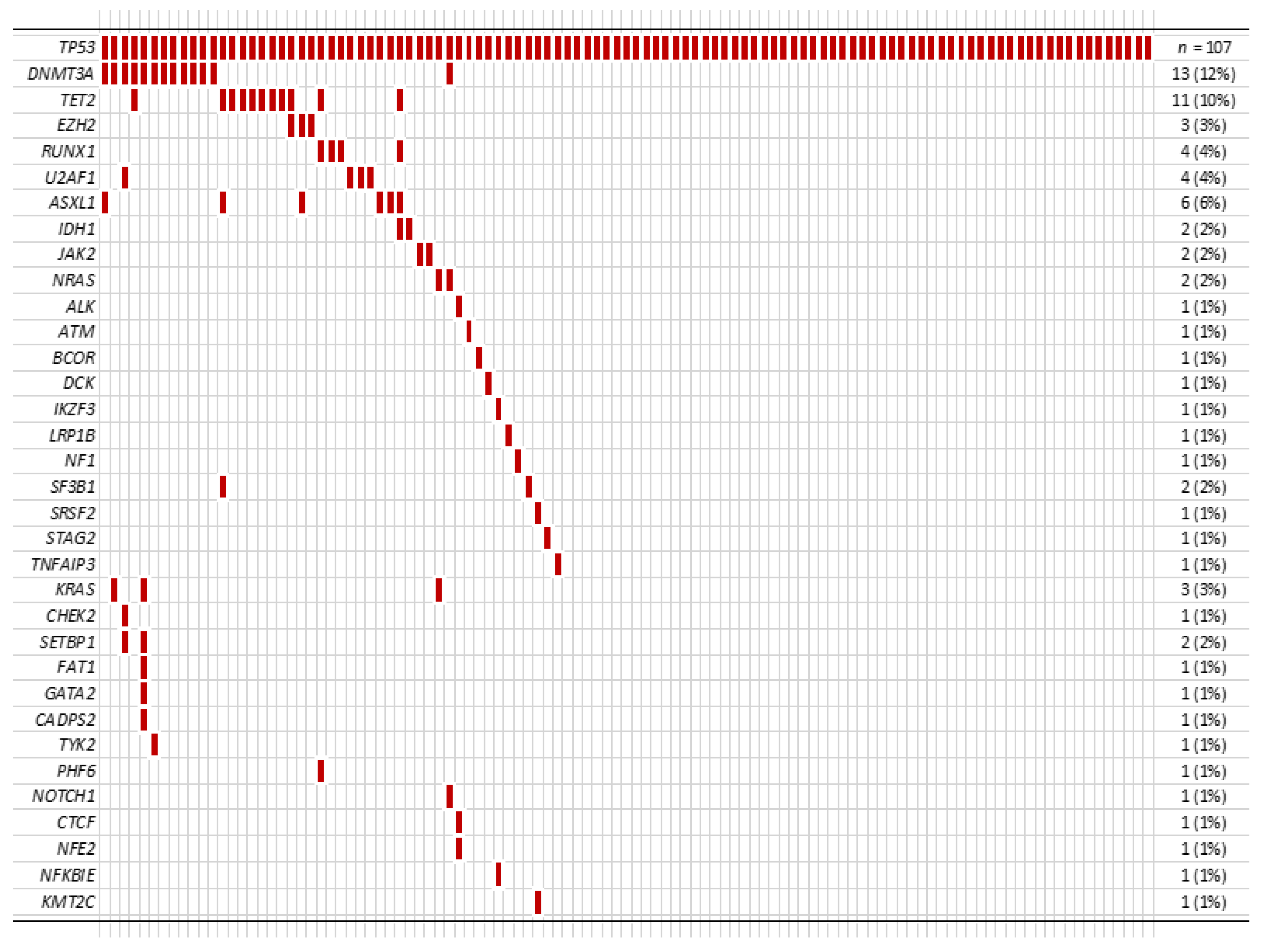
Among 107 patients, 47 patients (44%) had additional mutation(s) in addition to the TP53 mutation (Figure 2). In these patients with co-mutation(s), the median number of additional mutation(s) was 1 (range 1 to 6), and DNMT3A was the most frequent co-mutation (13/107, 12%) followed by TET2 (11/107, 10%).
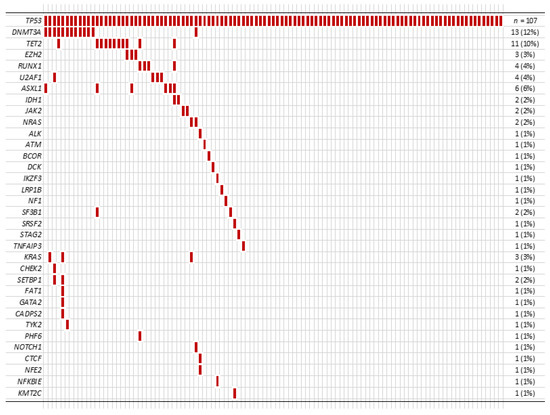
Figure 2.
Other co-mutations identified in our cohort.
3.2.3. Cytogenetic Abnormalities
Cytogenetic data were available for 104 patients, and 33 patients showed loss of 17p by conventional karyotype and/or FISH. Fifty patients had one TP53 mutation with no loss of 17p, 21 patients had two TP53 mutations with no loss of 17p, and 33 patients had one TP53 mutation with loss of 17p.
3.3. PHANTM Combined Phenotype Score
PHANTM combined phenotype score was calculated from the primary mutation in each patient. It ranged from −0.559 to 1.778 (median 1.00) (Table S1). PHANTM combined phenotype score, when analyzed as either continuous or as binary variable dichotomized at the median, was not significantly associated with the number of TP53 mutations or the presence of other co-mutations (Table 2).

Table 2.
PHANTM combined phenotype score (n = 107).
3.4. Therapy
During the follow up, 39 patients (36.4%) showed transformation to AML. Among these 39 patients who progressed to AML, 15 patients (38.5%) received stem cell transplant. Among 68 patients who did not progress to AML, 23 patients (33.8%) received stem cell transplant.
3.5. Outcome
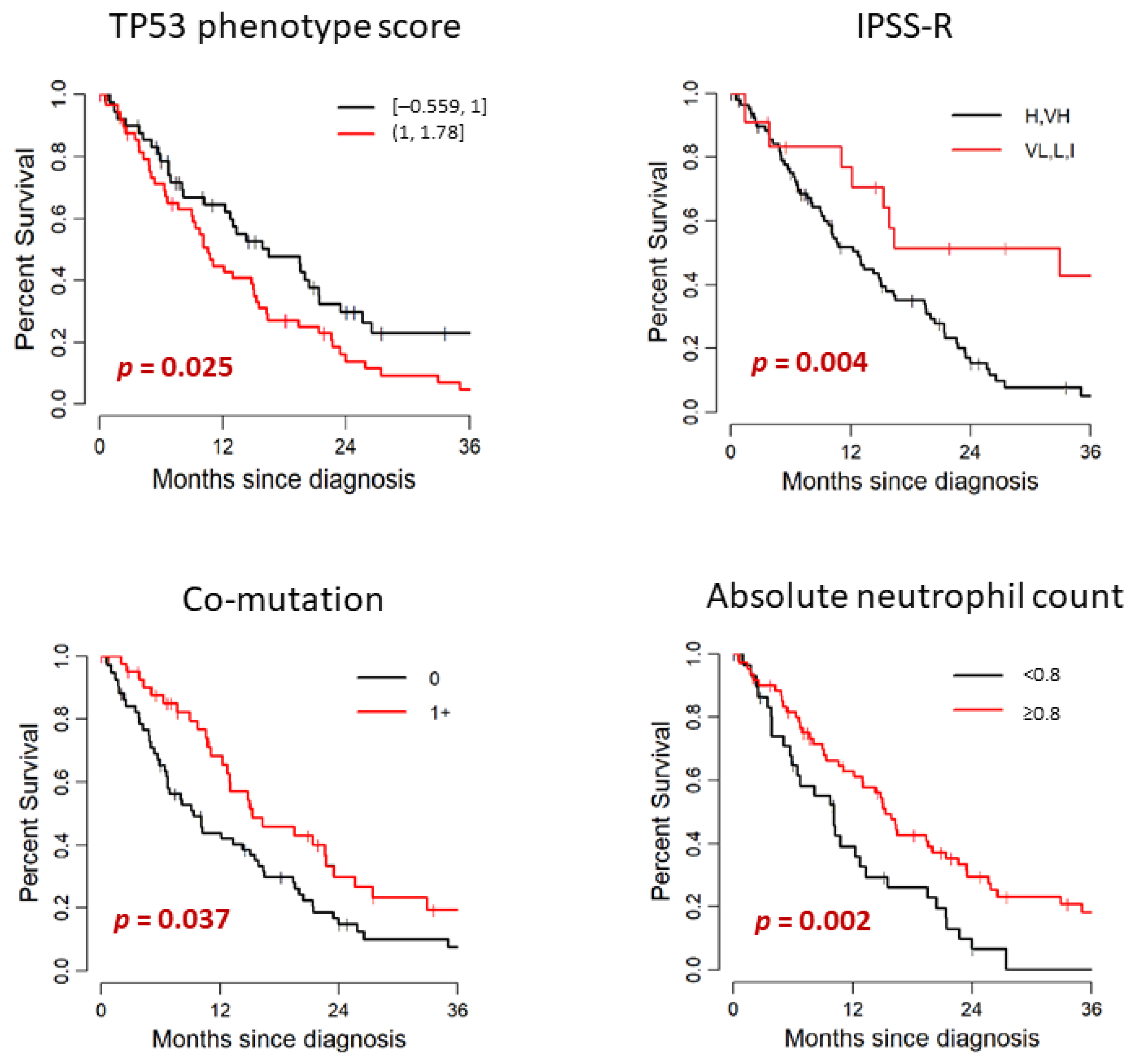
Patient’s outcome and its association with clinical and mutational characteristics are summarized in Table 3 and Figure 3. In the study cohort of 107 patients, 79 died. The median follow-up after diagnosis was 12.8 months (range: 0–50.5) among survivors. Median OS months (95% CI) was 14.84 (11.09–19.51). Median time from diagnosis to TP53 testing was 0.2 months (range: 0–40.7). TP53 testing was performed within 3 months of diagnosis for 97 patients (91%) and after 3 months for 10 patients (9%); the median time to TP53 testing for this subset was 9 months (range: 3.6–40.7). There was no significant difference in OS between patients with de novo MDS and therapy-related MDS (median OS: 15.07 and 10.59 months, respectively. p = 0.674). When we compared the WHO categories stratified by blast percentage, there was no significant difference in OS between MDS-EB-1/2 and MDS without excess blasts (median OS: 15.56 and 11.09 months, respectively. p = 0.806). The OS of lower risk IPSS-R groups (VL, L, I) was significantly longer than the IPSS-R higher risk groups (VH, H) (median OS: 32.93 and 12.76 months, respectively, p = 0.004). When we stratified by IPSS-R cytogenetic risk groups, the OS of lower risk groups (good, intermediate) showed tendency to have longer survival than the higher risk groups (poor, very poor); however, this finding was not statistically significant in our cohort (median OS: 23.52 and 13.03 months, respectively, p = 0.14). Cytopenia was also associated with poorer prognosis. This finding was statistically significant for the hemoglobin level (<10 vs. ≥10 g/dL, p = 0.036) and absolute neutrophil count (<0.8 vs. ≥0.8 × 109/L, p = 0.002) in our cohort. Higher hemoglobin level and higher absolute neutrophil count were significantly associated with better outcome.

Table 3.
Association of clinical and mutational characteristics with prognosis.
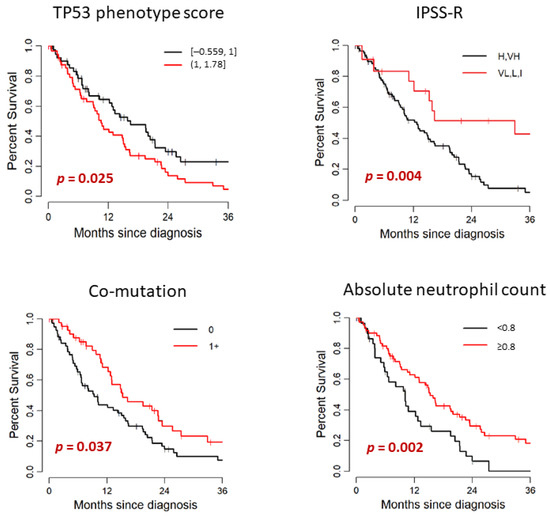
Figure 3.
Univariate analysis for association with OS.
When we analyzed the genetic data, median OS was significantly shorter for patients with PHANTM combined phenotype score above the median (>1) than that of the patients with scores below the median (≤1) (median OS: 10.59 and 16.51 months, respectively, p = 0.025). When analyzed as a continuous variable, higher PHANTM combined phenotype score was significantly associated with increased risk of death (HR (95%CI):1.77 (1.04–3.01), p = 0.035). Sixteen patients had PHANTM combined phenotype score of 1, and all of these patients had either frameshift mutation (six patients), in-frame deletion (three patients), or splice site mutation (seven patients). Therefore, we also evaluated the prognostic influence of PHANTM combined phenotype score by univariate analysis among these 3 groups; <1, 1 and >1. We observed the consistent findings (p = 0.027); however, due to the small number of patients in the middle group, the confidence interval of the median OS estimate was wide (Table S2). Patients with two TP53 mutations showed a tendency to have shorter survival than patients with one TP53 mutation (median OS: 8.16 versus 14.31 months, respectively, p = 0.094). TP53 VAF did not show statistically significant impact on prognosis both by either continuous or categorical analysis (<0.2 vs. 0.2–0.5 vs. >0.5). Locus of TP53 mutation and TP53 mutation type (missense vs. other) did not significantly differentiate patients on survival. The median OS was significantly longer for patients with 1+ co-mutations than that of the patients with no co-mutation (median OS: 15.33 and 9.38 months, respectively, p = 0.037). Patients who had one TP53 mutation and loss of chromosome 17p had shorter OS than that of the patients with one TP53 mutation with no loss of chromosome 17p (median OS: 12.76 and 16.38 months, respectively), and the patients with two TP53 mutations had the shortest OS among these three groups (OS: 8.16 months). However, these findings did not reach statistical significance (p = 0.07).
We did not see the statistical difference of prognosis between patients who received or not received allogeneic transplant (HR (95%CI): 0.82 (0.30–2.26), p = 0.71), or between patients who transformed or not transformed to AML (HR (95%CI): 0.58 (0.17–1.97), p = 0.39) in our cohort.
The results of multivariable analysis for association with OS are summarized in Table 4. PHANTM combined phenotype score as a categorical variable, dichotomized at the median (over 1 vs. equal or less than 1), was found to be an independent prognostic factor in the patients with TP53 mutated MDS (HR (95%CI): 1.62 (1.01–2.58, p = 0.044). Among other factors analyzed, IPSS-R showed independent prognostic significance (HR (95%CI): 0.42 (0.19–0.91), p = 0.028 for low vs. high risk groups). Having two TP53 mutations was also independently associated with increased risk of death (HR (95%CI): 1.84 (1.04–3.25), p = 0.037). Finding was not statistically significant when analysis was performed with PHANTM combined phenotype score as a continuous variable (HR (95%CI): 1.53 (0.87–2.71, p = 0.14).

Table 4.
Multivariate analysis for association with OS.
4. Discussion
Mutations in TP53 are observed in 5–10% of patients with MDS and are associated with adverse outcomes [8,9,10,27]. Preclinical models suggest that distinct mutation types in different TP53 mutation types in different TP53 gene domains may lead to different impacts on protein functionality [28,29,30,31]. Whether these functional differences translate into distinct clinical features and outcomes and how the type, number, and size of TP53 mutations influence the prognosis of patients with TP53-mutated MDS remains unclear. We applied the TP53 phenotypic scoring system created by Giacomelli et al. to annotate the P53 mutant function and analyzed its association with outcome. We identified that the median OS was significantly shorter for patients with PHANTM combined phenotype score above the median (>1) than that of the patients with scores below the median. When analyzed as a continuous variable, higher PHANTM combined phenotype score was associated with increased risk of death. By multivariable analysis, PHANTM combined phenotype score as a categorical variable, dichotomized at the median (over 1 vs. equal or less than 1), was found to be an independent prognostic factor in the patients with TP53 mutated MDS, along with other factors analyzed, including IPSS-R and having two TP53 mutations. Our data suggest that the functional information of TP53 mutations, represented by PHANTM combined phenotype score independently influence the clinical outcome in TP53-mutated myelodysplastic syndromes.
We applied the score of 1 for the cases with frameshift mutation, in-frame deletion, or splice site mutation, based on literatures that shows pathogenic features of these gene alterations [24,25]. In our cohort, 16 patients had PHANTM combined phenotype score of 1, and all of these patients had either frameshift mutation (six patients), in-frame deletion (three patients), or splice site mutation (seven patients). Therefore, we also evaluated the prognostic influence of PHANTM combined phenotype score by univariate analysis among these three groups: <1, 1, and >1. We observed the consistent findings (p = 0.027); however, due to the small number of patients in the middle group, the confidence interval of the median OS estimate was wide (Table S2). Analysis with larger cohort to obtain stable estimate is needed to confirm this finding in the future.
Detailed mechanisms of which factors of TP53 mutation have the most influence on prognosis are not well defined in patients with TP53-mutated MDS, including the types of TP53 mutation (missense, nonsense, or other), the number of TP53 mutations, or VAF of the TP53 mutation. One previous study showed that the types of TP53 mutation (missense, nonsense, or other) or location of TP53 mutations did not show significant association with OS, with only a trend to worse outcomes in patients with mutations not involving DNA-binding domain [32]. Another study performed in the post-transplant setting showed that patients with TP53 truncating mutations (frameshift, nonsense, or splice site) had poor survival compared to those with missense only or missense plus truncating mutations [7]. Previous studies also showed that there was no correlation between the number of TP53 mutations and OS [7,32]. In our study, based on the results of univariate analysis and previously published data, we chose IPSS-R, number of TP53 mutations, loss of 17p, and number of co-mutations as the variables for multivariate analysis along with PHANTM combines phenotype score [22,33]. Patients with one TP53 mutation had longer OS than patients with 2 TP53 mutations, and the finding was statistically significant only by multivariate analysis. Our study showed no significant correlation between the type or location of TP53 mutations and OS.
A recent study by Bernard et al. reported the importance of the allelic status of TP53 [33]. They reported that the multi-hit TP53 state (more than one gene mutation, mutation and deletion, mutation and copy-neutral loss of heterozygosity) in MDS underlies established associations with genome instability, treatment resistance, disease progression and dismal outcomes, indicating that consideration of TP53 allelic state is critical for diagnostic and prognostic precision in MDS [33]. The results of our study support this finding, since the patients with one TP53 mutation with no loss of chromosome 17p had longer OS than that of the patients with TP53 mutation and loss of chromosome 17p (median OS: 12.76 and 16.38 months, respectively), and the patients with 2 TP53 mutations had the shortest OS among these three groups (OS: 8.16 months). However, these findings did not reach statistical significance in our limited cohort of 107 patients (p = 0.07). This may reflect the relatively small size of our cohort or the inability to detect copy-neutral LOH in our study, which represented 20% of the TP53 multi-hit cases in the Bernard et al. study [33]. Although the prognostic significance of the TP53 VAF reported in several studies could be explained by a correlation between high VAF and multi-hit TP53 status, some studies, including ours, did not find TP53 VAF to correlate with outcome [7,10,32,34,35]. The VAF is influenced by sample quality (hemodilution) and the presence of normal background hematopoietic cells, thus it may not accurately reflect the single versus multi-hit status of the TP53 gene.
Our study also showed that co-mutation(s) of other oncogenic genes (other driver mutations), IPSS-R and cytopenias are significantly associated with prognosis. It has been previously reported that the total number of co-mutations differ among the TP53 allelic status [33]. Patients with monoallelic TP53 mutations frequently have several co-mutations, while patients with multi-hit TP53 state have fewer co-mutations [33]. Our study showed that the median OS was significantly longer for patients with 1+ co-mutations than that of the patients with no co-mutation (median OS: 15.33 and 9.38 months, respectively, p = 0.037). This is likely reflecting the comparison of OS between patients with monoallelic TP53 mutations and patients with multi-hit TP53 state. Blast percentage and preceding cytotoxic therapy have been reported to be associated with prognosis in TP53-mutated MDS; however, we did not see any difference in prognosis between MDS with excess blasts and other MDS, or between de novo and therapy-related disease in our cohort [32,36].
5. Conclusions
We found that the functional status of TP53 mutations, represented by PHANTM combined phenotype score, is independently associated with the clinical outcome in patients with TP53-mutated MDS. In patients with TP53-mutated MDS, higher PHANTM combined phenotype score is associated with poorer clinical outcome. Our findings suggest that TP53-mutated MDS is heterogeneous and that not all TP53 mutations harbor the same impact on prognosis. Use of the PHANTM combined score may help improve prognostic precision in MDS in addition to the previously reported TP53 allelic state.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13215502/s1, Table S1: TP53 primary mutations and PHANTM combined phenotype score of 107 patients, Table S2: Association of PHANTM combined phenotype score and prognosis: Univariate analysis among three groups.
Author Contributions
Conceptualization: M.Y., V.N., R.P.H. and V.M.K.; Data curation: A.Z.O. and H.M.; Formal analysis: M.H.; Methodology: M.Y., M.H., V.N., R.P.H. and V.M.K.; Resources: A.D., A.M.B. and V.M.K.; Software: M.H.; Validation: M.Y., M.H., M.E.A. and Y.L.; Writing—original draft: M.Y.; Writing—review and editing, A.M.B., V.N. and R.P.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of Memorial Sloan Kettering Cancer Center (16-1591, 9/11/2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available in Supplemental Table S1.
Acknowledgments
We thank Morgan J. Coleman, Charlene C. Kabel, and Harold K. Elias, at Memorial Sloan Kettering Cancer Center, New York, NY, for their support for data collection.
Conflicts of Interest
M.Y. received consultant fee from Janssen Pharmaceuticals. M.E.A. received consultant fees from Janssen Global Services, Bristol-Myers Squibb, and Astra Zeneca, and was a speaker of Biocartis, Invivoscribe, physician educational resources, Peerview institute for medical education, clinical care options, and RMEI medical education. A.D. received consultant fees from Roche, Physicians’ Education Resource, Seattle Genetics, Peerview Institute, Takeda, EUSA Pharma and research grants from National Cancer Institute, Roche and Takeda. A.M.B. is supported in part by an Evans MDS Young Investigator Award, and has consulted for Acceleron, Agios, Novartis, Celgene/BMS, Takeda, Keros Therapeutics and Taiho. All other authors declare no conflicts of interest.
References
- Hasserjian, R.P.; Orazi, A.; Brunning, R.D.; Germing, U.; Le Beau, M.M.; Porwit, A.; Baumann, I.; Hellstrom-Lindberg, E.; List, A.F.; Cazzola, M.; et al. Myeloysplastic syndromes. In WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, Revised, 4th ed.; Swerdlow, S.H., Campo, E., Harris, N.L., Jaffe, E.S., Pileri, S.A., Stein, H., Thiele, J., Arber, D.A., Hasserjian, R.P., Le Beau, M.M., Orazi, A., et al., Eds.; IARC: Lyon, Switzerland, 2017; pp. 97–120. [Google Scholar]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef]
- Hainaut, P.; Pfeifer, G.P. Somatic TP53 Mutations in the Era of Genome Sequencing. Cold Spring Harb. Perspect. Med. 2016, 6, a026179. [Google Scholar]
- Papaemmanuil, E.; Gerstung, M.; Malcovati, L.; Tauro, S.; Gundem, G.; Van Loo, P.; Yoon, C.J.; Ellis, P.; Wedge, D.C.; Pellagatti, A.; et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013, 122, 3616–3627. [Google Scholar] [CrossRef]
- Haase, D.; Stevenson, K.E.; Neuberg, D.; Maciejewski, J.P.; Nazha, A.; Sekeres, M.A.; Ebert, B.L.; Garcia-Manero, G.; Haferlach, C.; Haferlach, T.; et al. TP53 mutation status divides myelodysplastic syndromes with complex karyotypes into distinct prognostic subgroups. Leukemia 2019, 33, 1747–1758. [Google Scholar] [CrossRef] [Green Version]
- Bejar, R.; Stevenson, K.; Abdel-Wahab, O.; Galili, N.; Nilsson, B.; Garcia-Manero, G.; Kantarjian, H.; Raza, A.; Levine, R.L.; Neuberg, D.; et al. Clinical effect of point mutations in myelodysplastic syndromes. N. Engl. J. Med. 2011, 364, 2496–2506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindsley, R.C.; Saber, W.; Mar, B.G.; Redd, R.; Wang, T.; Haagenson, M.D.; Grauman, P.V.; Hu, Z.H.; Spellman, S.R.; Lee, S.J.; et al. Prognostic Mutations in Myelodysplastic Syndrome after Stem-Cell Transplantation. N. Engl. J. Med. 2017, 376, 536–547. [Google Scholar] [CrossRef] [Green Version]
- Montalban-Bravo, G.; Takahashi, K.; Patel, K.; Wang, F.; Xingzhi, S.; Nogueras, G.M.; Huang, X.; Pierola, A.A.; Jabbour, E.; Colla, S.; et al. Impact of the number of mutations in survival and response outcomes to hypomethylating agents in patients with myelodysplastic syndromes or myelodysplastic/myeloproliferative neoplasms. Oncotarget 2018, 9, 9714–9727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, K.; Patel, K.; Bueso-Ramos, C.; Zhang, J.; Gumbs, C.; Jabbour, E.; Kadia, T.; Andreff, M.; Konopleva, M.; DiNardo, C.; et al. Clinical implications of TP53 mutations in myelodysplastic syndromes treated with hypomethylating agents. Oncotarget 2016, 7, 14172–14187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sallman, D.A.; Komrokji, R.; Vaupel, C.; Cluzeau, T.; Geyer, S.M.; McGraw, K.L.; Al Ali, N.H.; Lancet, J.; McGinniss, M.J.; Nahas, S.; et al. Impact of TP53 mutation variant allele frequency on phenotype and outcomes in myelodysplastic syndromes. Leukemia 2016, 30, 666–673. [Google Scholar] [CrossRef]
- Haber, D.; Harlow, E. Tumour-suppressor genes: Evolving definitions in the genomic age. Nat. Genet. 1997, 16, 320–322. [Google Scholar] [CrossRef]
- Ewing, A.; Meynert, A.; Churchman, M.; Grimes, G.R.; Hollis, R.L.; Herrington, C.S.; Rye, T.; Bartos, C.; Croy, I.; Ferguson, M.; et al. Structural Variants at the BRCA1/2 Loci are a Common Source of Homologous Repair Deficiency in High-grade Serous Ovarian Carcinoma. Clin. Cancer Res. 2021, 27, 3201–3214. [Google Scholar] [CrossRef]
- Petitjean, A.; Mathe, E.; Kato, S.; Ishioka, C.; Tavtigian, S.V.; Hainaut, P.; Olivier, M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum. Mutat. 2007, 28, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Kato, S.; Han, S.Y.; Liu, W.; Otsuka, K.; Shibata, H.; Kanamaru, R.; Ishioka, C. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc. Natl. Acad. Sci. USA 2003, 100, 8424–8429. [Google Scholar] [CrossRef] [Green Version]
- Giacomelli, A.O.; Yang, X.; Lintner, R.E.; McFarland, J.M.; Duby, M.; Kim, J.; Howard, T.P.; Takeda, D.Y.; Ly, S.H.; Kim, E.; et al. Mutational processes shape the landscape of TP53 mutations in human cancer. Nat. Genet. 2018, 50, 1381–1387. [Google Scholar] [CrossRef]
- Milner, J.; Medcalf, E.A. Cotranslation of activated mutant p53 with wild type drives the wild-type p53 protein into the mutant conformation. Cell 1991, 65, 765–774. [Google Scholar] [CrossRef]
- Harvey, M.; Vogel, H.; Morris, D.; Bradley, A.; Bernstein, A.; Donehower, L.A. A mutant p53 transgene accelerates tumour development in heterozygous but not nullizygous p53-deficient mice. Nat. Genet. 1995, 9, 305–311. [Google Scholar] [CrossRef]
- De Vries, A.; Flores, E.R.; Miranda, B.; Hsieh, H.M.; van Oostrom, C.T.; Sage, J.; Jacks, T. Targeted point mutations of p53 lead to dominant-negative inhibition of wild-type p53 function. Proc. Natl. Acad. Sci. USA 2002, 99, 2948–2953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lang, G.A.; Iwakuma, T.; Suh, Y.A.; Liu, G.; Rao, V.A.; Parant, J.M.; Valentin-Vega, Y.A.; Terzian, T.; Caldwell, L.C.; Strong, L.C.; et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 2004, 119, 861–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freed-Pastor, W.A.; Prives, C. Mutant p53: One name, many proteins. Genes Dev. 2012, 26, 1268–1286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boettcher, S.; Miller, P.G.; Sharma, R.; McConkey, M.; Leventhal, M.; Krivtsov, A.V.; Giacomelli, A.O.; Wong, W.; Kim, J.; Chao, S.; et al. A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science 2019, 365, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, P.L.; Tuechler, H.; Schanz, J.; Sanz, G.; Garcia-Manero, G.; Sole, F.; Bennett, J.M.; Bowen, D.; Fenaux, P.; Dreyfus, F.; et al. Revised international prognostic scoring system for myelodysplastic syndromes. Blood 2012, 120, 2454–2465. [Google Scholar] [CrossRef]
- Shaffer, L.G.; McGowan-Jordan, J.; Schmid, M. An International System for Human Cytogenetic Nomenclature; Karger: Basel, Switzerland, 2013. [Google Scholar]
- Smeby, J.; Sveen, A.; Eilertsen, I.A.; Danielsen, S.A.; Hoff, A.M.; Eide, P.W.; Johannessen, B.; Hektoen, M.; Skotheim, R.; Guren, M.G.; et al. Transcriptional and functional consequences of TP53 splice mutations in colorectal cancer. Oncogenesis 2019, 8, 35. [Google Scholar] [CrossRef]
- Holmila, R.; Fouquet, C.; Cadranel, J.; Zalcman, G.; Soussi, T. Splice mutations in the p53 gene: Case report and review of the literature. Hum. Mutat. 2003, 21, 101–102. [Google Scholar] [CrossRef] [PubMed]
- Kalbfleisch, J.D.; Prentice, R.L. The Statistical Analysis of Failure Time Data, 2nd ed.; Wiley: Hoboken, NJ, USA, 2002. [Google Scholar]
- Haferlach, T.; Nagata, Y.; Grossmann, V.; Okuno, Y.; Bacher, U.; Nagae, G.; Schnittger, S.; Sanada, M.; Kon, A.; Alpermann, T.; et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014, 28, 241–247. [Google Scholar] [CrossRef] [Green Version]
- Kastenhuber, E.R.; Lowe, S.W. Putting p53 in Context. Cell 2017, 170, 1062–1078. [Google Scholar] [CrossRef] [Green Version]
- Muller, P.A.; Vousden, K.H. Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell 2014, 25, 304–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabapathy, K.; Lane, D.P. Therapeutic targeting of p53: All mutants are equal, but some mutants are more equal than others. Nat. Rev. Clin. Oncol. 2018, 15, 13–30. [Google Scholar] [CrossRef]
- Bieging, K.T.; Mello, S.S.; Attardi, L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 2014, 14, 359–370. [Google Scholar] [CrossRef] [Green Version]
- Montalban-Bravo, G.; Kanagal-Shamanna, R.; Benton, C.B.; Class, C.A.; Chien, K.S.; Sasaki, K.; Naqvi, K.; Alvarado, Y.; Kadia, T.M.; Ravandi, F.; et al. Genomic context and TP53 allele frequency define clinical outcomes in TP53-mutated myelodysplastic syndromes. Blood Adv. 2020, 4, 482–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernard, E.; Nannya, Y.; Hasserjian, R.P.; Devlin, S.M.; Tuechler, H.; Medina-Martinez, J.S.; Yoshizato, T.; Shiozawa, Y.; Saiki, R.; Malcovati, L.; et al. Implications of TP53 allelic state for genome stability, clinical presentation and outcomes in myelodysplastic syndromes. Nat. Med. 2020, 26, 1549–1556. [Google Scholar] [CrossRef]
- Wang, W.; Routbort, M.J.; Tang, Z.; Ok, C.Y.; Patel, K.P.; Daver, N.; Garcia-Manero, G.; Medeiros, L.J.; Wang, S.A. Characterization of TP53 mutations in low-grade myelodysplastic syndromes and myelodysplastic syndromes with a non-complex karyotype. Eur. J. Haematol. 2017, 99, 536–543. [Google Scholar] [CrossRef] [PubMed]
- Goel, S.; Hall, J.; Pradhan, K.; Hirsch, C.; Przychodzen, B.; Shastri, A.; Mantzaris, I.; Janakiram, M.; Battini, R.; Kornblum, N.; et al. High prevalence and allele burden-independent prognostic importance of p53 mutations in an inner-city MDS/AML cohort. Leukemia 2016, 30, 1793–1795. [Google Scholar] [CrossRef] [PubMed]
- Ok, C.Y.; Patel, K.P.; Garcia-Manero, G.; Routbort, M.J.; Peng, J.; Tang, G.; Goswami, M.; Young, K.H.; Singh, R.; Medeiros, L.J.; et al. TP53 mutation characteristics in therapy-related myelodysplastic syndromes and acute myeloid leukemia is similar to de novo diseases. J. Hematol. Oncol. 2015, 8, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).