Tris(dibenzylideneacetone)dipalladium(0) (Tris DBA) Abrogates Tumor Progression in Hepatocellular Carcinoma and Multiple Myeloma Preclinical Models by Regulating the STAT3 Signaling Pathway
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Cell Lines
2.3. MTT Assay
2.4. Annexin V Assay
2.5. Cell Cycle Analysis Using Flow Cytometry
2.6. Western Blotting
2.7. DNA Binding Assay
2.8. Immunocytochemistry for STAT3 Distribution
2.9. RNA Isolation and Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
2.10. Real-Time Quantitative PCR
2.11. Transfection with SHP2 siRNA, STAT3 siRNA, and STAT3C
2.12. In Vitro Migration Assay
2.13. Invasion Assay
2.14. In Vivo Acute Toxicity Studies
2.15. Xenograft MM Mouse Model
2.16. Immunohistochemical Analysis of MM Tumor Samples
2.17. Orthotopic HCC Tumor Model
2.18. Statistical Analysis
3. Results
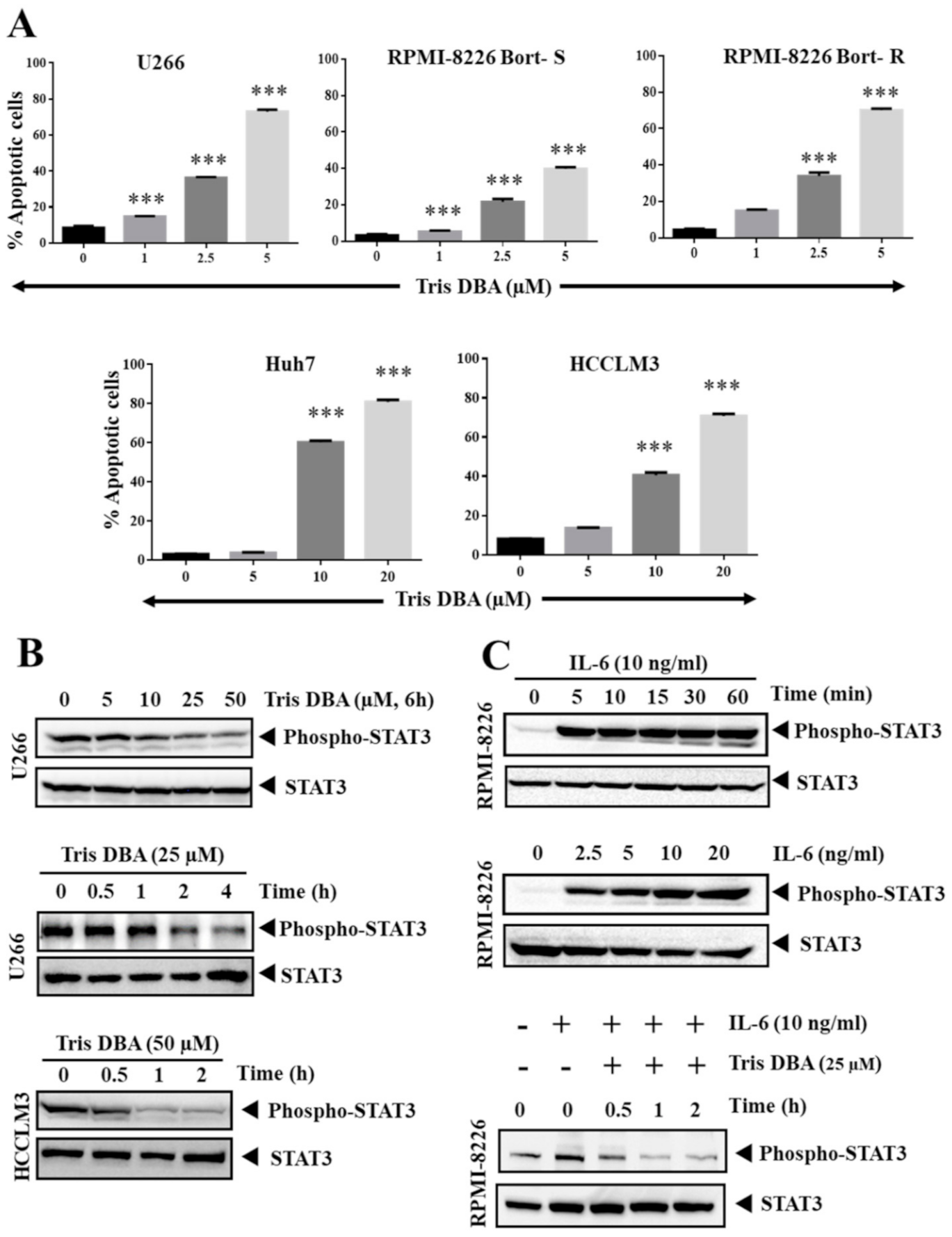
3.1. Tris DBA Decreased the Viability and Increased the subG1 Population of HCC and MM Cells
3.2. Tris DBA Induced Apoptosis of HCC and MM Cells
3.3. Tris DBA Altered STAT3 Phosphorylation in Tumor Cells
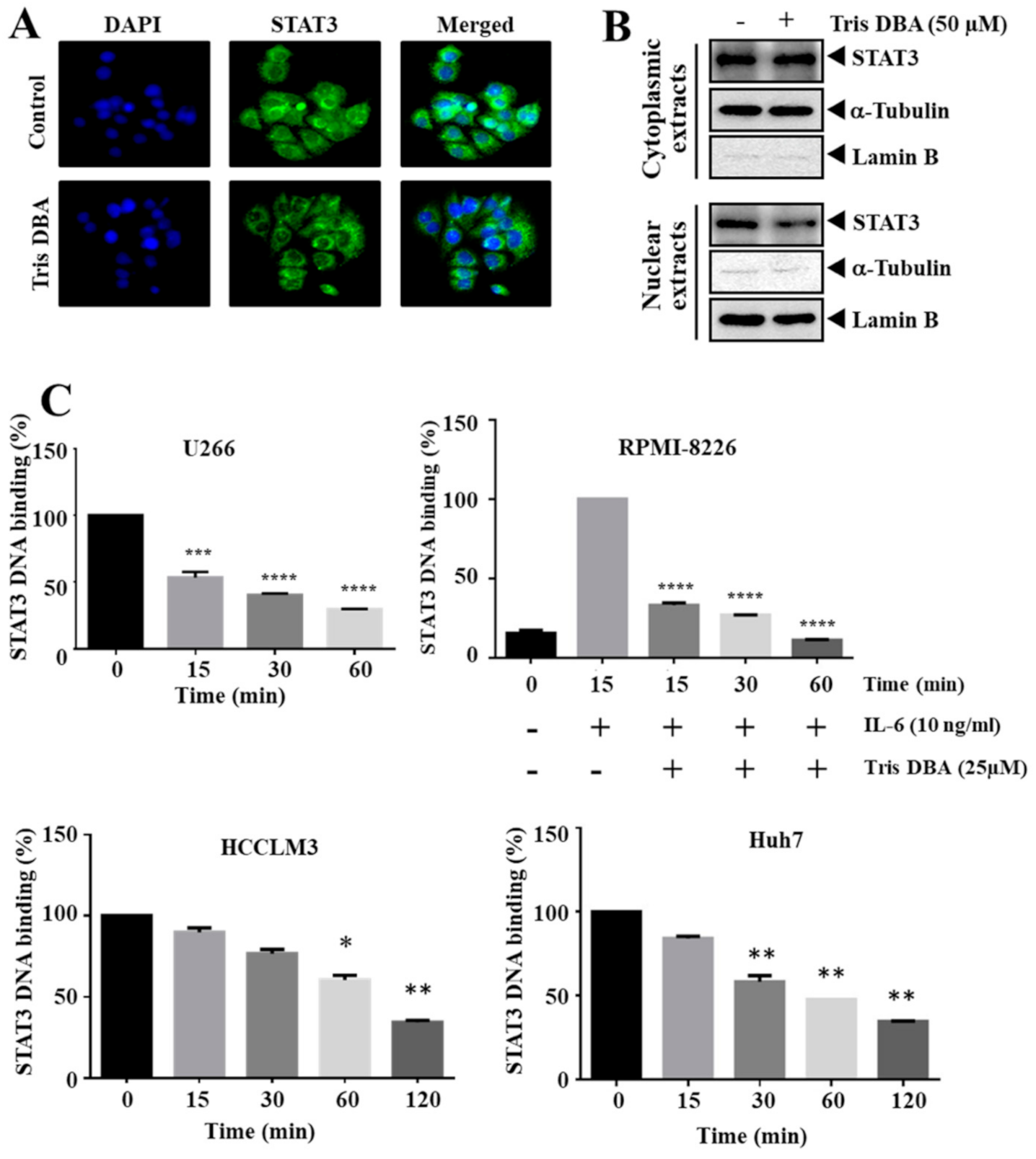
3.4. Tris DBA Decreased Nuclear Localization and Ablated the Binding of STAT3
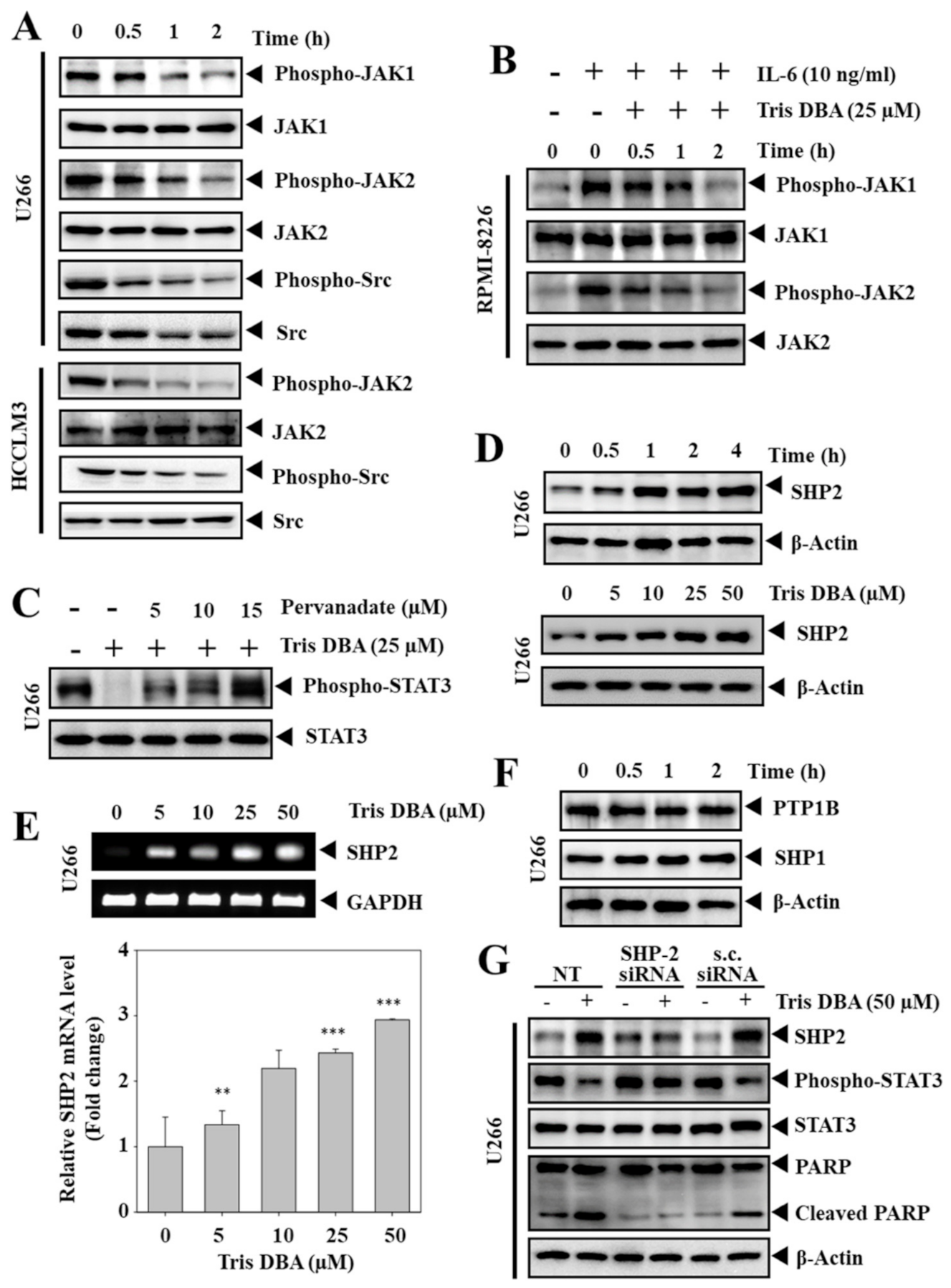
3.5. Tris DBA Inhibited the Phosphorylation of Upstream Kinases of STAT3 Signaling
3.6. Tris DBA Altered the Expression of SHP2
3.7. Depletion of SHP2 Reversed the Tris DBA Induced STAT3 Inhibition
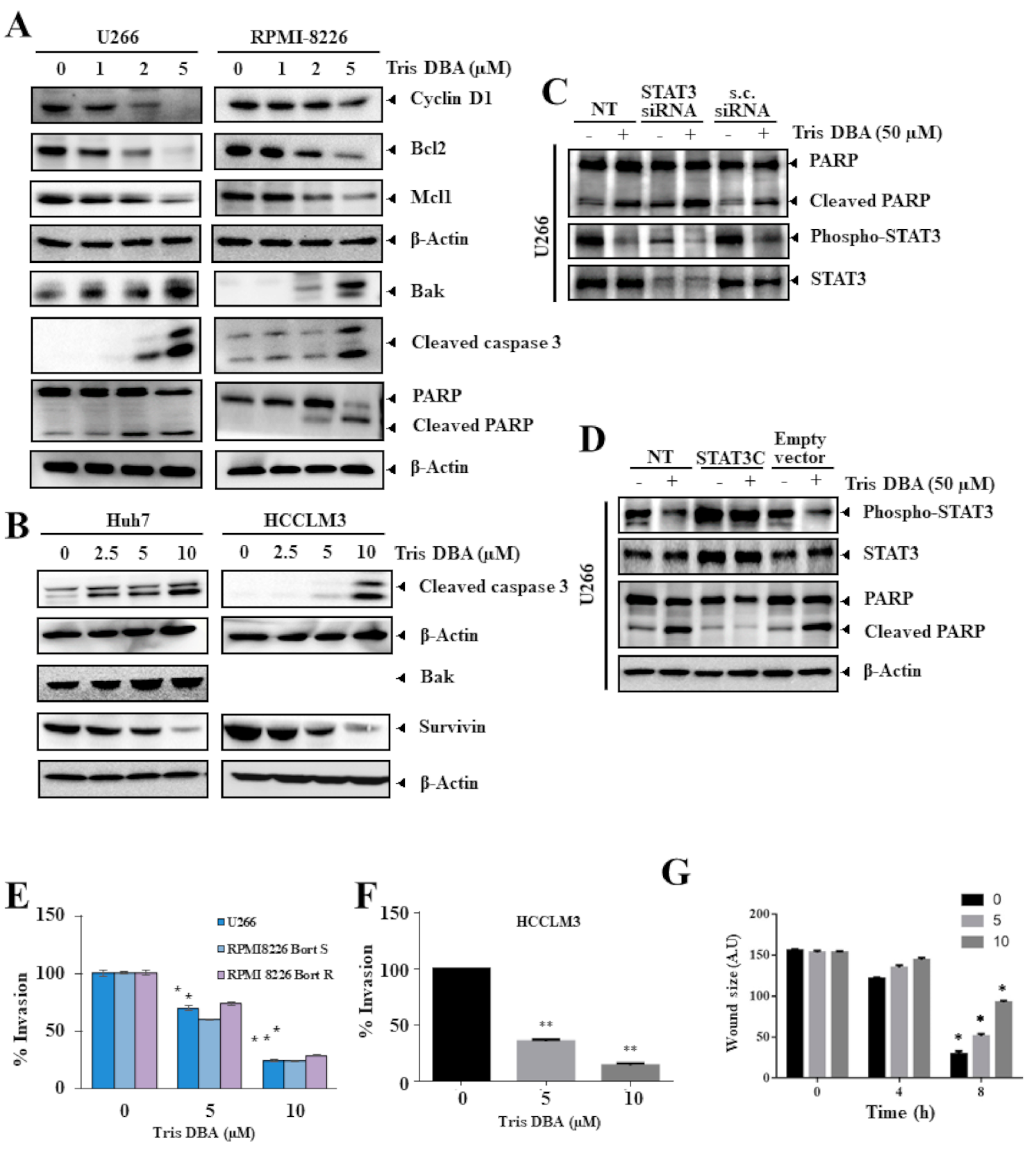
3.8. Tris DBA Altered the Expression of Oncogenic Proteins
3.9. Knock-Down or Overexpression of STAT3 Modulates Tris DBA-Promoted Apoptosis
3.10. Tris DBA Significantly Modulated the Migratory and Invasive Potential
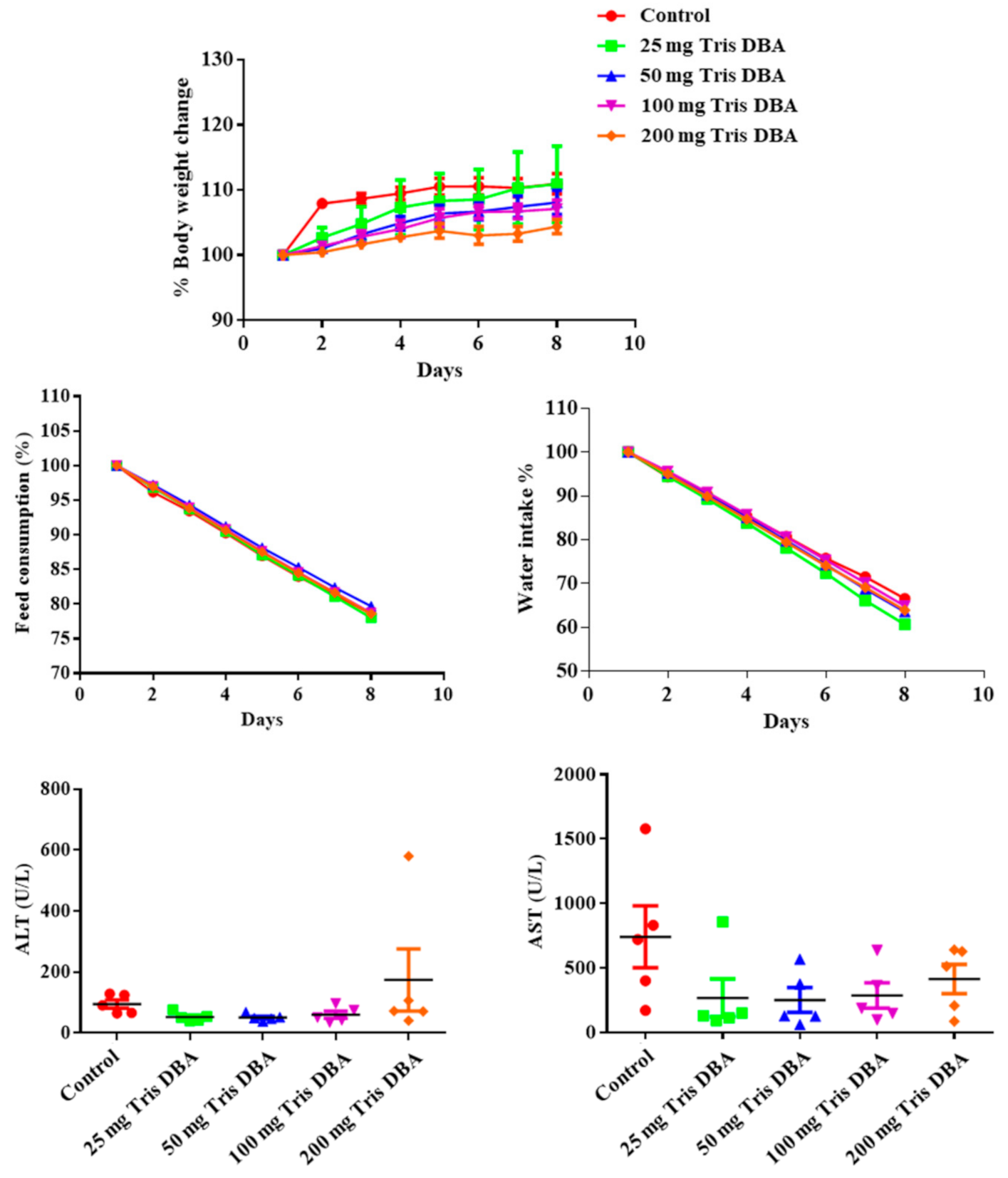
3.11. Tris DBA Did Not Display Toxicity in Preclinical Studies
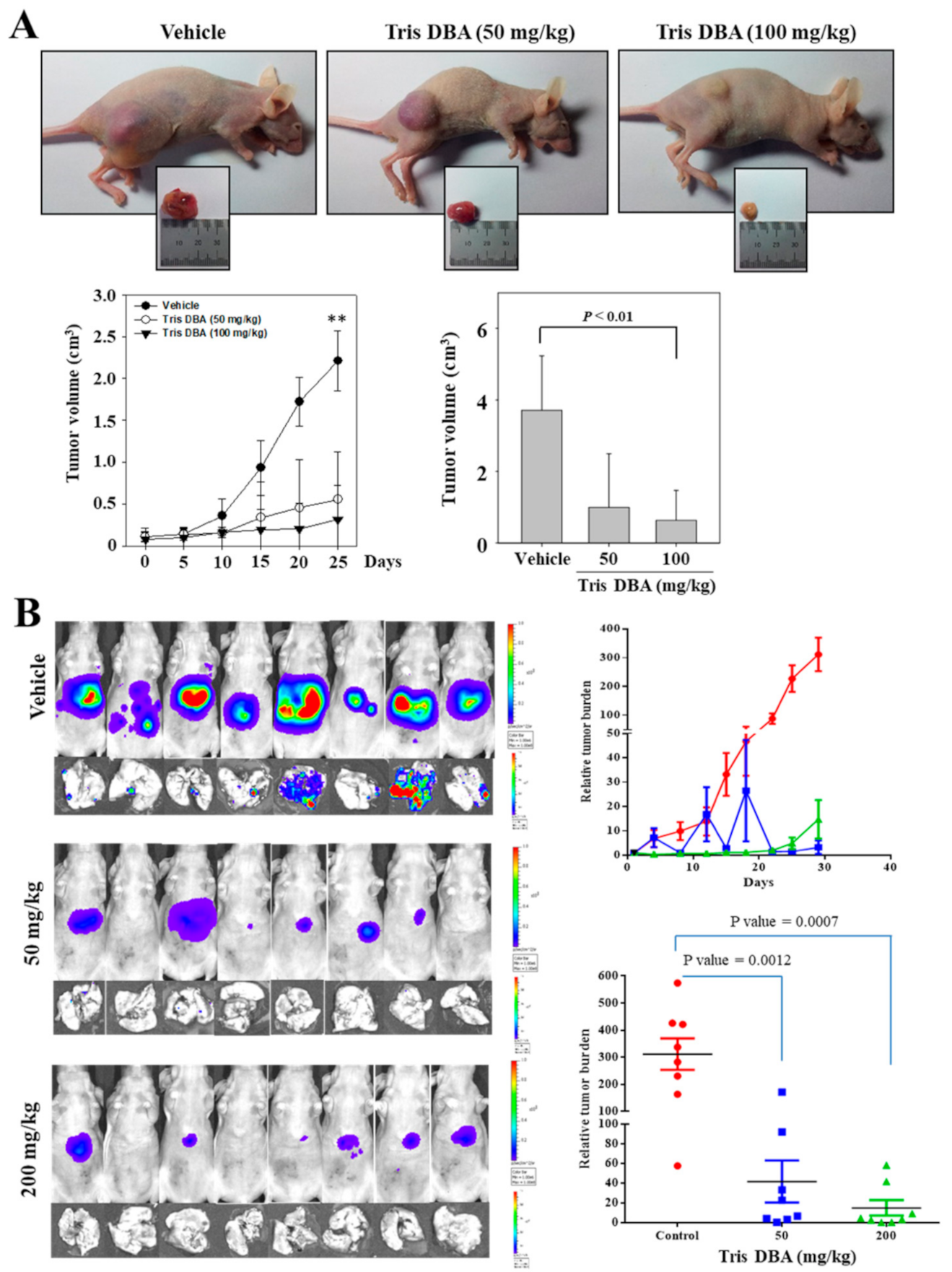
3.12. Tris DBA Imparted Antitumor Effect in Xenograft MM and Orthotopic HCC Mice Models
3.13. Tris DBA Reduced the Number of Ki67+ Cells in Tumor Tissues from the Xenograft MM Mouse Model
3.14. Tris DBA Repressed the Activation of STAT3 Signaling Proteins in Tumor Tissues
3.15. Tris DBA Modulated the Expression of Apoptosis-Related Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Rajendran, P.; Li, F.; Manu, K.A.; Shanmugam, M.K.; Loo, S.Y.; Kumar, A.P.; Sethi, G. γ-Tocotrienol is a novel inhibitor of constitutive and inducible STAT3 signalling pathway in human hepatocellular carcinoma: Potential role as an antiproliferative, pro-apoptotic and chemosensitizing agent. Br. J. Pharmacol. 2011, 163, 283–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balakumar, P.; Maung-U, K.; Jagadeesh, G. Prevalence and prevention of cardiovascular disease and diabetes mellitus. Pharmacol. Res. 2016, 113, 600–609. [Google Scholar] [CrossRef] [PubMed]
- Ghouri, Y.A.; Mian, I.; Rowe, J.H. Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. J. Carcinog. 2017, 16, 1. [Google Scholar] [CrossRef]
- Sarveazad, A.; Agah, S.; Babahajian, A.; Amini, N.; Bahardoust, M. Predictors of 5 year survival rate in hepatocellular carcinoma patients. J. Res. Med. Sci. 2019, 24, 86. [Google Scholar] [CrossRef] [PubMed]
- Mohan, C.D.; Bharathkumar, H.; Bulusu, K.C.; Pandey, V.; Rangappa, S.; Fuchs, J.E.; Shanmugam, M.K.; Dai, X.; Li, F.; Deivasigamani, A.; et al. Development of a novel azaspirane that targets the Janus kinase-signal transducer and activator of transcription (STAT) pathway in hepatocellular carcinoma in vitro and in vivo. J. Biol. Chem. 2014, 289, 34296–34307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohan, C.D.; Rangappa, S.; Nayak, S.C.; Sethi, G.; Rangappa, K.S. Paradoxical functions of long noncoding RNAs in modulating STAT3 signaling pathway in hepatocellular carcinoma. Biochim. Biophys. Acta (BBA)—Rev. Cancer 2021, 1876, 188574. [Google Scholar] [CrossRef]
- Rajendran, P.; Li, F.; Shanmugam, M.K.; Vali, S.; Abbasi, T.; Kapoor, S.; Ahn, K.S.; Kumar, A.P.; Sethi, G. Honokiol inhibits signal transducer and activator of transcription-3 signaling, proliferation, and survival of hepatocellular carcinoma cells via the protein tyrosine phosphatase SHP-1. J. Cell. Physiol. 2012, 227, 2184–2195. [Google Scholar] [CrossRef]
- Swamy, S.G.; Kameshwar, V.H.; Shubha, P.B.; Looi, C.Y.; Shanmugam, M.K.; Arfuso, F.; Dharmarajan, A.; Sethi, G.; Shivananju, N.S.; Bishayee, A. Targeting multiple oncogenic pathways for the treatment of hepatocellular carcinoma. Target. Oncol. 2017, 12, 1–10. [Google Scholar] [CrossRef]
- Hu, J.; Hu, W.X. Targeting signaling pathways in multiple myeloma: Pathogenesis and implication for treatments. Cancer Lett. 2018, 414, 214–221. [Google Scholar] [CrossRef]
- Arora, L.; Kumar, A.P.; Arfuso, F.; Chng, W.J.; Sethi, G. The Role of Signal Transducer and Activator of Transcription 3 (STAT3) and Its Targeted Inhibition in Hematological Malignancies. Cancers 2018, 10, 327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, C.D. Multiple myeloma. Cancer Imaging 2010, 10, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Kannaiyan, R.; Hay, H.S.; Rajendran, P.; Li, F.; Shanmugam, M.K.; Vali, S.; Abbasi, T.; Kapoor, S.; Sharma, A.; Kumar, A.P.; et al. Celastrol inhibits proliferation and induces chemosensitization through down-regulation of NF-κB and STAT3 regulated gene products in multiple myeloma cells. Br. J. Pharmacol. 2011, 164, 1506–1521. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Kim, C.; Lee, J.; Um, J.Y.; Sethi, G.; Ahn, K.S. Arctiin is a pharmacological inhibitor of STAT3 phosphorylation at tyrosine 705 residue and potentiates bortezomib-induced apoptotic and anti-angiogenic effects in human multiple myeloma cells. Phytomed. Int. J. Phytother. Phytopharm. 2019, 55, 282–292. [Google Scholar] [CrossRef]
- Lee, M.; Hirpara, J.L.; Eu, J.Q.; Sethi, G.; Wang, L.; Goh, B.C.; Wong, A.L. Targeting STAT3 and oxidative phosphorylation in oncogene-addicted tumors. Redox Biol. 2019, 25, 101073. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, C.; Ko, J.-H.; Jung, Y.Y.; Jung, S.H.; Kim, E.; Kong, M.; Chinnathambi, A.; Alahmadi, T.A.; Alharbi, S.A.; et al. Casticin inhibits growth and enhances ionizing radiation–induced apoptosis through the suppression of STAT3 signaling cascade. J. Cell. Biochem. 2019, 120, 9787–9798. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, C.; Baek, S.H.; Ko, J.H.; Lee, S.G.; Yang, W.M.; Um, J.Y.; Sethi, G.; Ahn, K.S. Capsazepine inhibits JAK/STAT3 signaling, tumor growth, and cell survival in prostate cancer. Oncotarget 2017, 8, 17700–17711. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, C.; Lee, S.-G.; Sethi, G.; Ahn, K.S. Ophiopogonin D, a Steroidal Glycoside Abrogates STAT3 Signaling Cascade and Exhibits Anti-Cancer Activity by Causing GSH/GSSG Imbalance in Lung Carcinoma. Cancers 2018, 10, 427. [Google Scholar] [CrossRef] [Green Version]
- Siveen, K.S.; Sikka, S.; Surana, R.; Dai, X.; Zhang, J.; Kumar, A.P.; Tan, B.K.; Sethi, G.; Bishayee, A. Targeting the STAT3 signaling pathway in cancer: Role of synthetic and natural inhibitors. Biochim. Biophys. Acta 2014, 1845, 136–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, A.L.A.; Hirpara, J.L.; Pervaiz, S.; Eu, J.Q.; Sethi, G.; Goh, B.C. Do STAT3 inhibitors have potential in the future for cancer therapy? Expert Opin. Investig. Drugs 2017, 26, 883–887. [Google Scholar] [CrossRef] [Green Version]
- Baek, S.H.; Ko, J.H.; Lee, H.; Jung, J.; Kong, M.; Lee, J.W.; Lee, J.; Chinnathambi, A.; Zayed, M.E.; Alharbi, S.A.; et al. Resveratrol inhibits STAT3 signaling pathway through the induction of SOCS-1: Role in apoptosis induction and radiosensitization in head and neck tumor cells. Phytomed. Int. J. Phytother. Phytopharm. 2016, 23, 566–577. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.H.; Lee, J.H.; Kim, C.; Ko, J.-H.; Ryu, S.-H.; Lee, S.-G.; Yang, W.M.; Um, J.-Y.; Chinnathambi, A.; Alharbi, S.A.; et al. Ginkgolic Acid C 17:1, Derived from Ginkgo biloba Leaves, Suppresses Constitutive and Inducible STAT3 Activation through Induction of PTEN and SHP-1 Tyrosine Phosphatase. Molecules 2017, 22, 276. [Google Scholar] [CrossRef] [PubMed]
- Chai, E.Z.; Siveen, K.S.; Shanmugam, M.K.; Arfuso, F.; Sethi, G. Analysis of the intricate relationship between chronic inflammation and cancer. Biochem. J. 2015, 468, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Loh, C.Y.; Arya, A.; Naema, A.F.; Wong, W.F.; Sethi, G.; Looi, C.Y. Signal Transducer and Activator of Transcription (STATs) Proteins in Cancer and Inflammation: Functions and Therapeutic Implication. Front. Oncol. 2019, 9, 48. [Google Scholar] [CrossRef] [Green Version]
- Mohan, C.D.; Rangappa, S.; Preetham, H.D.; Chandra Nayak, S.; Gupta, V.K.; Basappa, S.; Sethi, G.; Rangappa, K.S. Targeting STAT3 signaling pathway in cancer by agents derived from Mother Nature. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Rangappa, S.; Mohan, C.D.; Basappa, B.; Sethi, G.; Lin, Z.-X.; Rangappa, K.S.; Ahn, K.S. Brusatol, a Nrf2 Inhibitor Targets STAT3 Signaling Cascade in Head and Neck Squamous Cell Carcinoma. Biomolecules 2019, 9, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.H.; Mohan, C.D.; Basappa, S.; Rangappa, S.; Chinnathambi, A.; Alahmadi, T.A.; Alharbi, S.A.; Kumar, A.P.; Sethi, G.; Ahn, K.S.; et al. The IκB Kinase Inhibitor ACHP Targets the STAT3 Signaling Pathway in Human Non-Small Cell Lung Carcinoma Cells. Biomolecules 2019, 9, 875. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Mohan, C.D.; Shanmugam, M.K.; Rangappa, S.; Sethi, G.; Siveen, K.S.; Chinnathambi, A.; Alahmadi, T.A.; Alharbi, S.A.; Basappa, S.; et al. Vitexin abrogates invasion and survival of hepatocellular carcinoma cells through targeting STAT3 signaling pathway. Biochimie 2020, 175, 58–68. [Google Scholar] [CrossRef]
- Chai, E.Z.; Shanmugam, M.K.; Arfuso, F.; Dharmarajan, A.; Wang, C.; Kumar, A.P.; Samy, R.P.; Lim, L.H.; Wang, L.; Goh, B.C.; et al. Targeting transcription factor STAT3 for cancer prevention and therapy. Pharmacol. Ther. 2016, 162, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.M.; Li, F.; Rajendran, P.; Kumar, A.P.; Hui, K.M.; Sethi, G. Identification of beta-escin as a novel inhibitor of signal transducer and activator of transcription 3/Janus-activated kinase 2 signaling pathway that suppresses proliferation and induces apoptosis in human hepatocellular carcinoma cells. J. Pharmacol. Exp. Ther. 2010, 334, 285–293. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, C.; Sethi, G.; Ahn, K.S. Brassinin inhibits STAT3 signaling pathway through modulation of PIAS-3 and SOCS-3 expression and sensitizes human lung cancer xenograft in nude mice to paclitaxel. Oncotarget 2015, 6, 6386–6405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Ahn, K.S.; Kim, C.; Shanmugam, M.K.; Siveen, K.S.; Arfuso, F.; Samym, R.P.; Deivasigamanim, A.; Lim, L.H.; Wang, L.; et al. Nimbolide-Induced Oxidative Stress Abrogates STAT3 Signaling Cascade and Inhibits Tumor Growth in Transgenic Adenocarcinoma of Mouse Prostate Model. Antioxid. Redox Signal. 2016, 24, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, P.; Li, F.; Shanmugam, M.K.; Kannaiyan, R.; Goh, J.N.; Wong, K.F.; Wang, W.; Khin, E.; Tergaonkar, V.; Kumar, A.P.; et al. Celastrol suppresses growth and induces apoptosis of human hepatocellular carcinoma through the modulation of STAT3/JAK2 signaling cascade in vitro and in vivo. Cancer Prev. Res. 2012, 5, 631–643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bard-Chapeau, E.A.; Li, S.; Ding, J.; Zhang, S.S.; Zhu, H.H.; Princen, F.; Fang, D.D.; Han, T.; Bailly-Maitre, B.; Poli, V.; et al. Ptpn11/Shp2 Acts as a Tumor Suppressor in Hepatocellular Carcinogenesis. Cancer Cell 2011, 19, 629–639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierpont, C.G.; Mazza, M.C. Crystal and molecular structure of tris(dibenzylideneacetone)dipalladium(0). Inorg. Chem. 1974, 13, 1891–1895. [Google Scholar] [CrossRef]
- Bhandarkar, S.S.; Bromberg, J.; Carrillo, C.; Selvakumar, P.; Sharma, R.K.; Perry, B.N.; Govindarajan, B.; Fried, L.; Sohn, A.; Reddy, K.; et al. Tris (dibenzylideneacetone) dipalladium, a N-myristoyltransferase-1 inhibitor, is effective against melanoma growth in vitro and in vivo. Clin. Cancer Res. 2008, 14, 5743–5748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De la Puente, P.; Azab, F.; Muz, B.; Luderer, M.; Arbiser, J.; Azab, A.K. Tris DBA palladium overcomes hypoxia-mediated drug resistance in multiple myeloma. Leuk. Lymphoma 2016, 57, 1677–1686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kay, N.E.; Sassoon, T.; Secreto, C.; Sinha, S.; Shanafelt, T.D.; Ghosh, A.K.; Arbiser, J.L. Tris (dibenzylideneacetone) dipalladium: A small-molecule palladium complex is effective in inducing apoptosis in chronic lymphocytic leukemia B-cells. Leuk. Lymphoma 2016, 57, 2409–2416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diaz, B.; Ostapoff, K.T.; Toombs, J.E.; Lo, J.; Bonner, M.Y.; Curatolo, A.; Adsay, V.; Brekken, R.A.; Arbiser, J.L. Tris DBA palladium is highly effective against growth and metastasis of pancreatic cancer in an orthotopic model. Oncotarget 2016, 7, 51569–51580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rakesh, K.S.; Jagadish, S.; Swaroop, T.R.; Mohan, C.D.; Ashwini, N.; Harsha, K.B.; Zameer, F.; Girish, K.S.; Rangappa, K.S. Anti-Cancer Activity of 2,4-Disubstituted Thiophene Derivatives: Dual Inhibitors of Lipoxygenase and Cyclooxygenase. Med. Chem. 2015, 11, 462–472. [Google Scholar] [CrossRef]
- Rakesh, K.S.; Jagadish, S.; Balaji, K.S.; Zameer, F.; Swaroop, T.R.; Mohan, C.D.; Jayarama, S.; Rangappa, K.S. 3,5-Disubstituted Isoxazole Derivatives: Potential Inhibitors of Inflammation and Cancer. Inflammation 2016, 39, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Keerthy, H.K.; Garg, M.; Mohan, C.D.; Madan, V.; Kanojia, D.; Shobith, R.; Nanjundaswamy, S.; Mason, D.J.; Bender, A.; Basappa, B.; et al. Synthesis and characterization of novel 2-amino-chromene-nitriles that target Bcl-2 in acute myeloid leukemia cell lines. PLoS ONE 2014, 9, e107118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohan, C.D.; Anilkumar, N.C.; Rangappa, S.; Shanmugam, M.K.; Mishra, S.; Chinnathambi, A.; Alharbi, S.A.; Bhattacharjee, A.; Sethi, G.; Kumar, A.P.; et al. Novel 1,3,4-Oxadiazole Induces Anticancer Activity by Targeting NF-κB in Hepatocellular Carcinoma Cells. Front. Oncol. 2018, 8, 42. [Google Scholar] [CrossRef] [Green Version]
- Pandey, V.; Wang, B.; Mohan, C.D.; Raquib, A.R.; Rangappa, S.; Srinivasa, V.; Fuchs, J.E.; Girish, K.S.; Zhu, T.; Bender, A.; et al. Discovery of a small-molecule inhibitor of specific serine residue BAD phosphorylation. Proc. Natl. Acad. Sci. USA 2018, 115, E10505–E10514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bharathkumar, H.; Paricharak, S.; Dinesh, K.R.; Siveen, K.S.; Fuchs, J.E.; Rangappa, S.; Mohan, C.D.; Mohandas, N.; Kumar, A.P.; Sethi, G.; et al. Synthesis, biological evaluation and in silico and in vitro mode-of-action analysis of novel dihydropyrimidones targeting PPAR-γ. RSC Adv. 2014, 4, 45143–45146. [Google Scholar] [CrossRef] [Green Version]
- Sajith, A.M.; Narasimhamurthy, K.H.; Shanmugam, M.K.; Rangappa, S.; Chandra Nayak, S.; Chinnathambi, A.; Awad Alahmadi, T.; Ali Alharbi, S.; Haridas, K.R.; Reddy, E.K.; et al. Pyrimidine-2,4-dione targets STAT3 signaling pathway to induce cytotoxicity in hepatocellular carcinoma cells. Bioorg. Med. Chem. Lett. 2021, 50, 128332. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Lee, J.H.; Sethi, G.; Kim, C.; Baek, S.H.; Nam, D.; Chung, W.S.; Kim, S.H.; Shim, B.S.; Ahn, K.S. Bergamottin, a natural furanocoumarin obtained from grapefruit juice induces chemosensitization and apoptosis through the inhibition of STAT3 signaling pathway in tumor cells. Cancer Lett. 2014, 354, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Mohan, C.D.; Kim, C.; Siveen, K.S.; Manu, K.A.; Rangappa, S.; Chinnathambi, A.; Alharbi, S.A.; Rangappa, K.S.; Kumar, A.P.; Ahn, K.S. Crocetin Imparts Antiproliferative Activity via Inhibiting STAT3 Signaling in Hepatocellular Carcinoma. IUBMB Life 2021, 73. [Google Scholar] [CrossRef]
- Mohan, C.D.; Liew, Y.Y.; Jung, Y.Y.; Rangappa, S.; Preetham, H.D.; Chinnathambi, A.; Alahmadi, T.A.; Alharbi, S.A.; Lin, Z.-X.; Rangappa, K.S.; et al. Brucein D modulates MAPK signaling cascade to exert multi-faceted anti-neoplastic actions against breast cancer cells. Biochimie 2021, 182, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, A.; Pandey, V.; Mohan, C.D.; Chia, Y.T.; Rangappa, S.; Mathai, J.; Baburajeev, C.P.; Paricharak, S.; Mervin, L.H.; Bulusu, K.C.; et al. Novel Adamantanyl-Based Thiadiazolyl Pyrazoles Targeting EGFR in Triple-Negative Breast Cancer. ACS Omega 2016, 1, 1412–1424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baburajeev, C.P.; Dhananjaya Mohan, C.; Ananda, H.; Rangappa, S.; Fuchs, J.E.; Jagadish, S.; Sivaraman Siveen, K.; Chinnathambi, A.; Ali Alharbi, S.; Zayed, M.E.; et al. Development of Novel Triazolo-Thiadiazoles from Heterogeneous “Green” Catalysis as Protein Tyrosine Phosphatase 1B Inhibitors. Sci. Rep. 2015, 5, 14195. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Wang, L.; Deivasigamni, A.; Looi, C.Y.; Karthikeyan, C.; Trivedi, P.; Chinnathambi, A.; Alharbi, S.A.; Arfuso, F.; Dharmarajan, A.; et al. A novel benzimidazole derivative, MBIC inhibits tumor growth and promotes apoptosis via activation of ROS-dependent JNK signaling pathway in hepatocellular carcinoma. Oncotarget 2017, 8, 12831–12842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohan, C.D.; Bharathkumar, H.; Dukanya, D.; Rangappa, S.; Shanmugam, M.K.; Chinnathambi, A.; Alharbi, S.A.; Alahmadi, T.A.; Bhattacharjee, A.; Lobie, P.E.; et al. N-Substituted Pyrido-1,4-Oxazin-3-Ones Induce Apoptosis of Hepatocellular Carcinoma Cells by Targeting NF-κB Signaling Pathway. Front. Pharmacol. 2018, 9, 1125. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, M.K.; Rajendran, P.; Li, F.; Kim, C.; Sikka, S.; Siveen, K.S.; Kumar, A.P.; Ahn, K.S.; Sethi, G. Abrogation of STAT3 signaling cascade by zerumbone inhibits proliferation and induces apoptosis in renal cell carcinoma xenograft mouse model. Mol. Carcinog. 2015, 54, 971–985. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, N.B.; Mohan, C.D.; Basappa, S.; Pandey, V.; Rangappa, S.; Bharathkumar, H.; Kumar, A.P.; Lobie, P.E.; Rangappa, K.S. An azaspirane derivative suppresses growth and induces apoptosis of ER-positive and ER-negative breast cancer cells through the modulation of JAK2/STAT3 signaling pathway. Int. J. Oncol. 2016, 49, 1221–1229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, Y.Y.; Lee, J.H.; Nam, D.; Narula, A.S.; Namjoshi, O.A.; Blough, B.E.; Um, J.Y.; Sethi, G.; Ahn, K.S. Anti-myeloma Effects of Icariin Are Mediated Through the Attenuation of JAK/STAT3-Dependent Signaling Cascade. Front. Pharmacol. 2018, 9, 531. [Google Scholar] [CrossRef]
- Guo, C.; Yang, G.; Khun, K.; Kong, X.; Levy, D.; Lee, P.; Melamed, J. Activation of Stat3 in renal tumors. Am. J. Transl. Res. 2009, 1, 283–290. [Google Scholar] [PubMed]
- Lo, H.-W.; Cao, X.; Zhu, H.; Ali-Osman, F. Constitutively activated STAT3 frequently coexpresses with epidermal growth factor receptor in high-grade gliomas and targeting STAT3 sensitizes them to Iressa and alkylators. Clin. Cancer Res. 2008, 14, 6042–6054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, P.; Wu, D.; Zhao, L.; Huang, L.; Shen, G.; Huang, J.; Chai, Y. Prognostic role of STAT3 in solid tumors: A systematic review and meta-analysis. Oncotarget 2016, 7, 19863–19883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Huang, J.; Li, W.; Chen, Y.; Liu, X.; Wang, J. Meta-analysis of STAT3 and phospho-STAT3 expression and survival of patients with breast cancer. Oncotarget 2018, 9, 13060–13067. [Google Scholar] [CrossRef] [Green Version]
- Tong, M.; Wang, J.; Jiang, N.; Pan, H.; Li, D. Correlation between p-STAT3 overexpression and prognosis in lung cancer: A systematic review and meta-analysis. PLoS ONE 2017, 12, e0182282. [Google Scholar] [CrossRef] [Green Version]
- Ji, K.; Zhang, M.; Chu, Q.; Gan, Y.; Ren, H.; Zhang, L.; Wang, L.; Li, X.; Wang, W. The Role of p-STAT3 as a Prognostic and Clinicopathological Marker in Colorectal Cancer: A Systematic Review and Meta-Analysis. PLoS ONE 2016, 11, e0160125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shanmugam, M.K.; Lee, J.H.; Chai, E.Z.; Kanchi, M.M.; Kar, S.; Arfuso, F.; Dharmarajan, A.; Kumar, A.P.; Ramar, P.S.; Looi, C.Y.; et al. Cancer prevention and therapy through the modulation of transcription factors by bioactive natural compounds. Semin. Cancer Biol. 2016, 40–41, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Chonov, D.C.; Ignatova, M.M.K.; Ananiev, J.R.; Gulubova, M.V. IL-6 Activities in the Tumour Microenvironment. Part 1. Open Access Maced. J. Med. Sci. 2019, 7, 2391–2398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.; Lee, S.G.; Yang, W.M.; Arfuso, F.; Um, J.Y.; Kumar, A.P.; Bian, J.; Sethi, G.; Ahn, K.S. Formononetin-induced oxidative stress abrogates the activation of STAT3/5 signaling axis and suppresses the tumor growth in multiple myeloma preclinical model. Cancer Lett. 2018, 431, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.; Lee, H.J.; Kim, S.H.; Kim, T.; Han, T.Y.; Suh, Y.-G.; Chun, J.; Kim, S.Y.; Ahn, S.K. Novel Galiellalactone Analogues Can Target STAT3 Phosphorylation and Cause Apoptosis in Triple-Negative Breast Cancer. Biomolecules 2019, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohan, C.D.; Rangappa, S.; Nayak, S.C.; Jadimurthy, R.; Wang, L.; Sethi, G.; Garg, M.; Rangappa, K.S. Bacteria as a treasure house of secondary metabolites with anticancer potential. Semin. Cancer Biol. 2021, in press. [Google Scholar] [CrossRef]
- Anusha, S.; Anandakumar, B.S.; Mohan, C.D.; Nagabhushana, G.P.; Priya, B.S.; Rangappa, K.S.; Basappa, B.; Chandrappa, G.T. Preparation and use of combustion-derived Bi2O3 for the synthesis of heterocycles with anti-cancer properties by Suzuki-coupling reactions. RSC Adv. 2014, 4, 52181–52188. [Google Scholar] [CrossRef]
- Srinivas, V.; Mohan, C.D.; Baburajeev, C.P.; Rangappa, S.; Jagadish, S.; Fuchs, J.E.; Sukhorukov, A.Y.; Chandra, C.; Mason, D.J.; Kumar, K.S.S.; et al. Synthesis and characterization of novel oxazines and demonstration that they specifically target cyclooxygenase 2. Bioorg. Med. Chem. Lett. 2015, 25, 2931–2936. [Google Scholar] [CrossRef] [PubMed]
- Mohan, C.D.; Hari, S.; Preetham, H.D.; Rangappa, S.; Barash, U.; Ilan, N.; Nayak, S.C.; Gupta, V.K.; Basappa, B.; Vlodavsky, I.; et al. Targeting Heparanase in Cancer: Inhibition by Synthetic, Chemically Modified, and Natural Compounds. iScience 2019, 15, 360–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thushara, R.M.; Hemshekhar, M.; Basappa, B.; Kemparaju, K.; Rangappa, K.S.; Girish, K.S. Biologicals, platelet apoptosis and human diseases: An outlook. Crit. Rev. Oncol. Hematol. 2015, 93, 149–158. [Google Scholar] [CrossRef]
- Dai, X.; Ahn, K.S.; Kim, C.; Siveen, K.S.; Ong, T.H.; Shanmugam, M.K.; Li, F.; Shi, J.; Kumar, A.P.; Wang, L.Z.; et al. Ascochlorin, an isoprenoid antibiotic inhibits growth and invasion of hepatocellular carcinoma by targeting STAT3 signaling cascade through the induction of PIAS3. Mol. Oncol. 2015, 9, 818–833. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, C.; Kim, S.H.; Sethi, G.; Ahn, K.S. Farnesol inhibits tumor growth and enhances the anticancer effects of bortezomib in multiple myeloma xenograft mouse model through the modulation of STAT3 signaling pathway. Cancer Lett. 2015, 360, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, A.; Shanmugam, M.K.; Ong, T.H.; Li, F.; Perumal, E.; Chen, L.; Vali, S.; Abbasi, T.; Kapoor, S.; Ahn, K.S.; et al. Emodin inhibits growth and induces apoptosis in an orthotopic hepatocellular carcinoma model by blocking activation of STAT3. Br. J. Pharmacol. 2013, 170, 807–821. [Google Scholar] [CrossRef] [Green Version]
- Subramaniam, A.; Shanmugam, M.K.; Perumal, E.; Li, F.; Nachiyappan, A.; Dai, X.; Swamy, S.N.; Ahn, K.S.; Kumar, A.P.; Tan, B.K.; et al. Potential role of signal transducer and activator of transcription (STAT)3 signaling pathway in inflammation, survival, proliferation and invasion of hepatocellular carcinoma. Biochim. Biophys. Acta 2013, 1835, 46–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sethi, G.; Chatterjee, S.; Rajendran, P.; Li, F.; Shanmugam, M.K.; Wong, K.F.; Kumar, A.P.; Senapati, P.; Behera, A.K.; Hui, K.M.; et al. Inhibition of STAT3 dimerization and acetylation by garcinol suppresses the growth of human hepatocellular carcinoma in vitro and in vivo. Mol. Cancer 2014, 13, 66. [Google Scholar] [CrossRef] [Green Version]
- Harmer, D.; Falank, C.; Reagan, M.R. Interleukin-6 Interweaves the Bone Marrow Microenvironment, Bone Loss, and Multiple Myeloma. Front. Endocrinol. 2019, 9, 788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dabir, S.; Kluge, A.; Dowlati, A. The association and nuclear translocation of the PIAS3-STAT3 complex is ligand and time dependent. Mol. Cancer Res. 2009, 7, 1854–1860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.-F.; Lai, R. STAT3 in Cancer—Friend or Foe? Cancers 2014, 6, 1408–1440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, P.A.; Grandis, J.R. STAT3 signaling: Anticancer strategies and challenges. Mol. Interv. 2011, 11, 18–26. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Rajendran, P.; Sethi, G. Thymoquinone inhibits proliferation, induces apoptosis and chemosensitizes human multiple myeloma cells through suppression of signal transducer and activator of transcription 3 activation pathway. Br. J. Pharmacol. 2010, 161, 541–554. [Google Scholar] [CrossRef] [Green Version]
- Qi, C.; Han, T.; Tang, H.; Huang, K.; Min, J.; Li, J.; Ding, X.; Xu, Z. Shp2 Inhibits Proliferation of Esophageal Squamous Cell Cancer via Dephosphorylation of Stat3. Int. J. Mol. Sci. 2017, 18, 134. [Google Scholar] [CrossRef] [Green Version]
- Chong, P.S.Y.; Zhou, J.; Lim, J.S.L.; Hee, Y.T.; Chooi, J.-Y.; Chung, T.-H.; Tan, Z.T.; Zeng, Q.; Waller, D.D.; Sebag, M.; et al. IL6 Promotes a STAT3-PRL3 Feedforward Loop via SHP2 Repression in Multiple Myeloma. Cancer Res. 2019, 79, 4679–4688. [Google Scholar] [CrossRef]
- Chen, D.; Frezza, M.; Schmitt, S.; Kanwar, J.; Dou, Q.P. Bortezomib as the first proteasome inhibitor anticancer drug: Current status and future perspectives. Curr. Cancer Drug Targets 2011, 11, 239–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Zang, Y.; Sen, M.; Leeman-Neill, R.J.; Man, D.S.; Grandis, J.R.; Johnson, D.E. Bortezomib up-regulates activated signal transducer and activator of transcription-3 and synergizes with inhibitors of signal transducer and activator of transcription-3 to promote head and neck squamous cell carcinoma cell death. Mol. Cancer Ther. 2009, 8, 2211–2220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, M.; Zhu, L.; Somlo, G.; Hughes, A.; Zhou, B.; Yen, Y. Bortezomib therapeutic effect is associated with expression of FGFR3 in multiple myeloma cells. Anticancer Res. 2009, 29, 1–9. [Google Scholar]
- Mohan, C.D.; Srinivasa, V.; Rangappa, S.; Mervin, L.; Mohan, S.; Paricharak, S.; Baday, S.; Li, F.; Shanmugam, M.K.; Chinnathambi, A.; et al. Trisubstituted-Imidazoles Induce Apoptosis in Human Breast Cancer Cells by Targeting the Oncogenic PI3K/Akt/mTOR Signaling Pathway. PLoS ONE 2016, 11, e0153155. [Google Scholar] [CrossRef] [Green Version]
- Yeung, K.T.; Yang, J. Epithelial-mesenchymal transition in tumor metastasis. Mol. Oncol. 2017, 11, 28–39. [Google Scholar] [CrossRef] [Green Version]
- Yadav, A.; Kumar, B.; Datta, J.; Teknos, T.N.; Kumar, P. IL-6 promotes head and neck tumor metastasis by inducing epithelial-mesenchymal transition via the JAK-STAT3-SNAIL signaling pathway. Mol. Cancer Res. 2011, 9, 1658–1667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [Green Version]
- Cheng, G.Z.; Zhang, W.Z.; Sun, M.; Wang, Q.; Coppola, D.; Mansour, M.; Xu, L.M.; Costanzo, C.; Cheng, J.Q.; Wang, L.H. Twist is transcriptionally induced by activation of STAT3 and mediates STAT3 oncogenic function. J. Biol. Chem. 2008, 283, 14665–14673. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arora, L.; Mohan, C.D.; Yang, M.H.; Rangappa, S.; Deivasigamani, A.; Kumar, A.P.; Kunnumakkara, A.B.; Garg, M.; Chinnathambi, A.; Alharbi, S.A.; et al. Tris(dibenzylideneacetone)dipalladium(0) (Tris DBA) Abrogates Tumor Progression in Hepatocellular Carcinoma and Multiple Myeloma Preclinical Models by Regulating the STAT3 Signaling Pathway. Cancers 2021, 13, 5479. https://doi.org/10.3390/cancers13215479
Arora L, Mohan CD, Yang MH, Rangappa S, Deivasigamani A, Kumar AP, Kunnumakkara AB, Garg M, Chinnathambi A, Alharbi SA, et al. Tris(dibenzylideneacetone)dipalladium(0) (Tris DBA) Abrogates Tumor Progression in Hepatocellular Carcinoma and Multiple Myeloma Preclinical Models by Regulating the STAT3 Signaling Pathway. Cancers. 2021; 13(21):5479. https://doi.org/10.3390/cancers13215479
Chicago/Turabian StyleArora, Loukik, Chakrabhavi Dhananjaya Mohan, Min Hee Yang, Shobith Rangappa, Amudha Deivasigamani, Alan Prem Kumar, Ajaikumar B. Kunnumakkara, Manoj Garg, Arunachalam Chinnathambi, Sulaiman Ali Alharbi, and et al. 2021. "Tris(dibenzylideneacetone)dipalladium(0) (Tris DBA) Abrogates Tumor Progression in Hepatocellular Carcinoma and Multiple Myeloma Preclinical Models by Regulating the STAT3 Signaling Pathway" Cancers 13, no. 21: 5479. https://doi.org/10.3390/cancers13215479
APA StyleArora, L., Mohan, C. D., Yang, M. H., Rangappa, S., Deivasigamani, A., Kumar, A. P., Kunnumakkara, A. B., Garg, M., Chinnathambi, A., Alharbi, S. A., Alahmadi, T. A., Rangappa, K. S., Hui, K. M., Sethi, G., & Ahn, K. S. (2021). Tris(dibenzylideneacetone)dipalladium(0) (Tris DBA) Abrogates Tumor Progression in Hepatocellular Carcinoma and Multiple Myeloma Preclinical Models by Regulating the STAT3 Signaling Pathway. Cancers, 13(21), 5479. https://doi.org/10.3390/cancers13215479







