Predicting Tumor Budding Status in Cervical Cancer Using MRI Radiomics: Linking Imaging Biomarkers to Histologic Characteristics
Abstract
:Simple Summary
Abstract
1. Introduction
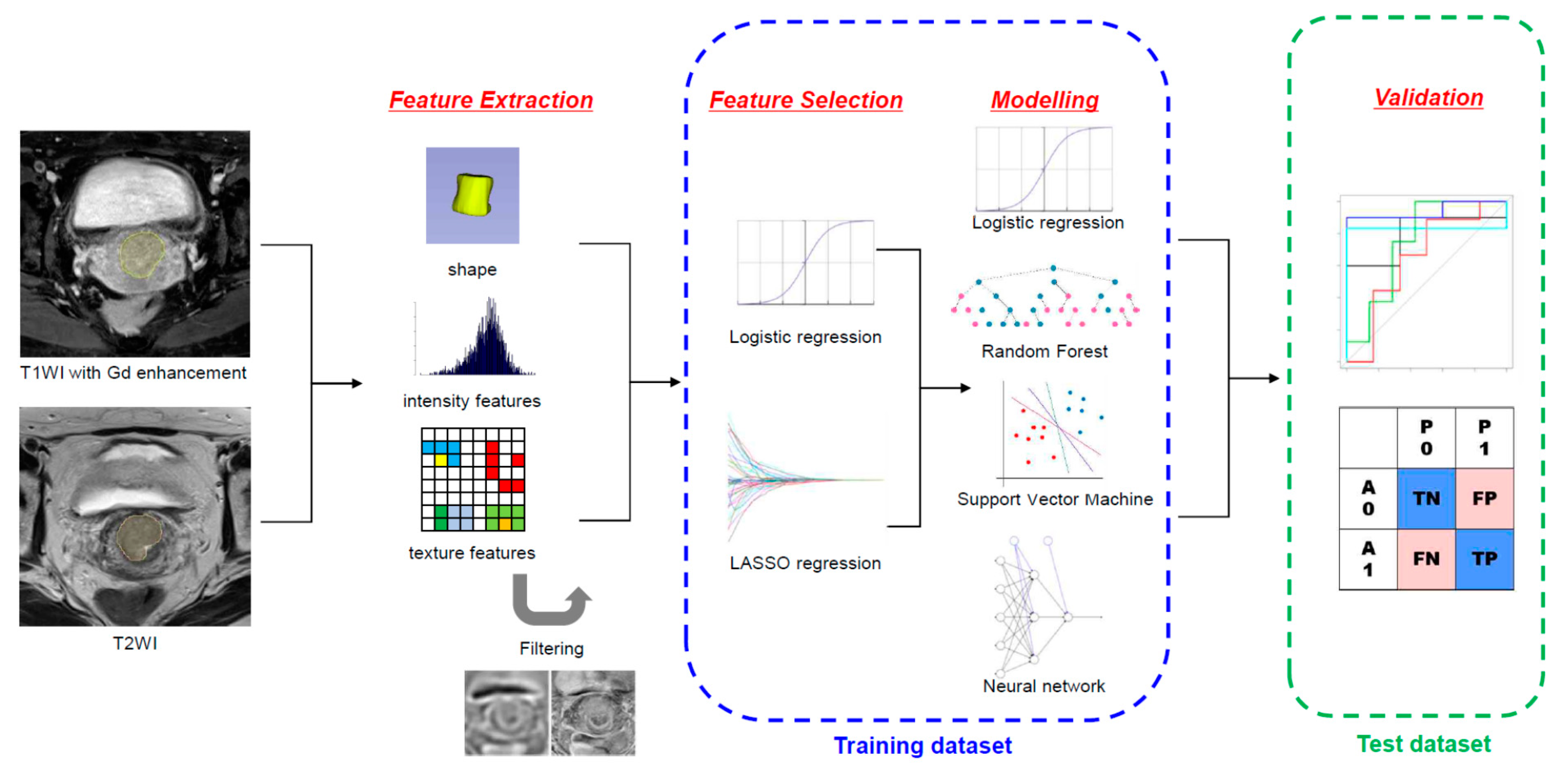
2. Materials and Methods
2.1. Study Population
2.2. Histopathological Evaluation
2.3. Image Acquisition
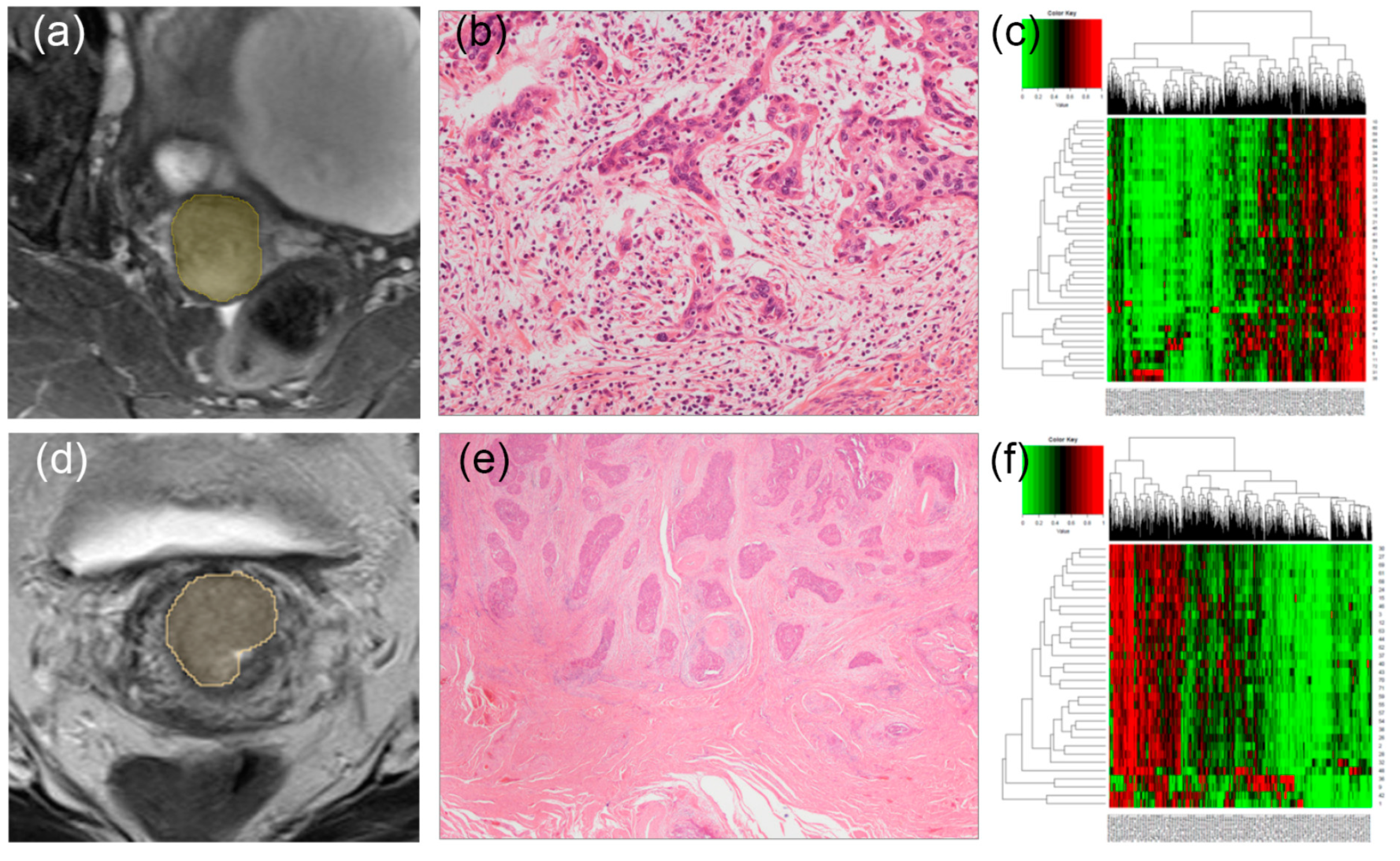
2.4. Segmentation and Preprocessing
2.5. Radiomic Feature Extraction and Selection
2.6. Statistical Analysis and Machine Learning (ML) Model Building
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Graham, R.P.; Vierkant, R.A.; Tillmans, L.S.; Wang, A.H.; Laird, P.W.; Weisenberger, D.J.; Lynch, C.F.; French, A.J.; Slager, S.L.; Raissian, Y.; et al. Tumor Budding in Colorectal Carcinoma: Confirmation of Prognostic Significance and Histologic Cutoff in a Population-based Cohort. Am. J. Surg. Pathol. 2015, 39, 1340–1346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koelzer, V.H.; Zlobec, I.; Lugli, A. Tumor budding in colorectal cancer--ready for diagnostic practice? Hum. Pathol. 2016, 47, 4–19. [Google Scholar] [CrossRef] [PubMed]
- Karamitopoulou, E.; Gloor, B. Tumour budding is a strong and independent prognostic factor in pancreatic cancer. Reply to comment. Eur. J. Cancer 2013, 49, 2458–2459. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Chong, G.O.; Park, J.Y.; Chung, D.; Lee, Y.H.; Lee, H.J.; Hong, D.G.; Han, H.S.; Lee, Y.S. Tumor budding in cervical cancer as a prognostic factor and its possible role as an additional intermediate-risk factor. Gynecol. Oncol. 2020, 159, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Chong, G.O.; Park, N.J.Y.; Han, H.S.; Cho, J.; Kim, M.G.; Choi, Y.; Yeo, J.Y.; Lee, Y.H.; Hong, D.G.; Park, J.Y. Intratumoral budding: A novel prognostic biomarker for tumor recurrence and a potential predictor of nodal metastasis in uterine cervical cancer. Eur. J. Surg. Oncol. 2021, in press. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [Green Version]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Patro, S.; Sahu, K.K. Normalization: A preprocessing stage. arXiv 2015, arXiv:1503.06462. [Google Scholar]
- Riedmiller, M.; Rprop, I. Rprop-Description and Implementation Details. 1994. Available online: http://www.inf.fu-berlin.de/lehre/WS06/Musterererkennung/Paper/rprop.pdf (accessed on 25 September 2021).
- Carter, J.V.; Pan, J.; Rai, S.N.; Galandiuk, S. ROC-ing along: Evaluation and interpretation of receiver operating characteristic curves. Surgery 2016, 159, 1638–1645. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, H.; Wang, S.; Dong, D.; Yin, F.; Chen, A.; Wang, S.; Zhao, G.; Fang, M.; Tian, J.; et al. MR-Based Radiomics Nomogram of Cervical Cancer in Prediction of the Lymph-Vascular Space Invasion preoperatively. J. Magn. Reson. Imaging 2019, 49, 1420–1426. [Google Scholar] [CrossRef] [PubMed]
- Hua, W.Q.; Xiao, T.H.; Jiang, X.R.; Liu, Z.Y.; Wang, M.Y.; Zheng, H.R.; Wang, S.S. Lymph-vascular space invasion prediction in cervical cancer: Exploring radiomics and deep learning multilevel features of tumor and peritumor tissue on multiparametric MRI. Biomed. Signal. Process. Control 2020, 58, 101869. [Google Scholar] [CrossRef]
- Zlobec, I.; Lugli, A. Tumour budding in colorectal cancer: Molecular rationale for clinical translation. Nat. Rev. Cancer 2018, 18, 203–204. [Google Scholar] [CrossRef]
- Zlobec, I.; Berger, M.D.; Lugli, A. Tumour budding and its clinical implications in gastrointestinal cancers. Br. J. Cancer 2020, 123, 700–708. [Google Scholar] [CrossRef]
- Lugli, A.; Zlobec, I.; Berger, M.D.; Kirsch, R.; Nagtegaal, I.D. Tumour budding in solid cancers. Nat. Rev. Clin. Oncol. 2021, 18, 101–115. [Google Scholar] [CrossRef]
- Slik, K.; Blom, S.; Turkki, R.; Valimaki, K.; Kurki, S.; Mustonen, H.; Haglund, C.; Carpen, O.; Kallioniemi, O.; Korkeila, E.; et al. Combined epithelial marker analysis of tumour budding in stage II colorectal cancer. J. Pathol. Clin. Res. 2019, 5, 63–78. [Google Scholar] [CrossRef]
- Nearchou, I.P.; Kajiwara, Y.; Mochizuki, S.; Harrison, D.J.; Caie, P.D.; Ueno, H. Novel Internationally Verified Method Reports Desmoplastic Reaction as the Most Significant Prognostic Feature For Disease-specific Survival in Stage II Colorectal Cancer. Am. J. Surg. Pathol. 2019, 43, 1239–1248. [Google Scholar] [CrossRef] [PubMed]
- Lee, V.W.K.; Chan, K.F. Tumor budding and poorly-differentiated cluster in prognostication in Stage II colon cancer. Pathol. Res. Pract. 2018, 214, 402–407. [Google Scholar] [CrossRef]
- Ueno, H.; Ishiguro, M.; Nakatani, E.; Ishikawa, T.; Uetake, H.; Matsuda, C.; Nakamoto, Y.; Kotake, M.; Kurachi, K.; Egawa, T.; et al. Prospective Multicenter Study on the Prognostic and Predictive Impact of Tumor Budding in Stage II Colon Cancer: Results From the SACURA Trial. J. Clin. Oncol. 2019, 37, 1886–1894. [Google Scholar] [CrossRef]
- Barel, F.; Cariou, M.; Saliou, P.; Kermarrec, T.; Auffret, A.; Samaison, L.; Bourhis, A.; Badic, B.; Jezequel, J.; Cholet, F.; et al. Histopathological factors help to predict lymph node metastases more efficiently than extra-nodal recurrences in submucosa invading pT1 colorectal cancer. Sci. Rep. 2019, 9, 8342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosch, S.L.; Teerenstra, S.; de Wilt, J.H.; Cunningham, C.; Nagtegaal, I.D. Predicting lymph node metastasis in pT1 colorectal cancer: A systematic review of risk factors providing rationale for therapy decisions. Endoscopy 2013, 45, 827–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogers, A.C.; Winter, D.C.; Heeney, A.; Gibbons, D.; Lugli, A.; Puppa, G.; Sheahan, K. Systematic review and meta-analysis of the impact of tumour budding in colorectal cancer. Br. J. Cancer 2016, 115, 831–840. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.X.; Zhang, Z.Z.; Zhao, G.; Zhao, E.H. Prognostic and pathological impact of tumor budding in gastric cancer: A systematic review and meta-analysis. World J. Gastrointest. Oncol. 2019, 11, 898–908. [Google Scholar] [CrossRef]
- Almangush, A.; Pirinen, M.; Heikkinen, I.; Makitie, A.A.; Salo, T.; Leivo, I. Tumour budding in oral squamous cell carcinoma: A meta-analysis. Br. J. Cancer 2018, 118, 577–586. [Google Scholar] [CrossRef] [Green Version]
- Lawlor, R.T.; Veronese, N.; Nottegar, A.; Malleo, G.; Smith, L.; Demurtas, J.; Cheng, L.; Wood, L.D.; Silvestris, N.; Salvia, R.; et al. Prognostic Role of High-Grade Tumor Budding in Pancreatic Ductal Adenocarcinoma: A Systematic Review and Meta-Analysis with a Focus on Epithelial to Mesenchymal Transition. Cancers 2019, 11, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almangush, A.; Karhunen, M.; Hautaniemi, S.; Salo, T.; Leivo, I. Prognostic value of tumour budding in oesophageal cancer: A meta-analysis. Histopathology 2016, 68, 173–182. [Google Scholar] [CrossRef]
- Yang, K.; Park, W.; Huh, S.J.; Bae, D.S.; Kim, B.G.; Lee, J.W. Clinical outcomes in patients treated with radiotherapy after surgery for cervical cancer. Radiat. Oncol. J. 2017, 35, 39–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado, G.; Bundy, B.; Zaino, R.; Sevin, B.U.; Creasman, W.T.; Major, F. Prospective surgical-pathological study of disease-free interval in patients with stage IB squamous cell carcinoma of the cervix: A Gynecologic Oncology Group study. Gynecol. Oncol. 1990, 38, 352–357. [Google Scholar] [CrossRef]
- Rotman, M.; Sedlis, A.; Piedmonte, M.R.; Bundy, B.; Lentz, S.S.; Muderspach, L.I.; Zaino, R.J. A phase III randomized trial of postoperative pelvic irradiation in Stage IB cervical carcinoma with poor prognostic features: Follow-up of a gynecologic oncology group study. Int. J. Radiat. Oncol. Biol. Phys. 2006, 65, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Sedlis, A.; Bundy, B.N.; Rotman, M.Z.; Lentz, S.S.; Muderspach, L.I.; Zaino, R.J. A randomized trial of pelvic radiation therapy versus no further therapy in selected patients with stage IB carcinoma of the cervix after radical hysterectomy and pelvic lymphadenectomy: A Gynecologic Oncology Group Study. Gynecol. Oncol. 1999, 73, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.Y.; Park, S.I.; Nam, B.H.; Cho, C.K.; Kim, K.; Kim, B.J.; Kim, M.H.; Choi, S.C.; Lee, E.D.; Lee, K.H. Is adjuvant chemoradiotherapy overtreatment in cervical cancer patients with intermediate risk factors? Int. J. Radiat. Oncol. Biol. Phys. 2011, 79, 794–799. [Google Scholar] [CrossRef]
- Li, Y.; Lu, L.; Xiao, M.; Dercle, L.; Huang, Y.; Zhang, Z.; Schwartz, L.H.; Li, D.; Zhao, B. CT Slice Thickness and Convolution Kernel Affect Performance of a Radiomic Model for Predicting EGFR Status in Non-Small Cell Lung Cancer: A Preliminary Study. Sci. Rep. 2018, 8, 17913. [Google Scholar] [CrossRef]
- Ganeshan, B.; Skogen, K.; Pressney, I.; Coutroubis, D.; Miles, K. Tumour heterogeneity in oesophageal cancer assessed by CT texture analysis: Preliminary evidence of an association with tumour metabolism, stage, and survival. Clin. Radiol. 2012, 67, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Ng, F.; Kozarski, R.; Ganeshan, B.; Goh, V. Assessment of tumor heterogeneity by CT texture analysis: Can the largest cross-sectional area be used as an alternative to whole tumor analysis? Eur. J. Radiol. 2013, 82, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Ganeshan, B.; Panayiotou, E.; Burnand, K.; Dizdarevic, S.; Miles, K. Tumour heterogeneity in non-small cell lung carcinoma assessed by CT texture analysis: A potential marker of survival. Eur. Radiol. 2012, 22, 796–802. [Google Scholar] [CrossRef] [PubMed]
- Ng, F.; Ganeshan, B.; Kozarski, R.; Miles, K.A.; Goh, V. Assessment of primary colorectal cancer heterogeneity by using whole-tumor texture analysis: Contrast-enhanced CT texture as a biomarker of 5-year survival. Radiology 2013, 266, 177–184. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Lu, J.; Gao, C.; Zeng, J.; Zhou, C.; Lai, X.; Cai, W.; Xu, M. Predicting the response to neoadjuvant chemotherapy for breast cancer: Wavelet transforming radiomics in MRI. BMC Cancer 2020, 20, 100. [Google Scholar] [CrossRef]
- Banerjee, I.; Malladi, S.; Lee, D.; Depeursinge, A.; Telli, M.; Lipson, J.; Golden, D.; Rubin, D.L. Assessing treatment response in triple-negative breast cancer from quantitative image analysis in perfusion magnetic resonance imaging. J. Med. Imaging (Bellingham) 2018, 5, 011008. [Google Scholar] [CrossRef]
| Characteristic | Training Set (n = 48) | Test Set (n = 26) | p-Value |
|---|---|---|---|
| Age (years) | 49.1 ± 11.8 | 51.4 ± 10.1 | 0.4 |
| FIGO stage (n, %) | 0.517 † | ||
| IB1 | 11 (22.9%) | 3 (6.3%) | |
| IB2 | 12 (25.0%) | 12 (25.0%) | |
| IB3 | 3 (6.3%) | 1 (2.1%) | |
| IIA | 1 (2.1%) | 0 (0.0%) | |
| IIB | 5 (10.4%) | 3 (6.3%) | |
| IIIC1 | 16 (33.3%) | 7 (14.6%) | |
| Histology (n, %) | 0.580 † | ||
| Squamous cell carcinoma | 36 (75.0%) | 18 (37.5%) | |
| Adenocarcinoma | 8 (16.7%) | 7 (14.6%) | |
| Adenosquamous carcinoma | 4 (8.3%) | 1 (2.1%) | |
| Tumor size (cm) | 2.8 ± 1.6 | 2.6 ± 1.0 | 0.417 |
| Lymphovascular invasion (n, %) | 23 (47.9%) | 11 (22.9%) | 0.828 |
| Deep stromal invasion (n, %) | 33 (68.8%) | 19 (39.6%) | 0.903 |
| Parametrial invasion (n, %) | 12 (25.0%) | 7 (14.6%) | >0.999 |
| Lymph node metastasis (n, %) | 16 (33.3%) | 7 (14.6%) | 0.76 |
| Intratumor budding | >0.999 | ||
| Intratumor budding counts | 7.8 ± 9.9 | 7.7 ± 10.3 | |
| Intratumoral budding (n, %) | |||
| >4 | 27 (56.3%) | 15 (31.3%) | |
| ≤4 | 21 (43.8%) | 11 (22.9%) |
| MR Sequence | Feature Name |
|---|---|
| T1 | log.sigma.1.0.mm.3D_glszm_ZonePercentage log.sigma.3.0.mm.3D_ngtdm_Contrast wavelet.LLH_gldm_LargeDependenceEmphasis wavelet.LHH_firstorder_Skewness wavelet.HLL_glszm_SizeZoneNonUniformityNormalized wavelet.HLH_gldm_LargeDependenceHighGrayLevelEmphasis wavelet.HHL_gldm_DependenceVariance. wavelet.LLL_glcm_InverseVariance. |
| T2 | log.sigma.3.0.mm.3D_ glcm_Imc2 wavelet.LLH_glszm_HighGrayLevelZoneEmphasis wavelet.LLH_glszm_LowGrayLevelZoneEmphasis wavelet.LLH_glszm_SmallAreaHighGrayLevelEmphasis wavelet.LHL_gldm_HighGrayLevelEmphasis wavelet.LHL_gldm_LowGrayLevelEmphasis wavelet.LHL_glrlm_HighGrayLevelRunEmphasis wavelet.LHL_glrlm_LowGrayLevelRunEmphasis wavelet.LHL_glszm_GrayLevelNonUniformityNormalized wavelet.LHH_firstorder_Kurtosis wavelet.HLH_glszm_ZoneEntropy wavelet.LLL_firstorder_Minimum |
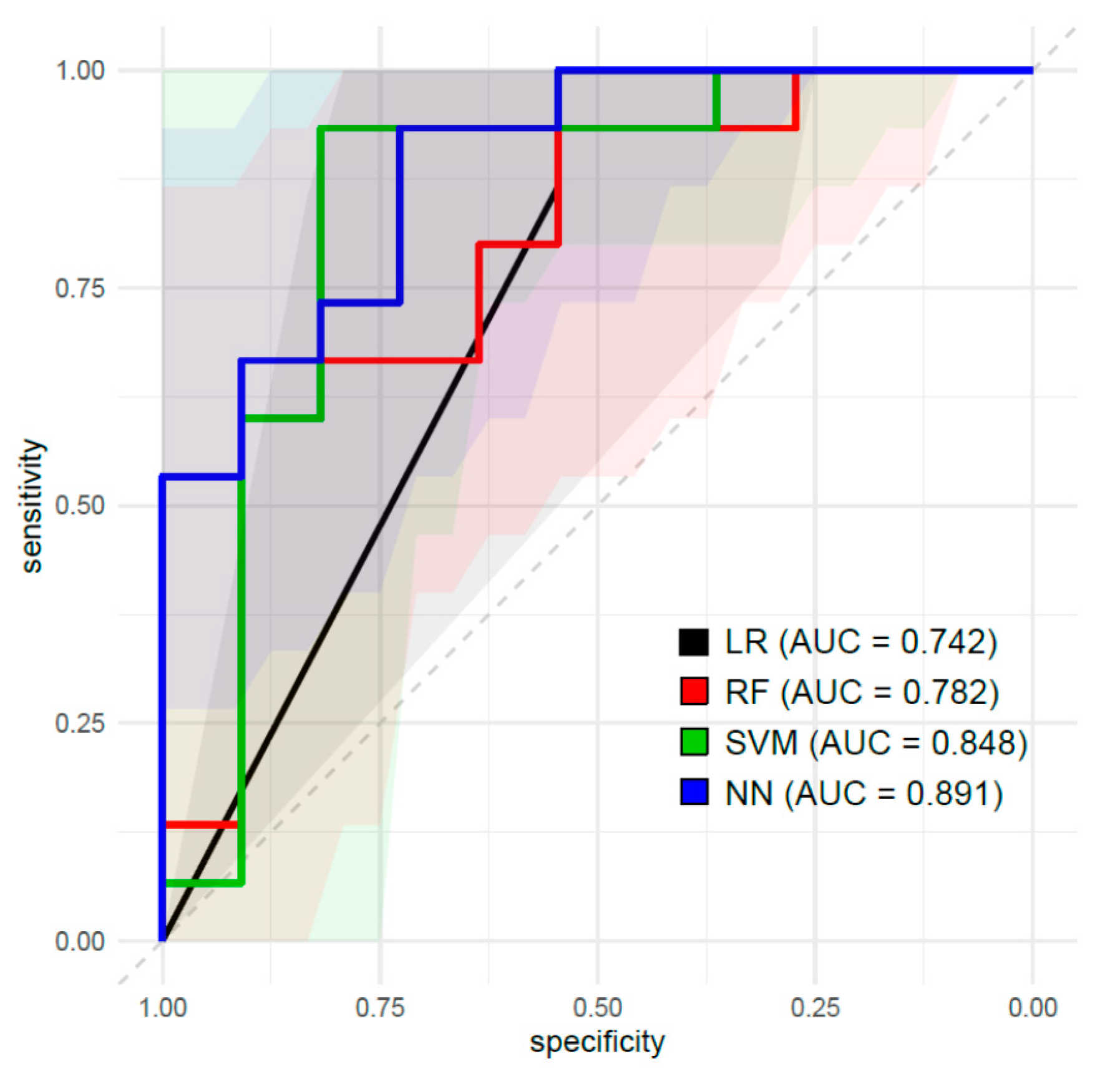
| Classifier | AUC (95% CI) | Accuracy (95% CI) | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|
| LR | 0.742 (0.572–0.907) | 0.769 (0.564–0.910) | 0.857 | 0.737 | 0.546 | 0.933 |
| RF | 0.782 (0.528–0.884) | 0.731 (0.522–0.884) | 0.750 | 0.722 | 0.546 | 0.867 |
| SVM | 0.849 (0.740–1.000) | 0.885 (0.699–0.976) | 0.900 | 0.875 | 0.818 | 0.933 |
| NN | 0.891 (0.768–1.000) | 0.731 (0.522–0.884) | 0.667 | 0.786 | 0.727 | 0.733 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chong, G.O.; Park, S.-H.; Park, N.J.-Y.; Bae, B.K.; Lee, Y.H.; Jeong, S.Y.; Kim, J.-C.; Park, J.Y.; Ando, Y.; Han, H.S. Predicting Tumor Budding Status in Cervical Cancer Using MRI Radiomics: Linking Imaging Biomarkers to Histologic Characteristics. Cancers 2021, 13, 5140. https://doi.org/10.3390/cancers13205140
Chong GO, Park S-H, Park NJ-Y, Bae BK, Lee YH, Jeong SY, Kim J-C, Park JY, Ando Y, Han HS. Predicting Tumor Budding Status in Cervical Cancer Using MRI Radiomics: Linking Imaging Biomarkers to Histologic Characteristics. Cancers. 2021; 13(20):5140. https://doi.org/10.3390/cancers13205140
Chicago/Turabian StyleChong, Gun Oh, Shin-Hyung Park, Nora Jee-Young Park, Bong Kyung Bae, Yoon Hee Lee, Shin Young Jeong, Jae-Chul Kim, Ji Young Park, Yu Ando, and Hyung Soo Han. 2021. "Predicting Tumor Budding Status in Cervical Cancer Using MRI Radiomics: Linking Imaging Biomarkers to Histologic Characteristics" Cancers 13, no. 20: 5140. https://doi.org/10.3390/cancers13205140
APA StyleChong, G. O., Park, S.-H., Park, N. J.-Y., Bae, B. K., Lee, Y. H., Jeong, S. Y., Kim, J.-C., Park, J. Y., Ando, Y., & Han, H. S. (2021). Predicting Tumor Budding Status in Cervical Cancer Using MRI Radiomics: Linking Imaging Biomarkers to Histologic Characteristics. Cancers, 13(20), 5140. https://doi.org/10.3390/cancers13205140






