WWC Proteins: Important Regulators of Hippo Signaling in Cancer
Simple Summary
Abstract
1. Introduction
1.1. Hippo Signaling and Cancer
1.2. WWC Proteins: Upstream Regulators of the Hippo Pathway
2. WWC Proteins and Cancer
2.1. WWC1
2.2. WWC2
2.3. WWC3
3. WWC Binding Proteins and Hippo Signaling in Cancer
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meng, Z.; Moroishi, T.; Guan, K.L. Mechanisms of Hippo pathway regulation. Genes Dev. 2016, 30, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Pan, D. The Hippo Signaling Pathway in Development and Disease. Dev. Cell 2019, 50, 264–282. [Google Scholar] [CrossRef] [PubMed]
- Maugeri-Saccà, M.; De Maria, R. The Hippo pathway in normal development and cancer. Pharmacol. Ther. 2018, 186, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Udan, R.S.; Kango-Singh, M.; Nolo, R.; Tao, C.; Halder, G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat. Cell Biol. 2003, 5, 914–920. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Huang, J.; Dong, J.; Pan, D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 2003, 114, 445–456. [Google Scholar] [CrossRef]
- Huang, J.; Wu, S.; Barrera, J.; Matthews, K.; Pan, D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 2005, 122, 421–434. [Google Scholar] [CrossRef]
- Dong, J.; Feldmann, G.; Huang, J.; Wu, S.; Zhang, N.; Comerford, S.A.; Gayyed, M.F.; Anders, R.A.; Maitra, A.; Pan, D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 2007, 130, 1120–1133. [Google Scholar] [CrossRef]
- Fulford, A.; Tapon, N.; Ribeiro, P.S. Upstairs, downstairs: Spatial regulation of Hippo signalling. Curr. Opin. Cell Biol. 2018, 51, 22–32. [Google Scholar] [CrossRef]
- Schneider, A.; Huentelman, M.J.; Kremerskothen, J.; Duning, K.; Spoelgen, R.; Nikolich, K. KIBRA: A New Gateway to Learning and Memory? Front. Aging Neurosci. 2010, 2, 4. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, S.; Wennmann, D.O.; Chen, Y.; Kremerskothen, J.; Dong, J. KIBRA: In the brain and beyond. Cell Signal. 2014, 26, 1392–1399. [Google Scholar] [CrossRef]
- Duning, K.; Schurek, E.M.; Schlüter, M.; Bayer, M.; Reinhardt, H.C.; Schwab, A.; Schaefer, L.; Benzing, T.; Schermer, B.; Saleem, M.A.; et al. KIBRA modulates directional migration of podocytes. J. Am. Soc. Nephrol. 2008, 19, 1891–1903. [Google Scholar] [CrossRef] [PubMed]
- Yoshihama, Y.; Sasaki, K.; Horikoshi, Y.; Suzuki, A.; Ohtsuka, T.; Hakuno, F.; Takahashi, S.; Ohno, S.; Chida, K. KIBRA suppresses apical exocytosis through inhibition of aPKC kinase activity in epithelial cells. Curr. Biol. 2011, 21, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Traer, C.J.; Rutherford, A.C.; Palmer, K.J.; Wassmer, T.; Oakley, J.; Attar, N.; Carlton, J.G.; Kremerskothen, J.; Stephens, D.J.; Cullen, P.J. SNX4 coordinates endosomal sorting of TfnR with dynein-mediated transport into the endocytic recycling compartment. Nat. Cell Biol. 2007, 9, 1370–1380. [Google Scholar] [CrossRef] [PubMed]
- Kremerskothen, J.; Plaas, C.; Büther, K.; Finger, I.; Veltel, S.; Matanis, T.; Liedtke, T.; Barnekow, A. Characterization of KIBRA, a novel WW domain-containing protein. Biochem. Biophys. Res. Commun. 2003, 300, 862–867. [Google Scholar] [CrossRef]
- Wennmann, D.O.; Schmitz, J.; Wehr, M.C.; Krahn, M.P.; Koschmal, N.; Gromnitza, S.; Schulze, U.; Weide, T.; Chekuri, A.; Skryabin, B.V.; et al. Evolutionary and molecular facts link the WWC protein family to Hippo signaling. Mol. Biol. Evol. 2014, 31, 1710–1723. [Google Scholar] [CrossRef]
- Duning, K.; Wennmann, D.O.; Bokemeyer, A.; Reissner, C.; Wersching, H.; Thomas, C.; Buschert, J.; Guske, K.; Franzke, V.; Flöel, A.; et al. Common exonic missense variants in the C2 domain of the human KIBRA protein modify lipid binding and cognitive performance. Transl. Psychiatry 2013, 3, e272. [Google Scholar] [CrossRef]
- Hermann, A.; Wennmann, D.O.; Gromnitza, S.; Edeling, M.; Van Marck, V.; Sudol, M.; Schaefer, L.; Duning, K.; Weide, T.; Pavenstädt, H.; et al. WW and C2 domain-containing proteins regulate hepatic cell differentiation and tumorigenesis through the hippo signaling pathway. Hepatology 2018, 67, 1546–1559. [Google Scholar] [CrossRef]
- Baumgartner, R.; Poernbacher, I.; Buser, N.; Hafen, E.; Stocker, H. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev. Cell 2010, 18, 309–316. [Google Scholar] [CrossRef]
- Genevet, A.; Wehr, M.C.; Brain, R.; Thompson, B.J.; Tapon, N. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev. Cell 2010, 18, 300–308. [Google Scholar] [CrossRef]
- Yu, J.; Zheng, Y.; Dong, J.; Klusza, S.; Deng, W.M.; Pan, D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev. Cell 2010, 18, 288–299. [Google Scholar] [CrossRef]
- Xiao, L.; Chen, Y.; Ji, M.; Dong, J. KIBRA regulates Hippo signaling activity via interactions with large tumor suppressor kinases. J. Biol. Chem. 2011, 286, 7788–7796. [Google Scholar] [CrossRef] [PubMed]
- Hilton, H.N.; Stanford, P.M.; Harris, J.; Oakes, S.R.; Kaplan, W.; Daly, R.J.; Ormandy, C.J. KIBRA interacts with discoidin domain receptor 1 to modulate collagen-induced signalling. Biochim. Biophys. Acta 2008, 1783, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Mussell, A.L.; Denson, K.E.; Shen, H.; Chen, Y.; Yang, N.; Frangou, C.; Zhang, J. Loss of KIBRA function activates EGFR signaling by inducing AREG. Oncotarget 2018, 9, 29975–29984. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tang, P. Genomic Pathology and Biomarkers in Breast Cancer. Crit. Rev. Oncog. 2017, 22, 411–426. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Xiang, L.; Li, T.; Bai, Z. Cancer Hallmarks, Biomarkers and Breast Cancer Molecular Subtypes. J. Cancer 2016, 7, 1281–1294. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Katsaros, D.; Biglia, N.; Shen, Y.; Fu, Y.; Tiirikainen, M.; Yu, H. Low expression of WWC1, a tumor suppressor gene, is associated with aggressive breast cancer and poor survival outcome. FEBS Open Bio 2019, 9, 1270–1280. [Google Scholar] [CrossRef]
- Rayala, S.K.; den Hollander, P.; Manavathi, B.; Talukder, A.H.; Song, C.; Peng, S.; Barnekow, A.; Kremerskothen, J.; Kumar, R. Essential role of KIBRA in co-activator function of dynein light chain 1 in mammalian cells. J. Biol. Chem. 2006, 281, 19092–19099. [Google Scholar] [CrossRef]
- Kumar, P.; Aggarwal, R. An overview of triple-negative breast cancer. Arch. Gynecol. Obstet. 2016, 293, 247–269. [Google Scholar] [CrossRef]
- Knight, J.F.; Sung, V.Y.C.; Kuzmin, E.; Couzens, A.L.; de Verteuil, D.A.; Ratcliffe, C.D.H.; Coelho, P.P.; Johnson, R.M.; Samavarchi-Tehrani, P.; Gruosso, T.; et al. KIBRA (WWC1) Is a Metastasis Suppressor Gene Affected by Chromosome 5q Loss in Triple-Negative Breast Cancer. Cell Rep. 2018, 22, 3191–3205. [Google Scholar] [CrossRef]
- Schelleckes, K.; Schmitz, B.; Ciarimboli, G.; Lenders, M.; Pavenstädt, H.J.; Herrmann, E.; Brand, S.M.; Brand, E. Promoter methylation inhibits expression of tumor suppressor KIBRA in human clear cell renal cell carcinoma. Clin. Epigenetics 2017, 9, 109. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, H.; Sun, H.; Chen, J.; Huang, D.; Han, X.; Ren, X.; Lin, S.; Fan, Q.; Tian, W.; et al. Association of peripheral blood leukocyte KIBRA methylation with gastric cancer risk: A case-control study. Cancer Med. 2018, 7, 2682–2690. [Google Scholar] [CrossRef] [PubMed]
- Yoshihama, Y.; Izumisawa, Y.; Akimoto, K.; Satoh, Y.; Mizushima, T.; Satoh, K.; Chida, K.; Takagawa, R.; Akiyama, H.; Ichikawa, Y.; et al. High expression of KIBRA in low atypical protein kinase C-expressing gastric cancer correlates with lymphatic invasion and poor prognosis. Cancer Sci. 2013, 104, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Wang, X.; Ma, L.; Guo, Z.; Liu, H.; Du, M.; Chu, H.; Wang, M.; Wang, Z.; Zhang, Z. Genetic variations in Hippo pathway genes influence bladder cancer risk in a Chinese population. Arch. Toxicol. 2020, 94, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Du, W.B.; Huang, Z.; Luo, L.; Tong, S.P.; Li, H.Q.; Li, X.; Tong, J.H.; Yao, Y.L.; Zhang, W.B.; Meng, Y. TCF19 aggravates the malignant progression of colorectal cancer by negatively regulating WWC1. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 655–663. [Google Scholar] [PubMed]
- Kim, S.C.; Shin, R.; Seo, H.Y.; Kim, M.; Park, J.W.; Jeong, S.Y.; Ku, J.L. Identification of a Novel Fusion Gene, FAM174A-WWC1, in Early-Onset Colorectal Cancer: Establishment and Characterization of Four Human Cancer Cell Lines from Early-Onset Colorectal Cancers. Transl. Oncol. 2019, 9, 1185–1195. [Google Scholar] [CrossRef]
- Basu-Roy, U.; Bayin, N.S.; Rattanakorn, K.; Han, E.; Placantonakis, D.G.; Mansukhani, A.; Basilico, C. Sox2 antagonizes the Hippo pathway to maintain stemness in cancer cells. Nat. Commun. 2015, 6, 6411. [Google Scholar] [CrossRef]
- Hill, V.K.; Dunwell, T.L.; Catchpoole, D.; Krex, D.; Brini, A.T.; Griffiths, M.; Craddock, C.; Maher, E.R.; Latif, F. Frequent epigenetic inactivation of KIBRA, an upstream member of the Salvador/Warts/Hippo (SWH) tumor suppressor network, is associated with specific genetic event in B-cell acute lymphocytic leukemia. Epigenetics 2011, 6, 326–332. [Google Scholar] [CrossRef]
- Shinawi, T.; Hill, V.; Dagklis, A.; Baliakas, P.; Stamatopoulos, K.; Agathanggelou, A.; Stankovic, T.; Maher, E.R.; Ghia, P.; Latif, F. KIBRA gene methylation is associated with unfavorable biological prognostic parameters in chronic lymphocytic leukemia. Epigenetics 2012, 7, 211–215. [Google Scholar] [CrossRef]
- Donato, E.; Biagioni, F.; Bisso, A.; Caganova, M.; Amati, B.; Campaner, S. YAP and TAZ are dispensable for physiological and malignant haematopoiesis. Leukemia 2018, 32, 2037–2040. [Google Scholar] [CrossRef]
- An, Y.; Zhang, Q.; Li, X.; Wang, Z.; Li, Y.; Tang, X. Upregulated microRNA miR-21 promotes the progression of lung adenocarcinoma through inhibition of KIBRA and the Hippo signaling pathway. Biomed. Pharmacother. 2018, 108, 1845–1855. [Google Scholar] [CrossRef]
- Stauffer, S.; Chen, X.; Zhang, L.; Chen, Y.; Dong, J. KIBRA promotes prostate cancer cell proliferation and motility. FEBS J. 2016, 283, 1800–1811. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.J.; Xue, W.; Peng, J.; Wang, Y.; Wie, L.; Yang, Z.; Zhu, H.H.; Fang, Y.X.; Gao, W.Q. Elevated expression of Par3 promotes prostate cancer metastasis by forming a Par3/aPKC/KIBRA complex and inactivating the hippo pathway. J. Exp. Clin. Cancer Res. 2017, 36, 139. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yan, S.; Chen, J.; Gan, C.; Chen, D.; Li, Y.; Wen, J.; Kremerskothen, J.; Chen, S.; Zhang, J.; et al. WWC2 is an independent prognostic factor and prevents invasion via Hippo signalling in hepatocellular carcinoma. J. Cell Mol. Med. 2017, 21, 3718–3729. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yin, W.; Liu, H. MicroRNA-10a promotes epithelial-to-mesenchymal transition and stemness maintenance of pancreatic cancer stem cells via upregulating the Hippo signaling pathway through WWC2 inhibition. J. Cell Biochem. 2020, 121, 4505–4521. [Google Scholar] [CrossRef]
- Wang, G.; Zhou, Y.; Chen, W.; Yang, Y.; Ye, J.; Ou, H.; Wu, H. miR-21-5p promotes lung adenocarcinoma cell proliferation, migration and invasion via targeting WWC2. Cancer Biomark. 2020, 28, 549–559. [Google Scholar] [CrossRef]
- Yuan, B.; Yang, J.; Gu, H.; Ma, C. Down-Regulation of LINC00460 Represses Metastasis of Colorectal Cancer via WWC2. Dig. Dis. Sci. 2020, 65, 442–456. [Google Scholar] [CrossRef]
- Frassanito, M.A.; Desantis, V.; Di Marzo, L.; Craparotta, I.; Beltrame, L.; Marchini, S.; Annese, T.; Visino, F.; Arciuli, M.; Saltarella, I.; et al. Bone marrow fibroblasts overexpress miR-27b and miR-214 in step with multiple myeloma progression, dependent on tumour cell-derived exosomes. J. Pathol. 2019, 247, 241–253. [Google Scholar] [CrossRef]
- Han, Q.; Lin, X.; Zhang, X.; Jiang, G.; Zhang, Y.; Miao, Y.; Rong, X.; Zheng, X.; Han, Y.; Han, X.; et al. WWC3 regulates the Wnt and Hippo pathways via Dishevelled proteins and large tumour suppressor 1, to suppress lung cancer invasion and metastasis. J. Pathol. 2017, 242, 435–447. [Google Scholar] [CrossRef]
- Han, Q.; Kremerskothen, J.; Lin, X.; Zhang, X.; Rong, X.; Zhang, D.; Wang, E. WWC3 inhibits epithelial-mesenchymal transition of lung cancer by activating Hippo-YAP signaling. OncoTargets Ther. 2018, 11, 2581–2591. [Google Scholar] [CrossRef]
- Han, Q.; Rong, X.; Wang, E.; Liu, S. WW and C2 domain-containing protein-3 promoted EBSS-induced apoptosis through inhibiting autophagy in non-small cell lung cancer cells. J. Thorac Dis. 2020, 12, 4205–4215. [Google Scholar] [CrossRef]
- Rong, X.; Han, Q.; Lin, X.; Kremerskothen, J.; Wang, E. FRMPD1 activates the Hippo pathway via interaction with WWC3 to suppress the proliferation and invasiveness of lung cancer cells. Cancer Manag. Res. 2019, 11, 3395–3410. [Google Scholar] [CrossRef] [PubMed]
- Klimowski, L.K.; Garcia, B.A.; Shabanowitz, J.; Hunt, D.F.; Virshup, D.M. Site-specific casein kinase 1epsilon-dependent phosphorylation of Dishevelled modulates beta-catenin signaling. FEBS J. 2006, 273, 4594–4602. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jiang, M.; Yao, Y.; Cai, Z. WWC3 Inhibits Glioma Cell Proliferation Through Suppressing the Wnt/β-Catenin Signaling Pathway. DNA Cell Biol. 2018, 37, 31–37. [Google Scholar] [CrossRef]
- Polakis, P. The many ways of Wnt in cancer. Curr. Opin. Genet. Dev. 2007, 17, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Liu, X.; Zheng, J.; Xue, Y.; Liu, L.; Ma, J.; Wang, P.; Yang, C.; Wang, D.; Shao, L.; et al. Interaction of BACH2 with FUS promotes malignant progression of glioma cells via the TSLNC8-miR-10b-5p-WWC3 pathway. Mol. Oncol. 2020, 14, 2936–2959. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Zhou, J. WWC3 downregulation correlates with poor prognosis and inhibition of Hippo signaling in human gastric cancer. OncoTargets Ther. 2017, 10, 2931–2942. [Google Scholar] [CrossRef]
- Stammnitz, M.R.; Coorens, T.H.H.; Gori, K.C.; Hayes, D.; Fu, B.; Wang, J.; Martin-Herranz, D.E.; Alexandrov, L.B.; Baez-Ortega, A.; Barthorpe, S.; et al. The Origins and Vulnerabilities of Two Transmissible Cancers in Tasmanian Devils. Cancer Cell 2018, 33, 607–619. [Google Scholar] [CrossRef]
- Meng, L.; Liu, S.; Liu, F.; Sang, M.; Ju, Y.; Fan, X.; Gu, L.; Li, Z.; Geng, C.; Sang, M. ZEB1-Mediated Transcriptional Upregulation of circWWC3 Promotes Breast Cancer Progression through Activating Ras Signaling Pathway. Mol. Ther. Nucleic Acids 2020, 22, 124–137. [Google Scholar] [CrossRef]
- Moleirinho, S.; Chang, N.; Sims, A.H.; Tilston-Lünel, A.M.; Angus, L.; Steele, A.; Boswell, V.; Barnett, S.C.; Ormandy, C.; Faratian, D.; et al. KIBRA exhibits MST-independent functional regulation of the Hippo signaling pathway in mammals. Oncogene 2013, 32, 1821–1830. [Google Scholar] [CrossRef]
- Moleirinho, S.; Guerrant, W.; Kissil, J.L. The Angiomotins-from discovery to function. FEBS Lett. 2014, 588, 2693–2703. [Google Scholar] [CrossRef]
- Mana-Capelli, S.; McCollum, D. Angiomotins stimulate LATS kinase autophosphorylation and act as scaffolds that promote Hippo signaling. J. Biol. Chem. 2018, 293, 18230–18241. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Kugler, J.M.; Cohen, S.M. DUB3 Deubiquitylating Enzymes Regulate Hippo Pathway Activity by Regulating the Stability of ITCH, LATS and AMOT Proteins. PLoS ONE 2017, 12, e0169587. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Shen, Y.; Yang, J.; Li, S.; Wang, B.; Chen, Z.; Li, P.; Liu, P.; Yang, J. Angiomotin Family Members: Oncogenes or Tumor Suppressors? Int. J. Biol. Sci. 2017, 3, 772–781. [Google Scholar] [CrossRef] [PubMed]
- Poernbacher, I.; Baumgartner, R.; Marada, S.K.; Edwards, K.; Stocker, H. Drosophila Pez acts in Hippo signaling to restrict intestinal stem cell proliferation. Curr. Biol. 2012, 22, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Michaloglou, C.; Lehmann, W.; Martin, T.; Delaunay, C.; Hueber, A.; Barys, L.; Niu, H.; Billy, E.; Wartmann, M.; Ito, M.; et al. The tyrosine phosphatase PTPN14 is a negative regulator of YAP activity. PLoS ONE 2013, 8, e61916. [Google Scholar] [CrossRef]
- Wilson, K.E.; Yang, N.; Mussell, A.L.; Zhang, J. The Regulatory Role of KIBRA and PTPN14 in Hippo Signaling and Beyond. Genes 2016, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Büther, K.; Plaas, C.; Barnekow, A.; Kremerskothen, J. KIBRA is a novel substrate for protein kinase Czeta. Biochem. Biophys. Res. Commun. 2004, 317, 703–707. [Google Scholar] [CrossRef]
- Reina-Campos, M.; Diaz-Meco, M.T.; Moscat, J. The Dual Roles of the Atypical Protein Kinase Cs in Cancer. Cancer Cell 2019, 36, 218–235. [Google Scholar] [CrossRef]
- Vogt-Eisele, A.; Krüger, C.; Duning, K.; Weber, D.; Spoelgen, R.; Pitzer, C.; Plaas, C.; Eisenhardt, G.; Meyer, A.; Vogt, G.; et al. KIBRA (KIdney/BRAin protein) regulates learning and memory and stabilizes Protein kinase Mζ. J. Neurochem. 2014, 128, 686–700. [Google Scholar] [CrossRef]
- Su, T.; Ludwig, M.Z.; Xu, J.; Fehon, R.G. Kibra and Merlin Activate the Hippo Pathway Spatially Distinct from and Independent of Expanded. Dev. Cell 2017, 40, 478–490.e3. [Google Scholar] [CrossRef]
- Mao, X.; Li, P.; Wang, Y.; Liang, Z.; Liu, J.; Li, J.; Jiang, Y.; Bao, G.; Li, L.; Zhu, B.; et al. CRB3 regulates contact inhibition by activating the Hippo pathway in mammary epithelial cells. Cell Death Dis. 2017, 8, e2546. [Google Scholar] [CrossRef] [PubMed]
- Snigdha, K.; Gangwani, K.S.; Lapalikar, G.V.; Singh, A.; Kango-Singh, M. Hippo Signaling in Cancer: Lessons from Drosophila models. Front. Cell Dev. Biol. 2019, 7, 85. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Iyer, J.; Chowdhury, A.; Ji, M.; Xiao, L.; Yang, S.; Chen, Y.; Tsai, M.Y.; Dong, J. KIBRA regulates aurora kinase activity and is required for precise chromosome alignment during mitosis. J. Biol. Chem. 2012, 287, 34069–34077. [Google Scholar] [CrossRef] [PubMed]
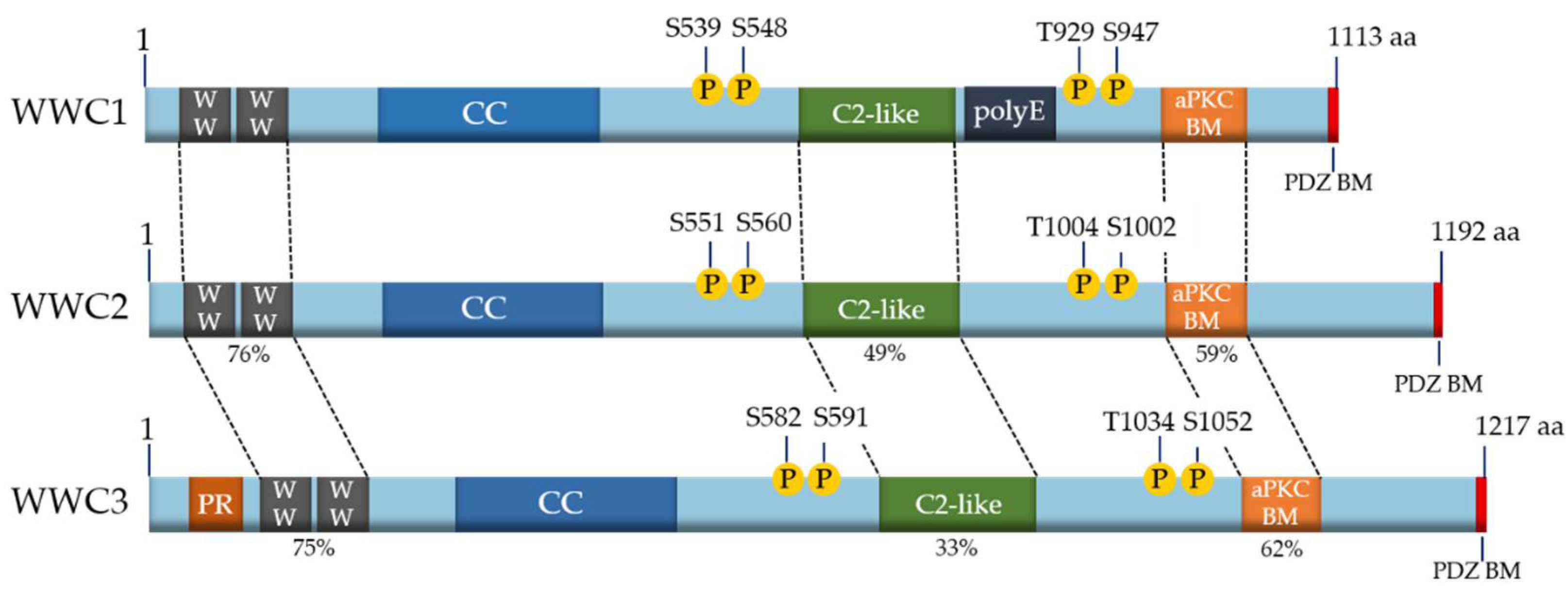
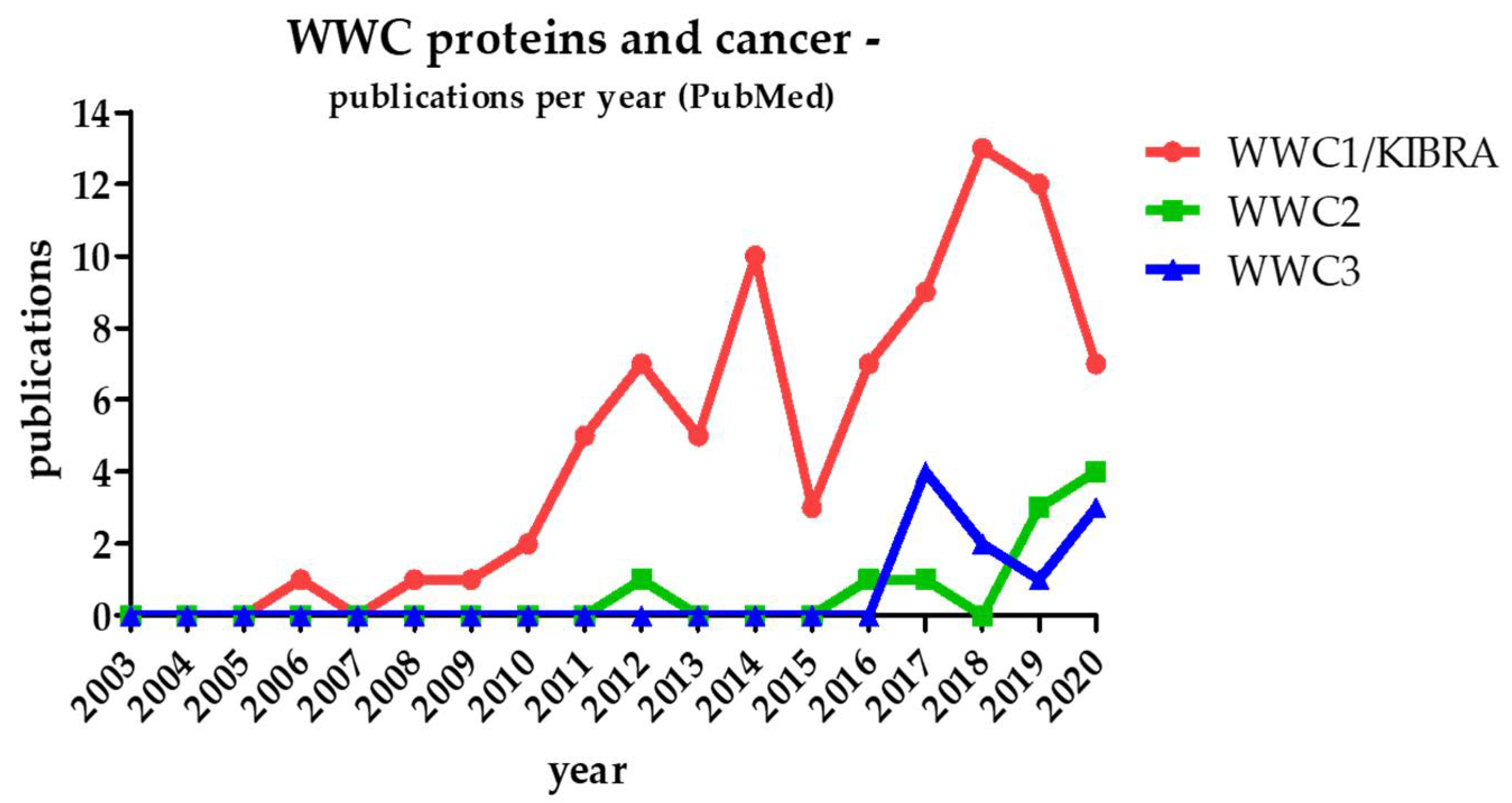
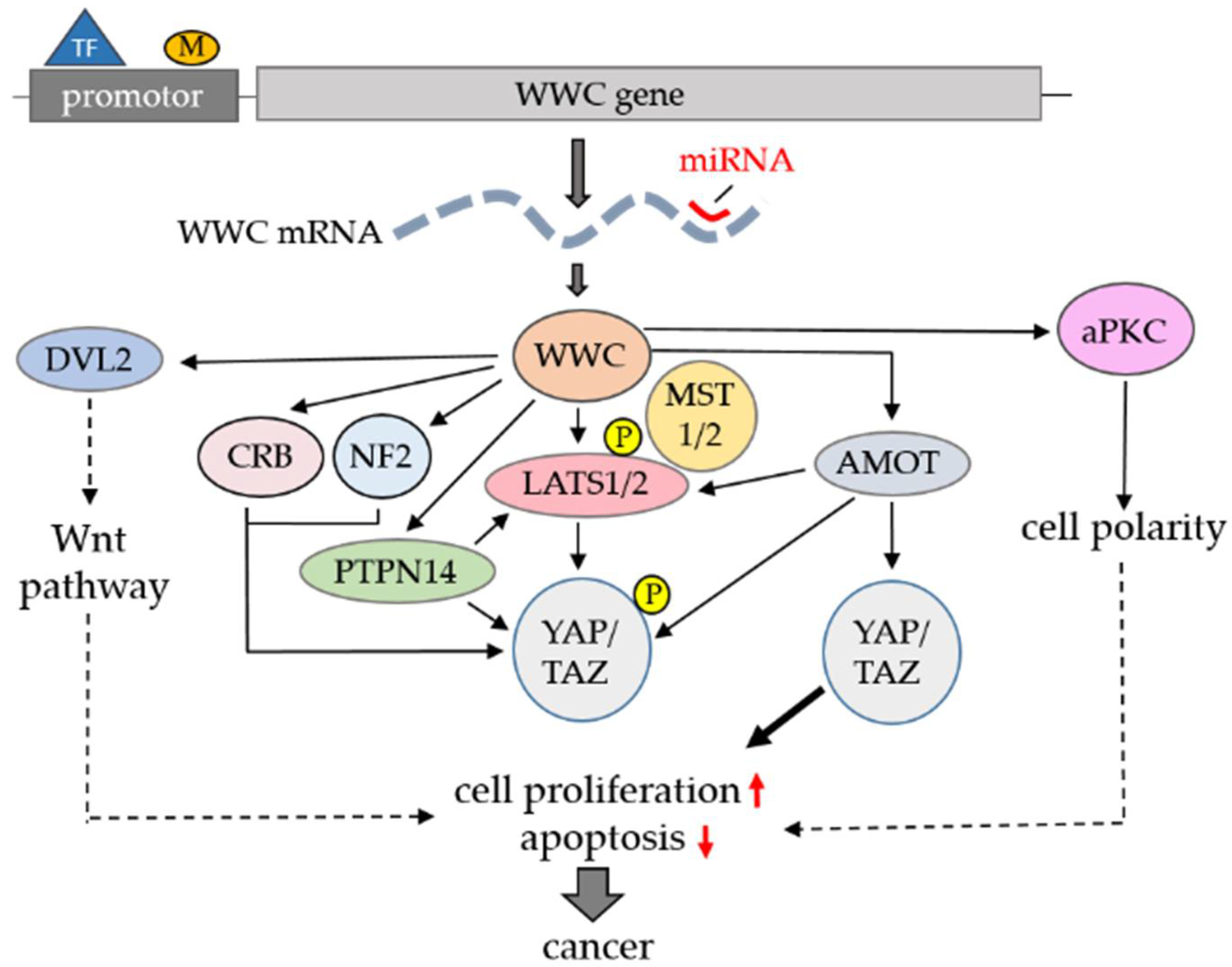
| Binding Partner | WW Binding Domain | Effect on Hippo Pathway | Link to Cancer [Reference] |
|---|---|---|---|
| LATS1/2 (WWC1/2/3) | WW domains | Activation | HCC [17] |
| AMOT (WWC1/2) | WW domains | Activation | HCC [17] |
| PTPN14 (WWC1) | WW domains | Activation | TNBC [66] |
| FRMPD1 (WWC3) | PDZ binding motif | Activation | NSCLC [51] |
| DVL2 (WWC3) | WW domains | Activation | NSCLC [48] |
| DDR1 (WWC1) | WW domains | Unknown | BC [22] |
| aPKC (WWC1) | aPKC binding motif | Inhibition | BC [22], GC [31] |
| WWC Family Member | Expression Level | Link to Cancer [Reference] |
|---|---|---|
| WWC1 | Low | BC [22,26,29] |
| WWC1 | High | BC [27] |
| WWC1 | Low | ccRCC [30] |
| WWC1 | High | GC [31,32] |
| WWC1 | High | BLC [33] |
| WWC1 | Low | CRC [34] |
| WWC1 | Low | OS [36] |
| WWC1 | Low | ALL [37] |
| WWC1 | Low | CLL [38] |
| WWC1 | Low | LUAD [40] |
| WWC1 | High | PC [41,42] |
| WWC2 | Low | HCC [17,43] |
| WWC2 | Low | PAC [44] |
| WWC2 | Low | LUAD [45] |
| WWC2 | Low | CRC [46] |
| WWC3 | Low | NSCLC [48,49] |
| WWC3 | Low | Glioma [53,55] |
| WWC3 | Low | GC [56] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Höffken, V.; Hermann, A.; Pavenstädt, H.; Kremerskothen, J. WWC Proteins: Important Regulators of Hippo Signaling in Cancer. Cancers 2021, 13, 306. https://doi.org/10.3390/cancers13020306
Höffken V, Hermann A, Pavenstädt H, Kremerskothen J. WWC Proteins: Important Regulators of Hippo Signaling in Cancer. Cancers. 2021; 13(2):306. https://doi.org/10.3390/cancers13020306
Chicago/Turabian StyleHöffken, Verena, Anke Hermann, Hermann Pavenstädt, and Joachim Kremerskothen. 2021. "WWC Proteins: Important Regulators of Hippo Signaling in Cancer" Cancers 13, no. 2: 306. https://doi.org/10.3390/cancers13020306
APA StyleHöffken, V., Hermann, A., Pavenstädt, H., & Kremerskothen, J. (2021). WWC Proteins: Important Regulators of Hippo Signaling in Cancer. Cancers, 13(2), 306. https://doi.org/10.3390/cancers13020306







