Resistance to Tyrosine Kinase Inhibitors in Chronic Myeloid Leukemia—From Molecular Mechanisms to Clinical Relevance
Abstract
Simple Summary
Abstract
1. Introduction
2. Molecular Mechanisms
2.1. BCR-ABL1 Mutational Landscape and Overexpression
2.2. DNA Damage Repair and Genomic Instability
2.3. Drug Transporters
2.4. Alternative Signaling Pathways
2.5. Stem Cell Metabolism and Pathways
2.6. Epigenetic Alterations
2.7. CML Microenvironment and Immunological Status
3. Methodologies to Access TKI Resistance
3.1. Molecular Approaches
3.2. Bioinformatics and Artificial Intelligence as Methodologies to Decipher Mechanisms of Action or Resistance to TKIs in Leukemia
4. Therapeutic Approaches against Resistance
4.1. BCR-ABL1 Targeted Therapies
4.2. Non-BCR-ABL1 Targeted Therapies
4.3. Epigenetic Modulators
4.4. Immunotherapies
4.5. Allogenic Stem Cell Transplantation
5. Patient’s Adhesion Impact on Resistant Process
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Baykal-Köse, S.; Acikgoz, E.; Yavuz, A.S.; Gönül Geyik, Ö.; Ateş, H.; Sezerman, O.U.; Özsan, G.H.; Yüce, Z. Adaptive phenotypic modulations lead to therapy resistance in chronic myeloid leukemia cells. PLoS ONE 2020, 15, e0229104. [Google Scholar] [CrossRef]
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2020 update on diagnosis, therapy and monitoring. Am. J. Hematol. 2020, 95, 691–709. [Google Scholar] [CrossRef]
- Surveillance, Epidemiology, and End Results (SEER) Program SEER*Stat Database: Incidence-SEER Research Data, 9 Registries, Nov 2020 Sub (1975-2018)-Linked To County Attributes-Time Dependent (1990–2018) Income/Rurality, 1969–2019 Counties; DCCPS, Surveillance Research Program; National Cancer Institute: Rockville, MD, USA. Available online: www.seer.cancer.gov (accessed on 25 September 2021).
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2016 update on diagnosis, therapy, and monitoring. Am. J. Hematol. 2016, 91, 252–265. [Google Scholar] [CrossRef]
- Soverini, S.; Mancini, M.; Bavaro, L.; Cavo, M.; Martinelli, G. Chronic myeloid leukemia: The paradigm of targeting oncogenic tyrosine kinase signaling and counteracting resistance for successful cancer therapy. Mol. Cancer 2018, 17, 49. [Google Scholar] [CrossRef] [PubMed]
- Heisterkamp, N.; Stephenson, J.R.; Groffen, J.; Hansen, P.F.; de Klein, A.; Bartram, C.R.; Grosveld, G. Localization of the c-abl oncogene adjacent to a translocation break point in chronic myelocytic leukaemia. Nature 1983, 306, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.-J.; Liu, Y.-F.; Xu, L.-Z.; Long, Z.-J.; Huang, D.; Yang, Y.; Liu, B.; Feng, J.-X.; Pan, Y.-J.; Yan, J.-S.; et al. The Philadelphia chromosome in leukemogenesis. Chin. J. Cancer 2016, 35, 48. [Google Scholar] [CrossRef] [PubMed]
- Laurent, E.; Talpaz, M.; Kantarjian, H.; Kurzrock, R. The BCR Gene and Philadelphia Chromosome-positive Leukemogenesis. Cancer Res. 2001, 61, 2343–2355. [Google Scholar] [PubMed]
- Reff, M.J.; Shillingburg, A.; Shah, B.; Elder, C.; Prescott, H.; Kennerly-Shah, J. Front-line use of tyrosine kinase inhibitors in chronic phase chronic myeloid leukemia: Practice considerations. J. Oncol. Pharm. Pract. 2019, 6, 156–174. [Google Scholar] [CrossRef]
- Savage, D.G.; Antman, K.H. Imatinib Mesylate—A New Oral Targeted Therapy. N. Engl. J. Med. 2002, 346, 683–693. [Google Scholar] [CrossRef]
- Manley, P.W.; Cowan-Jacob, S.W.; Buchdunger, E.; Fabbro, D.; Fendrich, G.; Furet, P.; Meyer, T.; Zimmermann, J. Imatinib: A selective tyrosine kinase inhibitor. Eur. J. Cancer 2002, 38, S19–S27. [Google Scholar] [CrossRef]
- Kantarjian, H.M.; Shah, N.P.; Cortes, J.E.; Baccarani, M.; Agarwal, M.B.; Undurraga, M.S.; Wang, J.; Kassack Ipiña, J.J.; Kim, D.-W.; Ogura, M.; et al. Dasatinib or imatinib in newly diagnosed chronic-phase chronic myeloid leukemia: 2-year follow-up from a randomized phase 3 trial (DASISION). Blood 2012, 119, 1123–1129. [Google Scholar] [CrossRef] [PubMed]
- Soverini, S.; Branford, S.; Nicolini, F.E.; Talpaz, M.; Deininger, M.W.N.; Martinelli, G.; Müller, M.C.; Radich, J.P.; Shah, N.P. Implications of BCR-ABL1 kinase domain-mediated resistance in chronic myeloid leukemia. Leuk. Res. 2014, 38, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Morozova, E.V.; Vlasova, Y.Y.; Pryanishnikova, M.V.; Lepik, K.V.; Afanasyev, B.V. Efficacy of Dasatinib in a CML Patient in Blast Crisis with F317L Mutation: A Case Report and Literature Review. Biomark. Insights 2015, 10, 43–47. [Google Scholar] [CrossRef] [PubMed]
- Baccarani, M.; Deininger, M.W.; Rosti, G.; Hochhaus, A.; Soverini, S.; Apperley, J.F.; Cervantes, F.; Clark, R.E.; Cortes, J.E.; Guilhot, F.; et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood 2013, 122, 872–884. [Google Scholar] [CrossRef]
- Eliasson, L.; Clifford, S.; Barber, N.; Marin, D. Exploring chronic myeloid leukemia patients’ reasons for not adhering to the oral anticancer drug imatinib as prescribed. Leuk. Res. 2011, 35, 626–630. [Google Scholar] [CrossRef]
- Andrade, A.R.; Leitão, D.D.S.; Paz, I.P.; Evangelista, T.R.; Mello, V.J.D.; Hamoy, M. Analysis of imatinib adherence in chronic myeloid leukemia: A retrospective study in a referral hospital in the Brazilian Amazon. Hematol. Transfus. Cell Ther. 2019, 41, 106–113. [Google Scholar] [CrossRef]
- Yaghmaie, M.; Yeung, C.C.S. Molecular Mechanisms of Resistance to Tyrosine Kinase Inhibitors. Curr. Hematol. Malig. Rep. 2019, 14, 395–404. [Google Scholar] [CrossRef]
- Banjar, H.; Adelson, D.; Brown, F.; Chaudhri, N. Intelligent Techniques Using Molecular Data Analysis in Leukaemia: An Opportunity for Personalized Medicine Support System. BioMed Res. Int. 2017, 2017, 3587309. [Google Scholar] [CrossRef]
- Sun, X.; Hu, B. Mathematical modeling and computational prediction of cancer drug resistance. Brief. Bioinform. 2017, 19, 1382–1399. [Google Scholar] [CrossRef]
- Apperley, J.F. Part I: Mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007, 8, 1018–1029. [Google Scholar] [CrossRef]
- Zhao, H.; Deininger, M.W. Declaration of Bcr-Abl1 independence. Leukemia 2020, 34, 2827–2836. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.B.; O’Hare, T.; Deininger, M.W. Mechanisms of Resistance to ABL Kinase Inhibition in Chronic Myeloid Leukemia and the Development of Next Generation ABL Kinase Inhibitors. Hematol. Oncol. Clin. N. Am. 2017, 31, 589–612. [Google Scholar] [CrossRef] [PubMed]
- Kaehler, M.; Cascorbi, I. Pharmacogenomics of Impaired Tyrosine Kinase Inhibitor Response: Lessons Learned From Chronic Myelogenous Leukemia. Front. Pharmacol. 2021, 12, 696960. [Google Scholar] [CrossRef] [PubMed]
- Deininger, M.W.; Shah, N.P.; Altman, J.K.; Berman, E.; Bhatia, R.; Bhatnagar, B.; DeAngelo, D.J.; Gotlib, J.; Hobbs, G.; Maness, L.; et al. Chronic Myeloid Leukemia, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2020, 18, 1385–1415. [Google Scholar] [CrossRef] [PubMed]
- Balabanov, S.; Braig, M.; Brümmendorf, T.H. Current aspects in resistance against tyrosine kinase inhibitors in chronic myelogenous leukemia. Drug Discov. Today Technol. 2014, 11, 89–99. [Google Scholar] [CrossRef]
- Meenakshi Sundaram, D.N.; Jiang, X.; Brandwein, J.M.; Valencia-Serna, J.; Remant, K.C.; Uludağ, H. Current outlook on drug resistance in chronic myeloid leukemia (CML) and potential therapeutic options. Drug Discov. Today 2019, 24, 1355–1369. [Google Scholar] [CrossRef]
- Vaidya, S.; Vundinti, B.R.; Shanmukhaiah, C.; Chakrabarti, P.; Ghosh, K. Evolution of BCR/ABL gene mutation in CML is time dependent and dependent on the pressure exerted by tyrosine kinase inhibitor. PLoS ONE 2015, 10, e0114828. [Google Scholar] [CrossRef]
- Diamond, J.M.; Melo, J.V. Mechanisms of resistance to BCR–ABL kinase inhibitors. Leuk. Lymphoma 2011, 52, 12–22. [Google Scholar] [CrossRef]
- Gorre, M.E.; Mohammed, M.; Ellwood, K.; Hsu, N.; Paquette, R.; Rao, P.N.; Sawyers, C.L. Clinical Resistance to STI-571 Cancer Therapy Caused by BCR-ABL Gene Mutation or Amplification. Science 2001, 293, 876–880. [Google Scholar] [CrossRef]
- Tan, F.H.; Putoczki, T.L.; Stylli, S.S.; Luwor, R.B. Ponatinib: A novel multi-tyrosine kinase inhibitor against human malignancies. Onco Targets 2019, 12, 635–645. [Google Scholar] [CrossRef]
- Braun, T.; Eide, C.; Druker, B. Response and Resistance to BCR-ABL1-Targeted Therapies. Cancer Cell 2020, 37, 530–542. [Google Scholar] [CrossRef]
- Cang, S.; Liu, D. P-loop mutations and novel therapeutic approaches for imatinib failures in chronic myeloid leukemia. J. Hematol. Oncol. 2008, 1, 15. [Google Scholar] [CrossRef]
- Redaelli, S.; Mologni, L.; Rostagno, R.; Piazza, R.; Magistroni, V.; Ceccon, M.; Viltadi, M.; Flynn, D.; Gambacorti-Passerini, C. Three novel patient-derived BCR/ABL mutants show different sensitivity to second and third generation tyrosine kinase inhibitors. Am. J. Hematol. 2012, 87, E125–E128. [Google Scholar] [CrossRef]
- Soverini, S.; Hochhaus, A.; Nicolini, F.E.; Gruber, F.; Lange, T.; Saglio, G.; Pane, F.; Müller, M.C.; Ernst, T.; Rosti, G.; et al. BCR-ABL kinase domain mutation analysis in chronic myeloid leukemia patients treated with tyrosine kinase inhibitors: Recommendations from an expert panel on behalf of European LeukemiaNet. Blood 2011, 118, 1208–1215. [Google Scholar] [CrossRef]
- Hochhaus, A.; Saglio, G.; Larson, R.A.; Kim, D.-W.; Etienne, G.; Rosti, G.; De Souza, C.; Kurokawa, M.; Kalaycio, M.E.; Hoenekopp, A.; et al. Nilotinib is associated with a reduced incidence of BCR-ABL mutations vs imatinib in patients with newly diagnosed chronic myeloid leukemia in chronic phase. Blood 2013, 121, 3703–3708. [Google Scholar] [CrossRef]
- Eide, C.A.; Zabriskie, M.S.; Savage Stevens, S.L.; Antelope, O.; Vellore, N.A.; Than, H.; Schultz, A.R.; Clair, P.; Bowler, A.D.; Pomicter, A.D.; et al. Combining the Allosteric Inhibitor Asciminib with Ponatinib Suppresses Emergence of and Restores Efficacy against Highly Resistant BCR-ABL1 Mutants. Cancer Cell 2019, 36, 431–443.e435. [Google Scholar] [CrossRef]
- Hochhaus, A.; Saussele, S.; Rosti, G.; Mahon, F.X.; Janssen, J.J.W.M.; Hjorth-Hansen, H.; Richter, J.; Buske, C. Chronic myeloid leukaemia: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2017, 28, 41–51. [Google Scholar] [CrossRef]
- Soverini, S.; Bavaro, L.; De Benedittis, C.; Martelli, M.; Iurlo, A.; Orofino, N.; Sica, S.; Sorà, F.; Lunghi, F.; Ciceri, F.; et al. Prospective assessment of NGS-detectable mutations in CML patients with nonoptimal response: The NEXT-in-CML study. Blood 2020, 135, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Radich, J. Structure, Function, and Resistance in Chronic Myeloid Leukemia. Cancer Cell 2014, 26, 305–306. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zabriskie, M.S.; Eide, C.A.; Tantravahi, S.K.; Vellore, N.A.; Estrada, J.; Nicolini, F.E.; Khoury, H.J.; Larson, R.A.; Konopleva, M.; Cortes, J.E.; et al. BCR-ABL1 Compound Mutations Combining Key Kinase Domain Positions Confer Clinical Resistance to Ponatinib in Ph Chromosome-Positive Leukemia. Cancer Cell 2014, 26, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Sigl, M.; Spoerl, S.; Schnittger, S.; Meissner, J.; Rummelt, C.; Peschel, C.; Duyster, J.; Ho, A.D.; von Bubnoff, N. Imatinib failure and response to dasatinib in a patient with chronic myeloid leukemia in blast crisis and a novel, nine-nucleotide BCR-ABL insertion mutation. Blood Cancer J. 2013, 3, e104–e105. [Google Scholar] [CrossRef][Green Version]
- Sherbenou, D.W.; Hantschel, O.; Kaupe, I.; Willis, S.; Bumm, T.; Turaga, L.P.; Lange, T.; Dao, K.-H.; Press, R.D.; Druker, B.J.; et al. BCR-ABL SH3-SH2 domain mutations in chronic myeloid leukemia patients on imatinib. Blood 2010, 116, 3278–3285. [Google Scholar] [CrossRef]
- Berman, E.; Jhanwar, S.; Hedvat, C.; Arcila, M.E.; Wahab, O.A.; Levine, R.; Maloy, M.; Ma, W.; Albitar, M. Resistance to imatinib in patients with chronic myelogenous leukemia and the splice variant BCR-ABL135INS. Leuk. Res. 2016, 49, 108–112. [Google Scholar] [CrossRef]
- Marcé, S.; Cortés, M.; Zamora, L.; Cabezón, M.; Grau, J.; Millá, F.; Feliu, E. A thirty-five nucleotides BCR-ABL1 insertion mutation of controversial significance confers resistance to imatinib in a patient with chronic myeloid leukemia (CML). Exp. Mol. Pathol. 2015, 99, 16–18. [Google Scholar] [CrossRef]
- Wylie, A.A.; Schoepfer, J.; Jahnke, W.; Cowan-Jacob, S.W.; Loo, A.; Furet, P.; Marzinzik, A.L.; Pelle, X.; Donovan, J.; Zhu, W.; et al. The allosteric inhibitor ABL001 enables dual targeting of BCR–ABL1. Nature 2017, 543, 733–737. [Google Scholar] [CrossRef]
- Mahon, F.X.; Deininger, M.W.N.; Schultheis, B.; Chabrol, J.; Reiffers, J.; Goldman, J.M.; Melo, J.V. Selection and characterization of BCR-ABL positive cell lines with differential sensitivity to the tyrosine kinase inhibitor STI571: Diverse mechanisms of resistance. Blood 2000, 96, 1070–1079. [Google Scholar] [CrossRef] [PubMed]
- Stetka, J.; Gursky, J.; Liñan Velasquez, J.; Mojzikova, R.; Vyhlidalova, P.; Vrablova, L.; Bartek, J.; Divoky, V. Role of DNA Damage Response in Suppressing Malignant Progression of Chronic Myeloid Leukemia and Polycythemia Vera: Impact of Different Oncogenes. Cancers 2020, 12, 903. [Google Scholar] [CrossRef] [PubMed]
- Podszywalow-Bartnicka, P.; Wolczyk, M.; Kusio-Kobialka, M.; Wolanin, K.; Skowronek, K.; Nieborowska-Skorska, M.; Dasgupta, Y.; Skorski, T.; Piwocka, K. Downregulation of BRCA1 protein in BCR-ABL1 leukemia cells depends on stress-triggered TIAR-mediated suppression of translation. Cell Cycle 2014, 13, 3727–3741. [Google Scholar] [CrossRef] [PubMed]
- Popp, H.D.; Kohl, V.; Naumann, N.; Flach, J.; Brendel, S.; Kleiner, H.; Weiss, C.; Seifarth, W.; Saussele, S.; Hofmann, W.-K.; et al. DNA Damage and DNA Damage Response in Chronic Myeloid Leukemia. Int. J. Mol. Sci. 2020, 21, 1177. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cortes, J.E.; Lin, P.; Beaty, M.W.; Ai, D.; Amin, H.M.; McDonnell, T.J.; Ok, C.Y.; Kantarjian, H.M.; Medeiros, L.J.; et al. Clinical and prognostic significance of 3q26.2 and other chromosome 3 abnormalities in CML in the era of tyrosine kinase inhibitors. Blood 2015, 126, 1699–1706. [Google Scholar] [CrossRef] [PubMed]
- Krishna Chandran, R.; Geetha, N.; Sakthivel, K.M.; Suresh Kumar, R.; Jagathnath Krishna, K.M.N.; Sreedharan, H. Impact of Additional Chromosomal Aberrations on the Disease Progression of Chronic Myelogenous Leukemia. Front. Oncol. 2019, 9, 88. [Google Scholar] [CrossRef]
- Fabarius, A.; Leitner, A.; Hochhaus, A.; Müller, M.C.; Hanfstein, B.; Haferlach, C.; Göhring, G.; Schlegelberger, B.; Jotterand, M.; Reiter, A.; et al. Impact of additional cytogenetic aberrations at diagnosis on prognosis of CML: Long-term observation of 1151 patients from the randomized CML Study IV. Blood 2011, 118, 6760–6768. [Google Scholar] [CrossRef] [PubMed]
- Skorski, T. Genetic Mechanisms of Chronic Myeloid Leukemia Blastic Transformation. Curr. Hematol. Malig. Rep. 2012, 7, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Bavaro, L.; Martelli, M.; Cavo, M.; Soverini, S. Mechanisms of Disease Progression and Resistance to Tyrosine Kinase Inhibitor Therapy in Chronic Myeloid Leukemia: An Update. Int. J. Mol. Sci. 2019, 20, 6141. [Google Scholar] [CrossRef] [PubMed]
- Głowacki, S.; Synowiec, E.; Szwed, M.; Toma, M.; Skorski, T.; Śliwiński, T. Relationship between Oxidative Stress and Imatinib Resistance in Model Chronic Myeloid Leukemia Cells. Biomolecules 2021, 11, 610. [Google Scholar] [CrossRef]
- Brady, N.; Gaymes, T.J.; Cheung, M.; Mufti, G.J.; Rassool, F.V. Increased error-prone NHEJ activity in myeloid leukemias is associated with DNA damage at sites that recruit key nonhomologous end-joining proteins. Cancer Res. 2003, 63, 1798–1805. [Google Scholar]
- Cramer, K.; Nieborowska-Skorska, M.; Koptyra, M.; Slupianek, A.; Penserga, E.T.P.; Eaves, C.J.; Aulitzky, W.; Skorski, T. BCR/ABL and Other Kinases from Chronic Myeloproliferative Disorders Stimulate Single-Strand Annealing, an Unfaithful DNA Double-Strand Break Repair. Cancer Res. 2008, 68, 6884–6888. [Google Scholar] [CrossRef]
- Deutsch, E.; Jarrousse, S.; Buet, D.E.; Dugray, A.; Bonnet, M.-L.; Vozenin-Brotons, M.-C.; Guilhot, F.O.; Turhan, A.G.; Feunteun, J.; Bourhis, J. Down-regulation of BRCA1 in BCR-ABL–expressing hematopoietic cells. Blood 2003, 101, 4583–4588. [Google Scholar] [CrossRef]
- Gaymes, T.J.; Mufti, G.J.; Rassool, F.V. Myeloid Leukemias Have Increased Activity of the Nonhomologous End-Joining Pathway and Concomitant DNA Misrepair that Is Dependent on the Ku70/86 Heterodimer. Cancer Res. 2002, 62, 2791–2797. [Google Scholar]
- Nowicki, M.O.; Falinski, R.; Koptyra, M.; Slupianek, A.; Stoklosa, T.; Gloc, E.; Nieborowska-Skorska, M.; Blasiak, J.; Skorski, T. BCR/ABL oncogenic kinase promotes unfaithful repair of the reactive oxygen species–dependent DNA double-strand breaks. Blood 2004, 104, 3746–3753. [Google Scholar] [CrossRef]
- Salles, D.; Mencalha, A.L.; Ireno, I.C.; Wiesmüller, L.; Abdelhay, E. BCR-ABL stimulates mutagenic homologous DNA double-strand break repair via the DNA-end-processing factor CtIP. Carcinogenesis 2010, 32, 27–34. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cramer-Morales, K.; Nieborowska-Skorska, M.; Scheibner, K.; Padget, M.; Irvine, D.A.; Sliwinski, T.; Haas, K.; Lee, J.; Geng, H.; Roy, D.; et al. Personalized synthetic lethality induced by targeting RAD52 in leukemias identified by gene mutation and expression profile. Blood 2013, 122, 1293–1304. [Google Scholar] [CrossRef] [PubMed]
- Dkhissi, F.; Aggoune, D.; Pontis, J.; Sorel, N.; Piccirilli, N.; LeCorf, A.; Guilhot, F.; Chomel, J.-C.; Ait-Si-Ali, S.; Turhan, A.G. The downregulation of BAP1 expression by BCR-ABL reduces the stability of BRCA1 in chronic myeloid leukemia. Exp. Hematol. 2015, 43, 775–780. [Google Scholar] [CrossRef] [PubMed]
- Piwocka, K.; Wolanin, K.; Kusio-Kobialka, M.; Podszywalow-Bartnicka, P. BCR-ABL Hits at Mitosis; Implications for Chromosomal Instability, Aneuploidy and Therapeutic Strategy; InTech Press: London, UK, 2011. [Google Scholar]
- Slupianek, A.; Falinski, R.; Znojek, P.; Stoklosa, T.; Flis, S.; Doneddu, V.; Pytel, D.; Synowiec, E.; Blasiak, J.; Bellacosa, A.; et al. BCR-ABL1 kinase inhibits uracil DNA glycosylase UNG2 to enhance oxidative DNA damage and stimulate genomic instability. Leukemia 2013, 27, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Wolanin, K.; Magalska, A.; Kusio-Kobialka, M.; Podszywalow-Bartnicka, P.; Vejda, S.; McKenna, S.L.; Mosieniak, G.; Sikora, E.; Piwocka, K. Expression of Oncogenic Kinase Bcr-Abl Impairs Mitotic Checkpoint and Promotes Aberrant Divisions and Resistance to Microtubule-Targeting Agents. Mol. Cancer Ther. 2010, 9, 1328–1338. [Google Scholar] [CrossRef][Green Version]
- Canitrot, Y.; Falinski, R.; Louat, T.; Laurent, G.; Cazaux, C.; Hoffmann, J.-S.B.; Lautier, D.; Skorski, T. p210 BCR/ABL kinase regulates nucleotide excision repair (NER) and resistance to UV radiation. Blood 2003, 102, 2632–2637. [Google Scholar] [CrossRef]
- Tobin, L.A.; Robert, C.; Rapoport, A.P.; Gojo, I.; Baer, M.R.; Tomkinson, A.E.; Rassool, F.V. Targeting abnormal DNA double-strand break repair in tyrosine kinase inhibitor-resistant chronic myeloid leukemias. Oncogene 2013, 32, 1784–1793. [Google Scholar] [CrossRef]
- Sallmyr, A.; Fan, J.; Rassool, F.V. Genomic instability in myeloid malignancies: Increased reactive oxygen species (ROS), DNA double strand breaks (DSBs) and error-prone repair. Cancer Lett. 2008, 270, 1–9. [Google Scholar] [CrossRef]
- Dinis, J.; Silva, V.; Gromicho, M.; Martins, C.; Laires, A.; Tavares, P.; Rendeiro, P.; Torres, F.; Rueff, J.; Rodrigues, A. DNA damage response in imatinib resistant chronic myeloid leukemia K562 cells. Leuk. Lymphoma 2012, 53, 2004–2014. [Google Scholar] [CrossRef]
- Tipping, A.J.; Deininger, M.W.; Goldman, J.M.; Melo, J.V. Comparative gene expression profile of chronic myeloid leukemia cells innately resistant to imatinib mesylate. Exp. Hematol. 2003, 31, 1073–1080. [Google Scholar] [CrossRef]
- De Lavallade, H.; Finetti, P.; Carbuccia, N.; Khorashad, J.S.; Charbonnier, A.; Foroni, L.; Apperley, J.F.; Vey, N.; Bertucci, F.; Birnbaum, D.; et al. A gene expression signature of primary resistance to imatinib in chronic myeloid leukemia. Leuk. Res. 2010, 34, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Frank, O.; Brors, B.; Fabarius, A.; Li, L.; Haak, M.; Merk, S.; Schwindel, U.; Zheng, C.; Müller, M.C.; Gretz, N.; et al. Gene expression signature of primary imatinib-resistant chronic myeloid leukemia patients. Leukemia 2006, 20, 1400–1407. [Google Scholar] [CrossRef] [PubMed]
- Villuendas, R.; Steegmann, J.L.; Pollán, M.; Tracey, L.; Granda, A.; Fernández-Ruiz, E.; Casado, L.F.; Martínez, J.; Martínez, P.; Lombardía, L.; et al. Identification of genes involved in imatinib resistance in CML: A gene-expression profiling approach. Leukemia 2006, 20, 1047–1054. [Google Scholar] [CrossRef] [PubMed]
- Neul, C.; Schaeffeler, E.; Sparreboom, A.; Laufer, S.; Schwab, M.; Nies, A.T. Impact of Membrane Drug Transporters on Resistance to Small-Molecule Tyrosine Kinase Inhibitors. Trends Pharmacol. Sci. 2016, 37, 904–932. [Google Scholar] [CrossRef]
- DeGorter, M.; Xia, C.; Yang, J.; Kim, R. Drug Transporters in Drug Efficacy and Toxicity. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 249–273. [Google Scholar] [CrossRef]
- Nigam, S.K. The SLC22 Transporter Family: A Paradigm for the Impact of Drug Transporters on Metabolic Pathways, Signaling, and Disease. Annu. Rev. Pharmacol. Toxicol. 2018, 58, 663–687. [Google Scholar] [CrossRef]
- Ben Hassine, I.; Gharbi, H.; Soltani, I.; Teber, M.; Farrah, A.; Ben Hadj Othman, H.; Amouri, H.; Bellaaj, H.; lakhal, R.B.; Romdhane, N.B.; et al. hOCT1 gene expression predict for optimal response to Imatinib in Tunisian patients with chronic myeloid leukemia. Cancer Chemother. Pharmacol. 2017, 79, 737–745. [Google Scholar] [CrossRef]
- da Cunha Vasconcelos, F.; Mauricio Scheiner, M.A.; Moellman-Coelho, A.; Mencalha, A.L.; Renault, I.Z.; Rumjanek, V.M.; Maia, R.C. Low ABCB1 and high OCT1 levels play a favorable role in the molecular response to imatinib in CML patients in the community clinical practice. Leuk. Res. 2016, 51, 3–10. [Google Scholar] [CrossRef]
- White, D.L.; Saunders, V.A.; Dang, P.; Engler, J.; Venables, A.; Zrim, S.; Zannettino, A.; Lynch, K.; Manley, P.W.; Hughes, T. Most CML patients who have a suboptimal response to imatinib have low OCT-1 activity: Higher doses of imatinib may overcome the negative impact of low OCT-1 activity. Blood 2007, 110, 4064–4072. [Google Scholar] [CrossRef]
- Yee, S.W.; Brackman, D.J.; Ennis, E.A.; Sugiyama, Y.; Kamdem, L.K.; Blanchard, R.; Galetin, A.; Zhang, L.; Giacomini, K.M. Influence of Transporter Polymorphisms on Drug Disposition and Response: A Perspective From the International Transporter Consortium. Clin. Pharmacol. Ther. 2018, 104, 803–817. [Google Scholar] [CrossRef] [PubMed]
- Chhikara, S.; Sazawal, S.; Seth, T.; Chaubey, R.; Singh, K.; Sharma, R.; Mishra, P.; Mahapatra, M.; Saxena, R. Molecular Response to Imatinib and Its Correlation with mRNA Expression Levels of Imatinib Influx Transporter (OCT1) in Indian Chronic Myeloid Leukemia Patients. Asian Pac. J. Cancer Prev. 2017, 18, 2043–2048. [Google Scholar] [CrossRef]
- Watkins, D.B.; Hughes, T.P.; White, D.L. OCT1 and imatinib transport in CML: Is it clinically relevant? Leukemia 2015, 29, 1960–1969. [Google Scholar] [CrossRef]
- Hu, S.; Franke, R.M.; Filipski, K.K.; Hu, C.; Orwick, S.J.; de Bruijn, E.A.; Burger, H.; Baker, S.D.; Sparreboom, A. Interaction of Imatinib with Human Organic Ion Carriers. Clin. Cancer Res. 2008, 14, 3141–3148. [Google Scholar] [CrossRef]
- Harrach, S.; Schmidt-Lauber, C.; Pap, T.; Pavenstädt, H.; Schlatter, E.; Schmidt, E.; Berdel, W.E.; Schulze, U.; Edemir, B.; Jeromin, S.; et al. MATE1 regulates cellular uptake and sensitivity to imatinib in CML patients. Blood Cancer J. 2016, 6, e470. [Google Scholar] [CrossRef]
- Alves, R.; Fonseca, A.R.; Goncalves, A.C.; Ferreira-Teixeira, M.; Lima, J.; Abrantes, A.M.; Alves, V.; Rodrigues-Santos, P.; Jorge, L.; Matoso, E.; et al. Drug transporters play a key role in the complex process of Imatinib resistance in vitro. Leuk Res. 2015, 39, 355–360. [Google Scholar] [CrossRef]
- Hiwase, D.K.; Saunders, V.; Hewett, D.; Frede, A.; Zrim, S.; Dang, P.; Eadie, L.; To, L.B.; Melo, J.; Kumar, S.; et al. Dasatinib Cellular Uptake and Efflux in Chronic Myeloid Leukemia Cells: Therapeutic Implications. Clin. Cancer Res. 2008, 14, 3881–3888. [Google Scholar] [CrossRef]
- Giannoudis, A.; Davies, A.; Lucas, C.M.; Harris, R.J.; Pirmohamed, M.; Clark, R.E. Effective dasatinib uptake may occur without human organic cation transporter 1 (hOCT1): Implications for the treatment of imatinib-resistant chronic myeloid leukemia. Blood 2008, 112, 3348–3354. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Li, S.; Chen, L. The physiological role of drug transporters. Protein Cell 2015, 6, 334–350. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, J.I.; Williams, R.T.; Henderson, M.J.; Norris, M.D.; Haber, M. ABC transporters as mediators of drug resistance and contributors to cancer cell biology. Drug Resist. Updates 2016, 26, 1–9. [Google Scholar] [CrossRef]
- Eadie, L.N.; Hughes, T.P.; White, D.L. Interaction of the Efflux Transporters ABCB1 and ABCG2 With Imatinib, Nilotinib, and Dasatinib. Clin. Pharmacol. Ther. 2014, 95, 294–306. [Google Scholar] [CrossRef] [PubMed]
- Eadie, L.N.; Dang, P.; Saunders, V.A.; Yeung, D.T.; Osborn, M.P.; Grigg, A.P.; Hughes, T.P.; White, D.L. The clinical significance of ABCB1 overexpression in predicting outcome of CML patients undergoing first-line imatinib treatment. Leukemia 2016, 31, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Maia, R.C.; Vasconcelos, F.C.; Souza, P.S.; Rumjanek, V.M. Towards Comprehension of the ABCB1/P-Glycoprotein Role in Chronic Myeloid Leukemia. Molecules 2018, 23, 119. [Google Scholar] [CrossRef] [PubMed]
- Eadie, L.N.; Hughes, T.P.; White, D.L. ABCB1 Overexpression Is a Key Initiator of Resistance to Tyrosine Kinase Inhibitors in CML Cell Lines. PLoS ONE 2016, 11, e0161470. [Google Scholar] [CrossRef] [PubMed]
- Hegedus, C.; Ozvegy-Laczka, C.; Apáti, A.; Magócsi, M.; Német, K.; Orfi, L.; Kéri, G.; Katona, M.; Takáts, Z.; Váradi, A.; et al. Interaction of nilotinib, dasatinib and bosutinib with ABCB1 and ABCG2: Implications for altered anti-cancer effects and pharmacological properties. Br. J. Pharmacol. 2009, 158, 1153–1164. [Google Scholar] [CrossRef]
- Zhou, S.; Schuetz, J.D.; Bunting, K.D.; Colapietro, A.-M.; Sampath, J.; Morris, J.J.; Lagutina, I.; Grosveld, G.C.; Osawa, M.; Nakauchi, H.; et al. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat. Med. 2001, 7, 1028–1034. [Google Scholar] [CrossRef]
- Sarkadi, B.; Özvegy-Laczka, C.; Német, K.; Váradi, A. ABCG2—A transporter for all seasons. FEBS Lett. 2004, 567, 116–120. [Google Scholar] [CrossRef]
- De Lima, L.T.; Vivona, D.; Bueno, C.T.; Hirata, R.D.C.; Hirata, M.H.; Luchessi, A.D.; de Castro, F.A.; Chauffaille, M.D.L.F.; Zanichelli, M.A.; Chiattone, C.S.; et al. Reduced ABCG2 and increased SLC22A1 mRNA expression are associated with imatinib response in chronic myeloid leukemia. Med. Oncol. 2014, 31, 851. [Google Scholar] [CrossRef]
- Rinaldetti, S.; Pfirrmann, M.; Manz, K.; Guilhot, J.; Dietz, C.; Panagiotidis, P.; Spiess, B.; Seifarth, W.; Fabarius, A.; Müller, M.; et al. Effect of ABCG2, OCT1, and ABCB1 (MDR1) Gene Expression on Treatment-Free Remission in a EURO-SKI Subtrial. Clin. Lymphoma Myeloma Leuk. 2018, 18, 266–271. [Google Scholar] [CrossRef]
- Eadie, L.N.; Dang, P.; Goyne, J.M.; Hughes, T.P.; White, D.L. ABCC6 plays a significant role in the transport of nilotinib and dasatinib, and contributes to TKI resistance in vitro, in both cell lines and primary patient mononuclear cells. PLoS ONE 2018, 13, e0192180. [Google Scholar] [CrossRef]
- Polillo, M.; Galimberti, S.; Baratè, C.; Petrini, M.; Danesi, R.; Di Paolo, A. Pharmacogenetics of BCR/ABL Inhibitors in Chronic Myeloid Leukemia. Int. J. Mol. Sci. 2015, 16, 22811–22829. [Google Scholar] [CrossRef]
- Ripperger, A.; Benndorf, R.A. The C421A (Q141K) polymorphism enhances the 3′-untranslated region (3′-UTR)-dependent regulation of ATP-binding cassette transporter ABCG2. Biochem. Pharmacol. 2016, 104, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Megías-Vericat, J.E.; Montesinos, P.; Herrero, M.J.; Moscardó, F.; Bosó, V.; Rojas, L.; Martínez-Cuadrón, D.; Hervás, D.; Boluda, B.; García-Robles, A.; et al. Impact of ABC single nucleotide polymorphisms upon the efficacy and toxicity of induction chemotherapy in acute myeloid leukemia. Leuk. Lymphoma 2017, 58, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Heyes, N.; Kapoor, P.; Kerr, I.D. Polymorphisms of the multidrug pump ABCG2: A systematic review of their effect on protein expression, function and drug pharmacokinetics. Drug Metab. Dispos. 2018, 46, 1886–1899. [Google Scholar] [CrossRef] [PubMed]
- Au, A.; Aziz Baba, A.; Goh, A.S.; Wahid Fadilah, S.A.; Teh, A.; Rosline, H.; Ankathil, R. Association of genotypes and haplotypes of multi-drug transporter genes ABCB1 and ABCG2 with clinical response to imatinib mesylate in chronic myeloid leukemia patients. Biomed. Pharmacother. 2014, 68, 343–349. [Google Scholar] [CrossRef]
- Kim, D.H.; Sriharsha, L.; Xu, W.; Kamel-Reid, S.; Liu, X.; Siminovitch, K.; Messner, H.A.; Lipton, J.H. Clinical Relevance of a Pharmacogenetic Approach Using Multiple Candidate Genes to Predict Response and Resistance to Imatinib Therapy in Chronic Myeloid Leukemia. Clin. Cancer Res. 2009, 15, 4750–4758. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.-P.; Zhao, X.-L.; Takahashi, N.; Angelini, S.; Dubashi, B.; Sun, L.; Xu, P. Trough concentration and ABCG2 polymorphism are better to predict imatinib response in chronic myeloid leukemia: A meta-analysis. Pharmacogenomics 2016, 18, 35–56. [Google Scholar] [CrossRef]
- Makhtar, S.M.; Husin, A.; Baba, A.A.; Ankathil, R. Genetic variations in influx transporter gene SLC22A1 are associated with clinical responses to imatinib mesylate among Malaysian chronic myeloid leukaemia patients. J. Genet. 2018, 97, 835–842. [Google Scholar] [CrossRef]
- Vaidya, S.; Ghosh, K.; Shanmukhaiah, C.; Vundinti, B.R. Genetic variations of hOCT1 gene and CYP3A4/A5 genes and their association with imatinib response in Chronic Myeloid Leukemia. Eur. J. Pharmacol. 2015, 765, 124–130. [Google Scholar] [CrossRef]
- Beretta, G.L.; Cassinelli, G.; Pennati, M.; Zuco, V.; Gatti, L. Overcoming ABC transporter-mediated multidrug resistance: The dual role of tyrosine kinase inhibitors as multitargeting agents. Eur. J. Med. Chem. 2017, 142, 271–289. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.-B.; Yu, W.-N.; Feng, J.-H.; Luo, J.-M. Structure and function of Gab2 and its role in cancer. Mol. Med. Rep. 2015, 12, 4007–4014. [Google Scholar] [CrossRef] [PubMed]
- Wohrle, F.U.; Halbach, S.; Aumann, K.; Schwemmers, S.; Braun, S.; Auberger, P.; Schramek, D.; Penninger, J.M.; Lasmann, S.; Werner, M.; et al. Gab2 signaling in chronic myeloid leukemia cells confers resistance to multiple Bcr-Abl inhibitors. Leukemia 2013, 27, 118–129. [Google Scholar] [CrossRef]
- Ahmed, W.; Van Etten, R.A. Signal Transduction in the Chronic Leukemias: Implications for Targeted Therapies. Curr. Hematol. Malig. Rep. 2013, 8, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Liu, P.; Wang, P.; Zhou, Z.; Fang, Q.; Wang, J. PKC-β/Alox5 axis activation promotes Bcr-Abl-independent TKI-resistance in chronic myeloid leukemia. J. Cell. Physiol. 2021, 236, 6312–6327. [Google Scholar] [CrossRef] [PubMed]
- Donato, N.J.; Wu, J.Y.; Stapley, J.; Gallick, G.; Lin, H.; Arlinghaus, R.; Talpaz, M. BCR-ABL independence and LYN kinase overexpression in chronic myelogenous leukemia cells selected for resistance to STI571. Blood 2003, 101, 690–698. [Google Scholar] [CrossRef] [PubMed]
- Li, S. Src-family kinases in the development and therapy of Philadelphia chromosome-positive chronic myeloid leukemia and acute lymphoblastic leukemia. Leuk. Lymphoma 2008, 49, 19–26. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Parsons, S.J.; Parsons, J.T. Src family kinases, key regulators of signal transduction. Oncogene 2004, 23, 7906–7909. [Google Scholar] [CrossRef]
- Warsch, W.; Walz, C.; Sexl, V. JAK of all trades: JAK2-STAT5 as novel therapeutic targets in BCR-ABL1+ chronic myeloid leukemia. Blood 2013, 122, 2167–2175. [Google Scholar] [CrossRef]
- Warsch, W.; Kollmann, K.; Eckelhart, E.; Fajmann, S.; Cerny-Reiterer, S.; Hölbl, A.; Gleixner, K.V.; Dworzak, M.; Mayerhofer, M.; Hoermann, G.; et al. High STAT5 levels mediate imatinib resistance and indicate disease progression in chronic myeloid leukemia. Blood 2011, 117, 3409–3420. [Google Scholar] [CrossRef]
- Nair, R.R.; Tolentino, J.H.; Hazlehurst, L.A. Role of STAT3 in Transformation and Drug Resistance in CML. Front. Oncol. 2012, 2, 30. [Google Scholar] [CrossRef]
- Bewry, N.N.; Nair, R.R.; Emmons, M.F.; Boulware, D.; Pinilla-Ibarz, J.; Hazlehurst, L.A. Stat3 contributes to resistance toward BCR-ABL inhibitors in a bone marrow microenvironment model of drug resistance. Mol. Cancer Ther. 2008, 7, 3169–3175. [Google Scholar] [CrossRef]
- Zhao, S.; Konopleva, M.; Cabreira-Hansen, M.; Xie, Z.; Hu, W.; Milella, M.; Estrov, Z.; Mills, G.B.; Andreeff, M. Inhibition of phosphatidylinositol 3-kinase dephosphorylates BAD and promotes apoptosis in myeloid leukemias. Leukemia 2004, 18, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Wagle, M.; Eiring, A.M.; Wongchenko, M.; Lu, S.; Guan, Y.; Wang, Y.; Lackner, M.; Amler, L.; Hampton, G.; Deininger, M.W.; et al. A role for FOXO1 in BCR–ABL1-independent tyrosine kinase inhibitor resistance in chronic myeloid leukemia. Leukemia 2016, 30, 1493–1501. [Google Scholar] [CrossRef] [PubMed]
- Quentmeier, H.; Eberth, S.; Romani, J.; Zaborski, M.; Drexler, H.G. BCR-ABL1-independent PI3Kinase activation causing imatinib-resistance. J. Hematol. Oncol. 2011, 4, 6. [Google Scholar] [CrossRef] [PubMed]
- Bai, D.; Ueno, L.; Vogt, P.K. Akt-mediated regulation of NFkappaB and the essentialness of NFkappaB for the oncogenicity of PI3K and Akt. Int. J. Cancer 2009, 125, 2863–2870. [Google Scholar] [CrossRef]
- Carter, B.Z.; Mak, D.H.; Schober, W.D.; Cabreira-Hansen, M.; Beran, M.; McQueen, T.; Chen, W.; Andreeff, M. Regulation of survivin expression through Bcr-Abl/MAPK cascade: Targeting survivin overcomes imatinib resistance and increases imatinib sensitivity in imatinib-responsive CML cells. Blood 2006, 107, 1555–1563. [Google Scholar] [CrossRef]
- Bernardo, P.S.; Lemos, L.G.T.; de Moraes, G.N.; Maia, R.C. Unraveling survivin expression in chronic myeloid leukemia: Molecular interactions and clinical implications. Blood Rev. 2020, 43, 100671. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Mak, P.Y.; Mu, H.; Mak, D.H.; Zeng, Z.; Cortes, J.; Liu, Q.; Andreeff, M.; Carter, B.Z. Combined inhibition of β-catenin and Bcr–Abl synergistically targets tyrosine kinase inhibitor-resistant blast crisis chronic myeloid leukemia blasts and progenitors in vitro and in vivo. Leukemia 2017, 31, 2065–2074. [Google Scholar] [CrossRef] [PubMed]
- Nieborowska-Skorska, M.; Flis, S.; Skorski, T. AKT-induced reactive oxygen species generate imatinib-resistant clones emerging from chronic myeloid leukemia progenitor cells. Leukemia 2014, 28, 2416–2418. [Google Scholar] [CrossRef]
- Blasiak, J.; Hoser, G.; Bialkowska-Warzecha, J.; Pawlowska, E.; Skorski, T. Reactive Oxygen Species and Mitochondrial DNA Damage and Repair in BCR-ABL1 Cells Resistant to Imatinib. BioRes Open Access 2015, 4, 334–342. [Google Scholar] [CrossRef]
- Skorski, T. Chronic myeloid leukemia cells refractory/resistant to tyrosine kinase inhibitors are genetically unstable and may cause relapse and malignant progression to the terminal disease state. Leuk. Lymphoma 2011, 52 (Suppl. 1), 23–29. [Google Scholar] [CrossRef]
- Koptyra, M.; Falinski, R.; Nowicki, M.O.; Stoklosa, T.; Majsterek, I.; Nieborowska-Skorska, M.; Blasiak, J.; Skorski, T. BCR/ABL kinase induces self-mutagenesis via reactive oxygen species to encode imatinib resistance. Blood 2006, 108, 319–327. [Google Scholar] [CrossRef]
- Branford, S.; Wang, P.; Yeung, D.T.; Thomson, D.; Purins, A.; Wadham, C.; Shahrin, N.H.; Marum, J.E.; Nataren, N.; Parker, W.T.; et al. Integrative genomic analysis reveals cancer-associated mutations at diagnosis of CML in patients with high-risk disease. Blood 2018, 132, 948–961. [Google Scholar] [CrossRef]
- Ko, T.K.; Javed, A.; Lee, K.L.; Pathiraja, T.N.; Liu, X.; Malik, S.; Soh, S.X.; Heng, X.T.; Takahashi, N.; Tan, J.H.J.; et al. An integrative model of pathway convergence in genetically heterogeneous blast crisis chronic myeloid leukemia. Blood 2020, 135, 2337–2353. [Google Scholar] [CrossRef] [PubMed]
- Holyoake, T.L.; Vetrie, D. The chronic myeloid leukemia stem cell: Stemming the tide of persistence. Blood 2017, 129, 1595–1606. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Rashkovan, M.; Ferrando, A. Metabolic dependencies and vulnerabilities in leukemia. Genes Dev. 2019, 33, 1460–1474. [Google Scholar] [CrossRef] [PubMed]
- Kuntz, E.M.; Baquero, P.; Michie, A.M.; Dunn, K.; Tardito, S.; Holyoake, T.L.; Helgason, G.V.; Gottlieb, E. Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nat. Med. 2017, 23, 1234–1240. [Google Scholar] [CrossRef]
- Zhang, H.; Li, H.; Xi, H.S.; Li, S. HIF1α is required for survival maintenance of chronic myeloid leukemia stem cells. Blood 2012, 119, 2595–2607. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z. The FoxO-Autophagy Axis in Health and Disease. Trends Endocrinol. Metab. 2019, 30, 658–671. [Google Scholar] [CrossRef] [PubMed]
- Heidel, F.H.; Bullinger, L.; Feng, Z.; Wang, Z.; Neff, T.A.; Stein, L.; Kalaitzidis, D.; Lane, S.W.; Armstrong, S.A. Genetic and Pharmacologic Inhibition β-Catenin Targets Imatinib-Resistant Leukemia Stem Cells in CML. Cell Stem Cell 2012, 10, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Naka, K.; Hoshii, T.; Muraguchi, T.; Tadokoro, Y.; Ooshio, T.; Kondo, Y.; Nakao, S.; Motoyama, N.; Hirao, A. TGF-β–FOXO signalling maintains leukaemia-initiating cells in chronic myeloid leukaemia. Nature 2010, 463, 676–680. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1. Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef]
- Bellodi, C.; Lidonnici, M.R.; Hamilton, A.; Helgason, G.V.; Soliera, A.R.; Ronchetti, M.; Galavotti, S.; Young, K.W.; Selmi, T.; Yacobi, R.; et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J. Clin. Investig. 2009, 119, 1109–1123. [Google Scholar] [CrossRef]
- Rubinsztein, D.C.; Codogno, P.; Levine, B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 2012, 11, 709–730. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Xu, N.; He, B.; Pan, C.; Lan, Y.; Zhou, H.; Liu, X. Inhibition of autophagy enhances the selective anti-cancer activity of tigecycline to overcome drug resistance in the treatment of chronic myeloid leukemia. J. Exp. Clin. Cancer Res. 2017, 36, 43. [Google Scholar] [CrossRef]
- Baquero, P.; Dawson, A.; Mukhopadhyay, A.; Kuntz, E.M.; Mitchell, R.; Olivares, O.; Ianniciello, A.; Scott, M.T.; Dunn, K.; Nicastri, M.C.; et al. Targeting quiescent leukemic stem cells using second generation autophagy inhibitors. Leukemia 2019, 33, 981–994. [Google Scholar] [CrossRef]
- Naka, K.; Jomen, Y.; Ishihara, K.; Kim, J.; Ishimoto, T.; Bae, E.-J.; Mohney, R.P.; Stirdivant, S.M.; Oshima, H.; Oshima, M.; et al. Dipeptide species regulate p38MAPK–Smad3 signalling to maintain chronic myelogenous leukaemia stem cells. Nat. Commun. 2015, 6, 8039. [Google Scholar] [CrossRef] [PubMed]
- Taya, Y.; Ota, Y.; Wilkinson, A.C.; Kanazawa, A.; Watarai, H.; Kasai, M.; Nakauchi, H.; Yamazaki, S. Depleting dietary valine permits nonmyeloablative mouse hematopoietic stem cell transplantation. Science 2016, 354, 1152–1155. [Google Scholar] [CrossRef] [PubMed]
- Hattori, A.; Tsunoda, M.; Konuma, T.; Kobayashi, M.; Nagy, T.; Glushka, J.; Tayyari, F.; McSkimming, D.; Kannan, N.; Tojo, A.; et al. Cancer progression by reprogrammed BCAA metabolism in myeloid leukaemia. Nature 2017, 545, 500–504. [Google Scholar] [CrossRef] [PubMed]
- Rothe, K.; Babaian, A.; Nakamichi, N.; Chen, M.; Chafe, S.C.; Watanabe, A.; Forrest, D.L.; Mager, D.L.; Eaves, C.J.; Dedhar, S.; et al. Integrin-Linked Kinase Mediates Therapeutic Resistance of Quiescent CML Stem Cells to Tyrosine Kinase Inhibitors. Cell Stem Cell 2020, 27, 110–124.e119. [Google Scholar] [CrossRef]
- Houtkooper, R.H.; Pirinen, E.; Auwerx, J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012, 13, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Abraham, A.; Qiu, S.; Chacko, B.K.; Li, H.; Paterson, A.; He, J.; Agarwal, P.; Shah, M.; Welner, R.; Darley-Usmar, V.M.; et al. SIRT1 regulates metabolism and leukemogenic potential in CML stem cells. J. Clin. Investig. 2019, 129, 2685–2701. [Google Scholar] [CrossRef]
- Röhrig, F.; Schulze, A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 2016, 16, 732–749. [Google Scholar] [CrossRef]
- Chen, Y.; Hu, Y.; Zhang, H.; Peng, C.; Li, S. Loss of the Alox5 gene impairs leukemia stem cells and prevents chronic myeloid leukemia. Nat. Genet. 2009, 41, 783–792. [Google Scholar] [CrossRef]
- Chen, Y.; Peng, C.; Abraham, S.A.; Shan, Y.; Guo, Z.; Desouza, N.; Cheloni, G.; Li, D.; Holyoake, T.L.; Li, S. Arachidonate 15-lipoxygenase is required for chronic myeloid leukemia stem cell survival. J. Clin. Investig. 2014, 124, 3847–3862. [Google Scholar] [CrossRef]
- Kim, T.; Tyndel, M.S.; Kim, H.J.; Ahn, J.-S.; Choi, S.H.; Park, H.J.; Kim, Y.-K.; Kim, S.Y.; Lipton, J.H.; Zhang, Z.; et al. Spectrum of somatic mutation dynamics in chronic myeloid leukemia following tyrosine kinase inhibitor therapy. Blood 2017, 129, 38–47. [Google Scholar] [CrossRef]
- Mitani, K.; Nagata, Y.; Sasaki, K.; Yoshida, K.; Chiba, K.; Tanaka, H.; Shiraishi, Y.; Miyano, S.; Makishima, H.; Nakamura, Y.; et al. Somatic mosaicism in chronic myeloid leukemia in remission. Blood 2016, 128, 2863–2866. [Google Scholar] [CrossRef][Green Version]
- Togasaki, E.; Takeda, J.; Yoshida, K.; Shiozawa, Y.; Takeuchi, M.; Oshima, M.; Saraya, A.; Iwama, A.; Yokote, K.; Sakaida, E.; et al. Frequent somatic mutations in epigenetic regulators in newly diagnosed chronic myeloid leukemia. Blood Cancer J. 2017, 7, e559. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Rinke, J.; Schafer, V.; Schnittger, S.; Kohlmann, A.; Obstfelder, E.; Kunert, C.; Ziermann, J.; Winkelmann, N.; Eigendorff, E.; et al. Molecular-defined clonal evolution in patients with chronic myeloid leukemia independent of the BCR-ABL status. Leukemia 2014, 28, 2292–2299. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, V.; Kohlmann, A.; Zenger, M.; Schindela, S.; Eder, C.; Weissmann, S.; Schnittger, S.; Kern, W.; Müller, M.C.; Hochhaus, A.; et al. A deep-sequencing study of chronic myeloid leukemia patients in blast crisis (BC-CML) detects mutations in 76.9% of cases. Leukemia 2011, 25, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Kunimoto, H.; Nakajima, H. Epigenetic dysregulation of hematopoietic stem cells and preleukemic state. Int. J. Hematol. 2017, 106, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Ji, T.; Chen, Q.; Tao, S.; Shi, Y.; Chen, Y.; Shen, L.; Wang, C.; Yu, L. The research progress of circular RNAs in hematological malignancies. Hematology 2019, 24, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Nie, Z.-Y.; Yao, M.; Yang, Z.; Yang, L.; Liu, X.-J.; Yu, J.; Ma, Y.; Zhang, N.; Zhang, X.-Y.; Liu, M.-H.; et al. De-regulated STAT5A/miR-202-5p/USP15/Caspase-6 regulatory axis suppresses CML cell apoptosis and contributes to Imatinib resistance. J. Exp. Clin. Cancer Res. 2020, 39, 17. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Mohrbacher, A.F.; Tsai, Y.C.; Groffen, J.; Heisterkamp, N.; Nichols, P.W.; Yu, M.C.; Luübbert, M.; Jones, P.A. Quantitative measure of c-abl andp15 methylation in chronic myelogenous leukemia: Biological implications. Blood 2000, 95, 2990–2992. [Google Scholar] [CrossRef] [PubMed]
- Avramouli, A.; Tsochas, S.; Mandala, E.; Katodritou, E.; Ioannou, M.; Ritis, K.; Speletas, M. Methylation status of RASSF1A in patients with chronic myeloid leukemia. Leuk. Res. 2009, 33, 1130–1132. [Google Scholar] [CrossRef] [PubMed]
- Dunwell, T.; Hesson, L.; Rauch, T.A.; Wang, L.; Clark, R.E.; Dallol, A.; Gentle, D.; Catchpoole, D.; Maher, E.R.; Pfeifer, G.P.; et al. A Genome-wide screen identifies frequently methylated genes in haematological and epithelial cancers. Mol. Cancer 2010, 9, 44. [Google Scholar] [CrossRef]
- Keramatinia, A.; Ahadi, A.; Akbari, M.E.; Mohseny, M.; Mosavi Jarahi, A.; Bahadori-Monfared, A.; Hashemi, M.; Moradi, A.; Mehrvar, N.; Kazemi, E.; et al. The roles of DNA epigenetics and clinical significance in Chronic Myeloid Leukemia: A review. Cell. Mol. Biol. 2018, 64, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Machova Polakova, K.; Koblihova, J.; Stopka, T. Role of Epigenetics in Chronic Myeloid Leukemia. Curr. Hematol. Malig. Rep. 2013, 8, 28–36. [Google Scholar] [CrossRef]
- Harada, H. MDS: Recent progress in molecular pathogenesis and clinical aspects. Jpn. J. Clin. Hematol. 2017, 58, 1941–1950. [Google Scholar] [CrossRef]
- Lernoux, M.; Schnekenburger, M.; Dicato, M.; Diederich, M. Epigenetic mechanisms underlying the therapeutic effects of HDAC inhibitors in chronic myeloid leukemia. Biochem. Pharmacol. 2020, 173, 113698. [Google Scholar] [CrossRef]
- Rampalli, S.; Pavithra, L.; Bhatt, A.; Kundu, T.K.; Chattopadhyay, S. Tumor suppressor SMAR1 mediates cyclin D1 repression by recruitment of the SIN3/histone deacetylase 1 complex. Mol. Cell Biol. 2005, 25, 8415–8429. [Google Scholar] [CrossRef]
- Feinstein, E.; Gale, R.P.; Reed, J.; Canaani, E. Expression of the normal p53 gene induces differentiation of K562 cells. Oncogene 1992, 7, 1853–1857. [Google Scholar]
- Agatheeswaran, S.; Pattnayak, N.C.; Chakraborty, S. Identification and functional characterization of the miRNA-gene regulatory network in chronic myeloid leukemia lineage negative cells. Sci. Rep. 2016, 6, 32493. [Google Scholar] [CrossRef]
- Srutova, K.; Curik, N.; Burda, P.; Savvulidi, F.; Silvestri, G.; Trotta, R.; Klamova, H.; Pecherkova, P.; Sovova, Z.; Koblihova, J.; et al. BCR-ABL1 mediated miR-150 downregulation through MYC contributed to myeloid differentiation block and drug resistance in chronic myeloid leukemia. Haematologica 2018, 103, 2016–2025. [Google Scholar] [CrossRef]
- Machová Poláková, K.; Lopotová, T.; Klamová, H.; Burda, P.; Trněný, M.; Stopka, T.; Moravcová, J. Expression patterns of microRNAs associated with CML phases and their disease related targets. Mol. Cancer 2011, 10, 41. [Google Scholar] [CrossRef]
- Ferreira, A.F.; Moura, L.G.; Tojal, I.; Ambrósio, L.; Pinto-Simões, B.; Hamerschlak, N.; Calin, G.A.; Ivan, C.; Covas, D.T.; Kashima, S.; et al. ApoptomiRs expression modulated by BCR–ABL is linked to CML progression and imatinib resistance. Blood Cells Mol. Dis. 2014, 53, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Prinsloo, A.; Pool, R.; Van Niekerk, C. Preliminary data on microRNA expression profiles in a group of South African patients diagnosed with chronic myeloid leukaemia. Mol. Clin. Oncol. 2017, 7, 386–390. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alves, R.; Gonçalves, A.C.; Jorge, J.; Marques, G.; Luís, D.; Ribeiro, A.B.; Freitas-Tavares, P.; Oliveiros, B.; Almeida, A.M.; Sarmento-Ribeiro, A.B. MicroRNA signature refine response prediction in CML. Sci. Rep. 2019, 9, 9666. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-Y.; Chen, K.-C.; Liu, H.-J.E.; Liu, A.-J.; Wang, K.-L.; Shih, C.-M. MicroRNA-1301-Mediated RanGAP1 Downregulation Induces BCR-ABL Nuclear Entrapment to Enhance Imatinib Efficacy in Chronic Myeloid Leukemia Cells. PLoS ONE 2016, 11, e0156260. [Google Scholar] [CrossRef]
- Chakraborty, C.; Sharma, A.R.; Patra, B.C.; Bhattacharya, M.; Sharma, G.; Lee, S.-S. MicroRNAs mediated regulation of MAPK signaling pathways in chronic myeloid leukemia. Oncotarget 2016, 7, 42683–42697. [Google Scholar] [CrossRef] [PubMed]
- Firatligil, B.; Biray Avci, C.; Baran, Y. miR-17 in imatinib resistance and response to tyrosine kinase inhibitors in chronic myeloid leukemia cells. J. BU ON 2013, 18, 437–441. [Google Scholar]
- Suresh, S.; McCallum, L.; Lu, W.; Lazar, N.; Perbal, B.; Irvine, A.E. MicroRNAs 130a/b are regulated by BCR-ABL and downregulate expression of CCN3 in CML. J. Cell Commun. Signal 2011, 5, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Chim, C.S.; Wong, K.Y.; Leung, C.Y.; Chung, L.P.; Hui, P.K.; Chan, S.Y.; Yu, L. Epigenetic inactivation of the hsa-miR-203 in haematological malignancies. J. Cell. Mol. Med. 2011, 15, 2760–2767. [Google Scholar] [CrossRef] [PubMed]
- Ho, M.S.H.; Medcalf, R.L.; Livesey, S.A.; Traianedes, K. The dynamics of adult haematopoiesis in the bone and bone marrow environment. Br. J. Haematol. 2015, 170, 472–486. [Google Scholar] [CrossRef]
- Torres-Barrera, P.; Mayani, H.; Chávez-González, A. Understanding the hematopoietic microenvironment in chronic myeloid leukemia: A concise review. Curr. Res. Transl. Med. 2021, 69, 103295. [Google Scholar] [CrossRef]
- Manier, S.; Sacco, A.; Leleu, X.; Ghobrial, I.M.; Roccaro, A.M. Bone Marrow Microenvironment in Multiple Myeloma Progression. J. Biomed. Biotechnol. 2012, 2012, 1–5. [Google Scholar] [CrossRef]
- Kumar, R.; Godavarthy, P.S.; Krause, D.S. The bone marrow microenvironment in health and disease at a glance. J. Cell Sci. 2018, 131, jcs201707. [Google Scholar] [CrossRef]
- Arrigoni, E.; Del Re, M.; Galimberti, S.; Restante, G.; Rofi, E.; Crucitta, S.; Baratè, C.; Petrini, M.; Danesi, R.; Di Paolo, A. Concise Review: Chronic Myeloid Leukemia: Stem Cell Niche and Response to Pharmacologic Treatment. Stem Cells Transl. Med. 2018, 7, 305–314. [Google Scholar] [CrossRef]
- Brück, O.; Blom, S.; Dufva, O.; Turkki, R.; Chheda, H.; Ribeiro, A.; Kovanen, P.; Aittokallio, T.; Koskenvesa, P.; Kallioniemi, O.; et al. Immune cell contexture in the bone marrow tumor microenvironment impacts therapy response in CML. Leukemia 2018, 32, 1643–1656. [Google Scholar] [CrossRef]
- Schepers, K.; Pietras, E.M.; Reynaud, D.; Flach, J.; Binnewies, M.; Garg, T.; Wagers, A.J.; Hsiao, E.C.; Passegué, E. Myeloproliferative Neoplasia Remodels the Endosteal Bone Marrow Niche into a Self-Reinforcing Leukemic Niche. Cell Stem Cell 2013, 13, 285–299. [Google Scholar] [CrossRef]
- Hazlehurst, L.A.; Dalton, W.S. Mechanisms Associated with cell Adhesion Mediated Drug Resistance (CAM-DR) in Hematopoietic Malignancies. Cancer Metastasis Rev. 2001, 20, 43–50. [Google Scholar] [CrossRef]
- Kumar, R.; Pereira, R.S.; Zanetti, C.; Minciacchi, V.R.; Merten, M.; Meister, M.; Niemann, J.; Dietz, M.S.; Rüssel, N.; Schnütgen, F.; et al. Specific, targetable interactions with the microenvironment influence imatinib-resistant chronic myeloid leukemia. Leukemia 2020, 34, 2087–2101. [Google Scholar] [CrossRef]
- Zhang, B.; Li, M.; McDonald, T.; Holyoake, T.L.; Moon, R.T.; Campana, D.; Shultz, L.; Bhatia, R. Microenvironmental protection of CML stem and progenitor cells from tyrosine kinase inhibitors through N-cadherin and Wnt–β-catenin signaling. Blood 2013, 121, 1824–1838. [Google Scholar] [CrossRef] [PubMed]
- Meads, M.B.; Hazlehurst, L.A.; Dalton, W.S. The Bone Marrow Microenvironment as a Tumor Sanctuary and Contributor to Drug Resistance. Clin. Cancer Res. 2008, 14, 2519–2526. [Google Scholar] [CrossRef]
- Mukaida, N.; Tanabe, Y.; Baba, T. Chemokines as a Conductor of Bone Marrow Microenvironment in Chronic Myeloid Leukemia. Int. J. Mol. Sci. 2017, 18, 1824. [Google Scholar] [CrossRef] [PubMed]
- Hoermann, G.; Greiner, G.; Valent, P. Cytokine Regulation of Microenvironmental Cells in Myeloproliferative Neoplasms. Mediat. Inflamm. 2015, 2015, 869242. [Google Scholar] [CrossRef]
- Muselli, F.; Peyron, J.-F.; Mary, D. Druggable Biochemical Pathways and Potential Therapeutic Alternatives to Target Leukemic Stem Cells and Eliminate the Residual Disease in Chronic Myeloid Leukemia. Int. J. Mol. Sci. 2019, 20, 5616. [Google Scholar] [CrossRef]
- Agarwal, P.; Isringhausen, S.; Li, H.; Paterson, A.J.; He, J.; Gomariz, Á.; Nagasawa, T.; Nombela-Arrieta, C.; Bhatia, R. Mesenchymal Niche-Specific Expression of Cxcl12 Controls Quiescence of Treatment-Resistant Leukemia Stem Cells. Cell Stem Cell 2019, 24, 769–784.e766. [Google Scholar] [CrossRef]
- Zhang, H.-M.; Li, Q.; Zhu, X.; Liu, W.; Hu, H.; Liu, T.; Cheng, F.; You, Y.; Zhong, Z.; Zou, P.; et al. miR-146b-5p within BCR-ABL1–Positive Microvesicles Promotes Leukemic Transformation of Hematopoietic Cells. Cancer Res. 2016, 76, 2901–2911. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, Y.; Yang, Y.; Chen, H.; Tu, H.; Li, J. Exosomes from Bone Marrow Microenvironment-Derived Mesenchymal Stem Cells Affect CML Cells Growth and Promote Drug Resistance to Tyrosine Kinase Inhibitors. Stem Cells Int. 2020, 2020, 8890201. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.; Yong, A.S.M. Immune Effector Recovery in Chronic Myeloid Leukemia and Treatment-Free Remission. Front. Immunol. 2017, 8, 469. [Google Scholar] [CrossRef]
- Hsieh, Y.-C.; Kirschner, K.; Copland, M. Improving outcomes in chronic myeloid leukemia through harnessing the immunological landscape. Leukemia 2021, 35, 1229–1242. [Google Scholar] [CrossRef] [PubMed]
- Inselmann, S.; Wang, Y.; Saussele, S.; Fritz, L.; Schütz, C.; Huber, M.; Liebler, S.; Ernst, T.; Cai, D.; Botschek, S.; et al. Development, Function, and Clinical Significance of Plasmacytoid Dendritic Cells in Chronic Myeloid Leukemia. Cancer Res. 2018, 78, 6223–6234. [Google Scholar] [CrossRef]
- Cayssials, E.; Guilhot, F. Chronic Myeloid Leukemia: Immunobiology and Novel Immunotherapeutic Approaches. BioDrugs 2017, 31, 143–149. [Google Scholar] [CrossRef]
- Christiansson, L.; Söderlund, S.; Svensson, E.; Mustjoki, S.; Bengtsson, M.; Simonsson, B.; Olsson-Strömberg, U.; Loskog, A.S.I. Increased Level of Myeloid-Derived Suppressor Cells, Programmed Death Receptor Ligand 1/Programmed Death Receptor 1, and Soluble CD25 in Sokal High Risk Chronic Myeloid Leukemia. PLoS ONE 2013, 8, e55818. [Google Scholar] [CrossRef]
- Mumprecht, S.; Schürch, C.; Schwaller, J.; Solenthaler, M.; Ochsenbein, A.F. Programmed death 1 signaling on chronic myeloid leukemia–specific T cells results in T-cell exhaustion and disease progression. Blood 2009, 114, 1528–1536. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Liu, J.; Shen, N.; Zhao, Z.; Cui, J.; Zhou, S.; Jiang, L.; Zhu, X.; Tang, L.; Liang, H.; et al. The interaction of tumor cells and myeloid-derived suppressor cells in chronic myelogenous leukemia. Leuk. Lymphoma 2020, 61, 128–137. [Google Scholar] [CrossRef]
- Mu, H.; Zhu, X.; Jia, H.; Zhou, L.; Liu, H. Combination Therapies in Chronic Myeloid Leukemia for Potential Treatment-Free Remission: Focus on Leukemia Stem Cells and Immune Modulation. Front. Oncol. 2021, 11, 1657. [Google Scholar] [CrossRef]
- Kwaśnik, P.; Giannopoulos, K. Treatment-Free Remission—A New Aim in the Treatment of Chronic Myeloid Leukemia. J. Pers. Med. 2021, 11, 697. [Google Scholar] [CrossRef]
- Hochhaus, A.; Baccarani, M.; Silver, R.T.; Schiffer, C.; Apperley, J.F.; Cervantes, F.; Clark, R.E.; Cortes, J.E.; Deininger, M.W.; Guilhot, F.; et al. European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia 2020, 34, 966–984. [Google Scholar] [CrossRef]
- Mughal, T.I.; Psaila, B.; DeAngelo, D.J.; Saglio, G.; Van Etten, R.A.; Radich, J.P. Interrogating the molecular genetics of chronic myeloproliferative malignancies for personalized management in 2021. Haematologica 2021, 106, 1787–1793. [Google Scholar] [CrossRef] [PubMed]
- Soverini, S.; Bernardi, S.; Galimberti, S. Molecular Testing in CML between Old and New Methods: Are We at a Turning Point? J. Clin. Med. 2020, 9, 3865. [Google Scholar] [CrossRef] [PubMed]
- Cumbo, C.; Anelli, L.; Specchia, G.; Albano, F. Monitoring of Minimal Residual Disease (MRD) in Chronic Myeloid Leukemia: Recent Advances. Cancer Manag. Res. 2020, 12, 3175–3189. [Google Scholar] [CrossRef] [PubMed]
- Hughes, T.P.; Ross, D.M. Moving treatment-free remission into mainstream clinical practice in CML. Blood 2016, 128, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.; Rea, D.; Lipton, J.H. Treatment-free remission with first- and second-generation tyrosine kinase inhibitors. Am. J. Hematol. 2019, 94, 346–357. [Google Scholar] [CrossRef]
- Hughes, T.P.; Ross, D.M. Remembrance of things past—Discontinuation of second-generation TKI therapy for CML. Nat. Rev. Clin. Oncol. 2017, 14, 201–202. [Google Scholar] [CrossRef]
- Brown, J.T.; Laosinchai-Wolf, W.; Hedges, J.B.; Watt, C.D.; Van Deerlin, V.M.; Fletcher, L.; Branford, S.; Labourier, E. Establishment of a standardized multiplex assay with the analytical performance required for quantitative measurement of BCR-ABL1 on the international reporting scale. Blood Cancer J. 2011, 1, e13. [Google Scholar] [CrossRef]
- Radich, J.P. Chronic myeloid leukemia: Global impact from a local laboratory. Cancer 2017, 123, 2594–2596. [Google Scholar] [CrossRef]
- Yan, D.; Pomicter, A.D.; O’Hare, T.; Deininger, M.W. ddeeper Than Deep: Can ddPCR Predict Successful Imatinib Cessation? Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 6561–6563. [Google Scholar] [CrossRef]
- Chung, H.J.; Hur, M.; Yoon, S.; Hwang, K.; Lim, H.S.; Kim, H.; Moon, H.W.; Yun, Y.M. Performance Evaluation of the QXDx BCR-ABL %IS Droplet Digital PCR Assay. Ann. Lab. Med. 2020, 40, 72–75. [Google Scholar] [CrossRef]
- Brown, J.T.; Beldorth, I.J.; Laosinchai-Wolf, W.; Fahey, M.E.; Jefferson, K.L.; Ruskin, A.K.; Roth, J.J.; Cai, L.; Watt, C.D.; Press, R.D.; et al. Analytical Validation of a Highly Sensitive, Multiplexed Chronic Myeloid Leukemia Monitoring System Targeting BCR-ABL1 RNA. J. Mol. Diagn. 2019, 21, 718–733. [Google Scholar] [CrossRef] [PubMed]
- Scott, S.; Cartwright, A.; Tapley, A.; Boeckx, N.; Cayuela, J.-M.; Corner, A.; Dulucq, S.; Galimberti, S.; Lauricella, C.; Rose, S.; et al. Digital Pcr for the Measurement of Bcr-Abl1 in Cml: A New Dawn? EHA Library: The Hague, The Netherlands, 2020. [Google Scholar]
- Kizilors, A.; Crisà, E.; Lea, N.; Passera, R.; Mian, S.; Anwar, J.; Best, S.; Nicolini, F.E.; Ireland, R.; Aldouri, M.; et al. Effect of low-level BCR-ABL1 kinase domain mutations identified by next-generation sequencing in patients with chronic myeloid leukaemia: A population-based study. Lancet Haematol. 2019, 6, e276–e284. [Google Scholar] [CrossRef]
- Cayuela, J.M.; Chomel, J.C.; Coiteux, V.; Dulucq, S.; Escoffre-Barbe, M.; Etancelin, P.; Etienne, G.; Hayette, S.; Millot, F.; Nibourel, O.; et al. Recommendations from the French CML Study Group (Fi-LMC) for BCR-ABL1 kinase domain mutation analysis in chronic myeloid leukemia. Bull. Du Cancer 2020, 107, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Soverini, S.; Abruzzese, E.; Bocchia, M.; Bonifacio, M.; Galimberti, S.; Gozzini, A.; Iurlo, A.; Luciano, L.; Pregno, P.; Rosti, G.; et al. Next-generation sequencing for BCR-ABL1 kinase domain mutation testing in patients with chronic myeloid leukemia: A position paper. J. Hematol. Oncol. 2019, 12, 131. [Google Scholar] [CrossRef] [PubMed]
- Soverini, S.; Martelli, M.; Bavaro, L.; De Benedittis, C.; Iurlo, A.; Galimberti, S.; Pregno, P.; Bonifacio, M.; Lunghi, F.; Castagnetti, F.; et al. Detection of Actionable BCR-ABL1 Kinase Domain (KD) Mutations in Chronic Myeloid Leukemia (CML) Patients with Failure and Warning Response to Tyrosine Kinase Inhibitors (TKIs): Potential Impact of Next-Generation Sequencing (NGS) and Droplet Digital PCR (ddPCR) on Clinical Decision Making. Blood 2019, 134, 661. [Google Scholar] [CrossRef]
- Salah, H.T.; Muhsen, I.N.; Salama, M.E.; Owaidah, T.; Hashmi, S.K. Machine learning applications in the diagnosis of leukemia: Current trends and future directions. Int. J. Lab. Hematol. 2019, 41, 717–725. [Google Scholar] [CrossRef]
- Haferlach, T.; Kohlmann, A.; Wieczorek, L.; Basso, G.; Kronnie, G.T.; Béné, M.C.; De Vos, J.; Hernández, J.M.; Hofmann, W.K.; Mills, K.I.; et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: Report from the International Microarray Innovations in Leukemia Study Group. J. Clin. Oncol. 2010, 28, 2529–2537. [Google Scholar] [CrossRef]
- De Las Rivas, J.; Alonso-López, D.; Arroyo, M.M. Chapter Nine-Human Interactomics: Comparative Analysis of Different Protein Interaction Resources and Construction of a Cancer Protein–Drug Bipartite Network. In Advances in Protein Chemistry and Structural Biology; Donev, R., Ed.; Academic Press: Cambridge, MA, USA, 2018; Volume 111, pp. 263–282. [Google Scholar]
- Arroyo, M.M.; Berral-González, A.; Bueno-Fortes, S.; Alonso-López, D.; Rivas, J.D.L. Mining Drug-Target Associations in Cancer: Analysis of Gene Expression and Drug Activity Correlations. Biomolecules 2020, 10, 667. [Google Scholar] [CrossRef]
- Lee, S.-I.; Celik, S.; Logsdon, B.A.; Lundberg, S.M.; Martins, T.J.; Oehler, V.G.; Estey, E.H.; Miller, C.P.; Chien, S.; Dai, J.; et al. A machine learning approach to integrate big data for precision medicine in acute myeloid leukemia. Nat. Commun. 2018, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Hauser, R.G.; Esserman, D.; Beste, L.A.; Ong, S.Y.; Colomb, D.G., Jr.; Bhargava, A.; Wadia, R.; Rose, M.G. A Machine Learning Model to Successfully Predict Future Diagnosis of Chronic Myelogenous Leukemia With Retrospective Electronic Health Records Data. Am. J. Clin. Pathol. 2021. [Google Scholar] [CrossRef]
- Sasaki, K.; Jabbour, E.J.; Ravandi, F.; Konopleva, M.; Borthakur, G.; Wierda, W.G.; Daver, N.; Takahashi, K.; Naqvi, K.; DiNardo, C.; et al. The LEukemia Artificial Intelligence Program (LEAP) in chronic myeloid leukemia in chronic phase: A model to improve patient outcomes. Am. J. Hematol. 2021, 96, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, D.G.; Tsimberidou, A.M. Dasatinib in chronic myeloid leukemia: A review. Ther. Clin. Risk Manag. 2009, 5, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Caldemeyer, L.; Dugan, M.; Edwards, J.; Akard, L. Long-Term Side Effects of Tyrosine Kinase Inhibitors in Chronic Myeloid Leukemia. Curr. Hematol. Malig. Rep. 2016, 11, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Sacha, T.; Saglio, G. Nilotinib in the treatment of chronic myeloid leukemia. Future Oncol. 2018, 15, 953–965. [Google Scholar] [CrossRef] [PubMed]
- Eskazan, A.E.; Keskin, D. Radotinib and its clinical potential in chronic-phase chronic myeloid leukemia patients: An update. Ther. Adv. Hematol. 2017, 8, 237–243. [Google Scholar] [CrossRef]
- Puttini, M.; Coluccia, A.M.L.; Boschelli, F.; Cleris, L.; Marchesi, E.; Donella-Deana, A.; Ahmed, S.; Redaelli, S.; Piazza, R.; Magistroni, V.; et al. In vitro and In vivo Activity of SKI-606, a Novel Src-Abl Inhibitor, against Imatinib-Resistant Bcr-Abl+ Neoplastic Cells. Cancer Res. 2006, 66, 11314–11322. [Google Scholar] [CrossRef]
- Cortes, J.E.; Kantarjian, H.M.; Brümmendorf, T.H.; Kim, D.-W.; Turkina, A.G.; Shen, Z.-X.; Pasquini, R.; Khoury, H.J.; Arkin, S.; Volkert, A.; et al. Safety and efficacy of bosutinib (SKI-606) in chronic phase Philadelphia chromosome–positive chronic myeloid leukemia patients with resistance or intolerance to imatinib. Blood 2011, 118, 4567–4576. [Google Scholar] [CrossRef]
- O’Hare, T.; Shakespeare, W.C.; Zhu, X.; Eide, C.A.; Rivera, V.M.; Wang, F.; Adrian, L.T.; Zhou, T.; Huang, W.-S.; Xu, Q.; et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell 2009, 16, 401–412. [Google Scholar] [CrossRef]
- Garcia-Gutiérrez, V.; Luna, A.; Alonso-Dominguez, J.M.; Estrada, N.; Boque, C.; Xicoy, B.; Giraldo, P.; Angona, A.; Alvarez-Larrán, A.; Sanchez-Guijo, F.; et al. Safety and efficacy of asciminib treatment in chronic myeloid leukemia patients in real-life clinical practice. Blood Cancer J. 2021, 11, 16. [Google Scholar] [CrossRef]
- Schoepfer, J.; Jahnke, W.; Berellini, G.; Buonamici, S.; Cotesta, S.; Cowan-Jacob, S.W.; Dodd, S.; Drueckes, P.; Fabbro, D.; Gabriel, T.; et al. Discovery of Asciminib (ABL001), an Allosteric Inhibitor of the Tyrosine Kinase Activity of BCR-ABL1. J. Med. Chem. 2018, 61, 8120–8135. [Google Scholar] [CrossRef]
- Eadie, L.N.; Saunders, V.A.; Branford, S.; White, D.L.; Hughes, T.P. The new allosteric inhibitor asciminib is susceptible to resistance mediated by ABCB1 and ABCG2 overexpression in vitro. Oncotarget 2018, 9. [Google Scholar] [CrossRef]
- Zhan, J.-Y.; Ma, J.; Zheng, Q.-C. Molecular dynamics investigation on the Asciminib resistance mechanism of I502L and V468F mutations in BCR-ABL. J. Mol. Graph. Model. 2019, 89, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Westerweel, P.E.; te Boekhorst, P.A.W.; Levin, M.-D.; Cornelissen, J.J. New Approaches and Treatment Combinations for the Management of Chronic Myeloid Leukemia. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef]
- Rossari, F.; Minutolo, F.; Orciuolo, E. Past, present, and future of Bcr-Abl inhibitors: From chemical development to clinical efficacy. J. Hematol. Oncol. 2018, 11, 84. [Google Scholar] [CrossRef]
- Cortes, J.; Lang, F. Third-line therapy for chronic myeloid leukemia: Current status and future directions. J. Hematol. Oncol. 2021, 14, 44. [Google Scholar] [CrossRef]
- Ivanova, E.S.; Tatarskiy, V.V.; Yastrebova, M.A.; Khamidullina, A.I.; Shunaev, A.V.; Kalinina, A.A.; Zeifman, A.A.; Novikov, F.N.; Dutikova, Y.V.; Chilov, G.G.; et al. PF 114, a novel selective inhibitor of BCR ABL tyrosine kinase, is a potent inducer of apoptosis in chronic myelogenous leukemia cells. Int. J. Oncol. 2019, 55, 289–297. [Google Scholar] [CrossRef]
- Liu, X.; Wang, G.; Yan, X.; Qiu, H.; Min, P.; Wu, M.; Tang, C.; Zhang, F.; Tang, Q.; Zhu, S.; et al. Preclinical development of HQP1351, a multikinase inhibitor targeting a broad spectrum of mutant KIT kinases, for the treatment of imatinib-resistant gastrointestinal stromal tumors. Cell Biosci. 2019, 9, 88. [Google Scholar] [CrossRef]
- Mian, A.; Rafiei, A.; Haberbosch, I.; Zeifman, A.; Titov, I.; Stroylov, V.; Metodieva, A.; Stroganov, O.; Novikov, F.; Brill, B.; et al. PF-114, a potent and selective inhibitor of native and mutated BCR/ABL is active against Philadelphia chromosome-positive (Ph+) leukemias harboring the T315I mutation. Leukemia 2015, 29. [Google Scholar] [CrossRef] [PubMed]
- Massimino, M.; Stella, S.; Tirrò, E.; Romano, C.; Pennisi, M.S.; Puma, A.; Manzella, L.; Zanghì, A.; Stagno, F.; Di Raimondo, F.; et al. Non ABL-directed inhibitors as alternative treatment strategies for chronic myeloid leukemia. Mol. Cancer 2018, 17, 56. [Google Scholar] [CrossRef] [PubMed]
- Million, R.P.; Van Etten, R.A. The Grb2 binding site is required for the induction of chronic myeloid leukemia-like disease in mice by the Bcr/Abl tyrosine kinase. Blood 2000, 96, 664–670. [Google Scholar] [CrossRef] [PubMed]
- Ohanian, M.; Tari Ashizawa, A.; Garcia-Manero, G.; Pemmaraju, N.; Kadia, T.; Jabbour, E.; Ravandi, F.; Borthakur, G.; Andreeff, M.; Konopleva, M.; et al. Liposomal Grb2 antisense oligodeoxynucleotide (BP1001) in patients with refractory or relapsed haematological malignancies: A single-centre, open-label, dose-escalation, phase 1/1b trial. Lancet Haematol. 2018, 5, e136–e146. [Google Scholar] [CrossRef]
- Ohanian, M.; Lin, T.L.; Craig, M.; Agrawal, A.; Halka, K.; Ashizawa, A.T.; Cortes, J.E.; Roboz, G.J. A phase II study of BP1001 (liposomal Grb2 antisense oligonucleotide) in patients with hematologic malignancies. J. Clin. Oncol. 2020, 38, TPS7561. [Google Scholar] [CrossRef]
- Agarwal, P.; Bhatia, R. Influence of Bone Marrow Microenvironment on Leukemic Stem Cells: Breaking Up an Intimate Relationship. In Advances in Cancer Research; Paul, B.F., Kenneth, D.T., Eds.; Academic Press: Cambridge MA, USA, 2015; Volume 127, pp. 227–252. [Google Scholar]
- Agarwal, S.; Hartz, A.M.S.; Elmquist, W.F.; Bauer, B. Breast cancer resistance protein and P-glycoprotein in brain cancer: Two gatekeepers team up. Curr. Pharm. Des. 2011, 17, 2793–2802. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, G.; Iacobucci, I.; Paolini, S.; Ottaviani, E. Farnesyltransferase inhibition in hematologic malignancies: The clinical experience with tipifarnib. Clin. Adv. Hematol. Oncol. 2008, 6, 303–310. [Google Scholar] [PubMed]
- Cortes, J.; Quintás-Cardama, A.; Garcia-Manero, G.; O’Brien, S.; Jones, D.; Faderl, S.; Ebarb, T.; Giles, F.; Thomas, D.; Kantarjian, H. Phase 1 study of tipifarnib in combination with imatinib for patients with chronic myelogenous leukemia in chronic phase after imatinib failure. Cancer 2007, 110, 2000–2006. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.; Jabbour, E.; Daley, G.Q.; O’Brien, S.; Verstovsek, S.; Ferrajoli, A.; Koller, C.; Zhu, Y.; Statkevich, P.; Kantarjian, H. Phase 1 study of lonafarnib (SCH 66336) and imatinib mesylate in patients with chronic myeloid leukemia who have failed prior single-agent therapy with imatinib. Cancer 2007, 110, 1295–1302. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, S. Lonafarnib: First Approval. Drugs 2021, 81, 283–289. [Google Scholar] [CrossRef]
- Ruvolo, P.P.; Zhou, L.; Watt, J.C.; Ruvolo, V.R.; Burks, J.K.; Jiffar, T.; Kornblau, S.; Konopleva, M.; Andreeff, M. Targeting PKC-mediated signal transduction pathways using enzastaurin to promote apoptosis in acute myeloid leukemia-derived cell lines and blast cells. J. Cell. Biochem. 2011, 112, 1696–1707. [Google Scholar] [CrossRef]
- Ma, L.; Shan, Y.; Bai, R.; Xue, L.; Eide, C.A.; Ou, J.; Zhu, L.J.; Hutchinson, L.; Cerny, J.; Khoury, H.J.; et al. A therapeutically targetable mechanism of BCR-ABL–independent imatinib resistance in chronic myeloid leukemia. Sci. Transl. Med. 2014, 6, 252ra121. [Google Scholar] [CrossRef]
- Sweet, K.; Hazlehurst, L.; Sahakian, E.; Powers, J.; Nodzon, L.; Kayali, F.; Hyland, K.; Nelson, A.; Pinilla-Ibarz, J. A phase I clinical trial of ruxolitinib in combination with nilotinib in chronic myeloid leukemia patients with molecular evidence of disease. Leuk. Res. 2018, 74, 89–96. [Google Scholar] [CrossRef]
- Singh, P.; Kumar, V.; Gupta, S.K.; Kumari, G.; Verma, M. Combating TKI resistance in CML by inhibiting the PI3K/Akt/mTOR pathway in combination with TKIs: A review. Med. Oncol. 2021, 38, 10. [Google Scholar] [CrossRef]
- Alves, R.; Gonçalves, A.C.; Jorge, J.; Alves, J.; Alves da Silva, A.; Freitas-Tavares, P.; Nascimento Costa, J.M.; Almeida, A.M.; Sarmento-Ribeiro, A.B. Everolimus in combination with Imatinib overcomes resistance in Chronic myeloid leukaemia. Med. Oncol. 2019, 36, 30. [Google Scholar] [CrossRef]
- Burchert, A.; Wang, Y.; Cai, D.; von Bubnoff, N.; Paschka, P.; Müller-Brüsselbach, S.; Ottmann, O.G.; Duyster, J.; Hochhaus, A.; Neubauer, A. Compensatory PI3-kinase/Akt/mTor activation regulates imatinib resistance development. Leukemia 2005, 19, 1774–1782. [Google Scholar] [CrossRef]
- Yee, K.W.L.; Zeng, Z.; Konopleva, M.; Verstovsek, S.; Ravandi, F.; Ferrajoli, A.; Thomas, D.; Wierda, W.; Apostolidou, E.; Albitar, M.; et al. Phase I/II Study of the Mammalian Target of Rapamycin Inhibitor Everolimus (RAD001) in Patients with Relapsed or Refractory Hematologic Malignancies. Clin. Cancer Res. 2006, 12, 5165–5173. [Google Scholar] [CrossRef]
- Bibi, S.; Arslanhan, M.D.; Langenfeld, F.; Jeanningros, S.; Cerny-Reiterer, S.; Hadzijusufovic, E.; Tchertanov, L.; Moriggl, R.; Valent, P.; Arock, M. Co-operating STAT5 and AKT signaling pathways in chronic myeloid leukemia and mastocytosis: Possible new targets of therapy. Haematologica 2014, 99, 417–429. [Google Scholar] [CrossRef]
- Mihalyova, J.; Jelinek, T.; Growkova, K.; Hrdinka, M.; Simicek, M.; Hajek, R. Venetoclax: A new wave in hematooncology. Exp. Hematol. 2018, 61, 10–25. [Google Scholar] [CrossRef] [PubMed]
- Okabe, S.; Tauchi, T.; Tanaka, Y.; Ohyashiki, K. Anti-Leukemic Effects of Venetoclax on Philadelphia Chromosome Positive Leukemia Cells. Blood 2016, 128, 5428. [Google Scholar] [CrossRef]
- Maiti, A.; Franquiz, M.J.; Ravandi, F.; Cortes, J.E.; Jabbour, E.J.; Sasaki, K.; Marx, K.; Daver, N.G.; Kadia, T.M.; Konopleva, M.Y.; et al. Venetoclax and BCR-ABL Tyrosine Kinase Inhibitor Combinations: Outcome in Patients with Philadelphia Chromosome-Positive Advanced Myeloid Leukemias. Acta Haematol. 2020, 143, 567–573. [Google Scholar] [CrossRef]
- Konopleva, M.; Martinelli, G.; Daver, N.; Papayannidis, C.; Wei, A.; Higgins, B.; Ott, M.; Mascarenhas, J.; Andreeff, M. MDM2 inhibition: An important step forward in cancer therapy. Leukemia 2020, 34, 2858–2874. [Google Scholar] [CrossRef] [PubMed]
- Saiki, A.Y.; Caenepeel, S.; Yu, D.; Lofgren, J.A.; Osgood, T.; Robertson, R.; Canon, J.; Su, C.; Jones, A.; Zhao, X.; et al. MDM2 antagonists synergize broadly and robustly with compounds targeting fundamental oncogenic signaling pathways. Oncotarget 2014, 5, 2030–2043. [Google Scholar] [CrossRef]
- Pal, I.; Safari, M.; Jovanovic, M.; Bates, S.E.; Deng, C. Targeting Translation of mRNA as a Therapeutic Strategy in Cancer. Curr. Hematol. Malig. Rep. 2019, 14, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, V.; Plunkett, W.; Cortes, J.E. Omacetaxine: A Protein Translation Inhibitor for Treatment of Chronic Myelogenous Leukemia. Clin. Cancer Res. 2014, 20, 1735–1740. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.; Lipton, J.H.; Rea, D.; Digumarti, R.; Chuah, C.; Nanda, N.; Benichou, A.-C.; Craig, A.R.; Michallet, M.; Nicolini, F.E.; et al. Phase 2 study of subcutaneous omacetaxine mepesuccinate after TKI failure in patients with chronic-phase CML with T315I mutation. Blood 2012, 120, 2573–2580. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.; Digumarti, R.; Parikh, P.M.; Wetzler, M.; Lipton, J.H.; Hochhaus, A.; Craig, A.R.; Benichou, A.C.; Nicolini, F.E.; Kantarjian, H.M.; et al. Phase 2 study of subcutaneous omacetaxine mepesuccinate for chronic-phase chronic myeloid leukemia patients resistant to or intolerant of tyrosine kinase inhibitors. Am. J. Hematol. 2013, 88, 350–354. [Google Scholar] [CrossRef] [PubMed]
- Irvine, D.A.; Zhang, B.; Kinstrie, R.; Tarafdar, A.; Morrison, H.; Campbell, V.L.; Moka, H.A.; Ho, Y.; Nixon, C.; Manley, P.W.; et al. Deregulated hedgehog pathway signaling is inhibited by the smoothened antagonist LDE225 (Sonidegib) in chronic phase chronic myeloid leukaemia. Sci. Rep. 2016, 6, 25476. [Google Scholar] [CrossRef]
- Okabe, S.; Tauchi, T.; Ohyashiki, K. GDC-0449, the Small Molecule Inhibitor of Hedgehog-Gli Pathway for the Treatment of BCR-ABL Positive Leukemia Cells and In Combination with Dasatinib. Blood 2010, 116, 2136. [Google Scholar] [CrossRef]
- Rousselot, P.; Prost, S.; Guilhot, J.; Roy, L.; Etienne, G.; Legros, L.; Charbonnier, A.; Coiteux, V.; Cony-Makhoul, P.; Huguet, F.; et al. Pioglitazone together with imatinib in chronic myeloid leukemia: A proof of concept study. Cancer 2016, 123, 1791–1799. [Google Scholar] [CrossRef]
- Prost, S.; Relouzat, F.; Spentchian, M.; Ouzegdouh, Y.; Saliba, J.; Massonnet, G.; Beressi, J.-P.; Verhoeyen, E.; Raggueneau, V.; Maneglier, B.; et al. Erosion of the chronic myeloid leukaemia stem cell pool by PPARγ agonists. Nature 2015, 525, 380–383. [Google Scholar] [CrossRef]
- Fenaux, P.; Mufti, G.J.; Hellstrom-Lindberg, E.; Santini, V.; Finelli, C.; Giagounidis, A.; Schoch, R.; Gattermann, N.; Sanz, G.; List, A.; et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: A randomised, open-label, phase III study. Lancet Oncol. 2009, 10, 223–232. [Google Scholar] [CrossRef]
- Pleyer, L.; Burgstaller, S.; Stauder, R.; Girschikofsky, M.; Sill, H.; Schlick, K.; Thaler, J.; Halter, B.; Machherndl-Spandl, S.; Zebisch, A.; et al. Azacitidine front-line in 339 patients with myelodysplastic syndromes and acute myeloid leukaemia: Comparison of French-American-British and World Health Organization classifications. J. Hematol. Oncol. 2016, 9, 39. [Google Scholar] [CrossRef]
- Kantarjian, H.; Issa, J.-P.J.; Rosenfeld, C.S.; Bennett, J.M.; Albitar, M.; DiPersio, J.; Klimek, V.; Slack, J.; de Castro, C.; Ravandi, F.; et al. Decitabine improves patient outcomes in myelodysplastic syndromes: Results of a phase III randomized study. Cancer 2006, 106, 1794–1803. [Google Scholar] [CrossRef]
- Dombret, H.; Seymour, J.F.; Butrym, A.; Wierzbowska, A.; Selleslag, D.; Jang, J.H.; Kumar, R.; Cavenagh, J.; Schuh, A.C.; Candoni, A.; et al. International phase 3 study of azacitidine vs. conventional care regimens in older patients with newly diagnosed AML with >30% blasts. Blood 2015, 126, 291–299. [Google Scholar] [CrossRef]
- Ritchie, E.K.; Feldman, E.J.; Christos, P.J.; Rohan, S.D.; Lagassa, C.B.; Ippoliti, C.; Scandura, J.M.; Carlson, K.; Roboz, G.J. Decitabine in patients with newly diagnosed and relapsed acute myeloid leukemia. Leuk. Lymphoma 2013, 54, 2003–2007. [Google Scholar] [CrossRef]
- Kantarjian, H.M.; O’Brien, S.; Cortes, J.; Giles, F.J.; Faderl, S.; Issa, J.-P.; Garcia-Manero, G.; Rios, M.B.; Shan, J.; Andreeff, M.; et al. Results of decitabine (5-aza-2′deoxycytidine) therapy in 130 patients with chronic myelogenous leukemia. Cancer 2003, 98, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Issa, J.-P.J.; Garcia-Manero, G.; Giles, F.J.; Mannari, R.; Thomas, D.; Faderl, S.; Bayar, E.; Lyons, J.; Rosenfeld, C.S.; Cortes, J.; et al. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2′-deoxycytidine (decitabine) in hematopoietic malignancies. Blood 2004, 103, 1635–1640. [Google Scholar] [CrossRef] [PubMed]
- Oki, Y.; Kantarjian, H.M.; Gharibyan, V.; Jones, D.; O’Brien, S.; Verstovsek, S.; Cortes, J.; Morris, G.M.; Garcia-Manero, G.; Issa, J.-P.J. Phase II study of low-dose decitabine in combination with imatinib mesylate in patients with accelerated or myeloid blastic phase of chronic myelogenous leukemia. Cancer 2007, 109, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Mithraprabhu, S.; Grigoriadis, G.; Khong, T.; Spencer, A. Deactylase inhibition in myeloproliferative neoplasms. Investig. New Drugs 2010, 28 (Suppl. 1), S50–S57. [Google Scholar] [CrossRef] [PubMed]
- Di Costanzo, A.; Del Gaudio, N.; Migliaccio, A.; Altucci, L. Epigenetic drugs against cancer: An evolving landscape. Arch. Toxicol. 2014, 88, 1651–1668. [Google Scholar] [CrossRef]
- Cardoso, B.A.; Ramos, T.L.; Belo, H.; Vilas-Boas, F.; Real, C.; Almeida, A.M. Vorinostat synergizes with antioxidant therapy to target myeloproliferative neoplasms. Exp. Hematol. 2019, 72, 60–71.e11. [Google Scholar] [CrossRef]
- Nimmanapalli, R.; Fuino, L.; Stobaugh, C.; Richon, V.; Bhalla, K. Cotreatment with the histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) enhances imatinib-induced apoptosis of Bcr-Abl–positive human acute leukemia cells. Blood 2003, 101, 3236–3239. [Google Scholar] [CrossRef]
- Fiskus, W.; Pranpat, M.; Balasis, M.; Bali, P.; Estrella, V.; Kumaraswamy, S.; Rao, R.; Rocha, K.; Herger, B.; Lee, F.; et al. Cotreatment with Vorinostat (Suberoylanilide Hydroxamic Acid) Enhances Activity of Dasatinib (BMS-354825) against Imatinib Mesylate–Sensitive or Imatinib Mesylate–Resistant Chronic Myelogenous Leukemia Cells. Clin. Cancer Res. 2006, 12, 5869–5878. [Google Scholar] [CrossRef]
- Vonka, V. Immunotherapy of chronic myeloid leukemia: Present state and future prospects. Immunotherapy 2010, 2, 227–241. [Google Scholar] [CrossRef]
- Bilich, T.; Nelde, A.; Bichmann, L.; Roerden, M.; Salih, H.R.; Kowalewski, D.J.; Schuster, H.; Tsou, C.-C.; Marcu, A.; Neidert, M.C.; et al. The HLA ligandome landscape of chronic myeloid leukemia delineates novel T-cell epitopes for immunotherapy. Blood 2019, 133, 550–565. [Google Scholar] [CrossRef]
- Egan, D.; Radich, J. When to consider allogeneic transplant for chronic myeloid leukemia. Adv. Cell Gene Ther. 2020, 3, e85. [Google Scholar] [CrossRef]
- Hochhaus, A.; Breccia, M.; Saglio, G.; García-Gutiérrez, V.; Réa, D.; Janssen, J.; Apperley, J. Expert opinion—Management of chronic myeloid leukemia after resistance to second-generation tyrosine kinase inhibitors. Leukemia 2020, 34, 1495–1502. [Google Scholar] [CrossRef]
- DeFilipp, Z.; Ancheta, R.; Liu, Y.; Hu, Z.-H.; Gale, R.P.; Snyder, D.; Schouten, H.C.; Kalaycio, M.; Hildebrandt, G.C.; Ustun, C.; et al. Maintenance Tyrosine Kinase Inhibitors Following Allogeneic Hematopoietic Stem Cell Transplantation for Chronic Myelogenous Leukemia: A Center for International Blood and Marrow Transplant Research Study. Biol. Blood Marrow Transplant. 2020, 26, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Yassine, F.; Reljic, T.; Alhaj Moustafa, M.; Iqbal, M.; Murthy, H.S.; Kumar, A.; Kharfan-Dabaja, M.A. Efficacy of allogeneic hematopoietic cell transplantation in patients with chronic phase CML resistant or intolerant to tyrosine kinase inhibitors. Hematol. Oncol. Stem Cell Ther. 2021. [Google Scholar] [CrossRef] [PubMed]
- Al-Dewik, N.I.; Morsi, H.M.; Samara, M.M.; Ghasoub, R.S.; Gnanam, C.C.; Bhaskaran, S.K.; Nashwan, A.J.; Al-Jurf, R.M.; Ismail, M.A.; Alsharshani, M.M.; et al. Is Adherence to Imatinib Mesylate Treatment among Patients with Chronic Myeloid Leukemia Associated with Better Clinical Outcomes in Qatar? Clin. Med. Insights Oncol. 2016, 10, CMO.S32822. [Google Scholar] [CrossRef]
- Chaudri, N.A. Adherence to Long-term Therapies Evidence for Action. Ann. Saudi Med. 2004, 24, 221–222. [Google Scholar] [CrossRef]
- Noens, L.; van Lierde, M.-A.; De Bock, R.; Verhoef, G.; Zachée, P.; Berneman, Z.; Martiat, P.; Mineur, P.; Van Eygen, K.; MacDonald, K.; et al. Prevalence, determinants, and outcomes of nonadherence to imatinib therapy in patients with chronic myeloid leukemia: The ADAGIO study. Blood 2009, 113, 5401–5411. [Google Scholar] [CrossRef]
- Marin, D.; Bazeos, A.; Mahon, F.-X.; Eliasson, L.; Milojkovic, D.; Bua, M.; Apperley, J.F.; Szydlo, R.; Desai, R.; Kozlowski, K.; et al. Adherence is the critical factor for achieving molecular responses in patients with chronic myeloid leukemia who achieve complete cytogenetic responses on imatinib. J. Clin. Oncol. 2010, 28, 2381–2388. [Google Scholar] [CrossRef] [PubMed]
- Darkow, T.; Henk, H.J.; Thomas, S.K.; Feng, W.; Baladi, J.-F.; Goldberg, G.A.; Hatfield, A.; Cortes, J. Treatment Interruptions and Non-Adherence with Imatinib and Associated Healthcare Costs. PharmacoEconomics 2007, 25, 481–496. [Google Scholar] [CrossRef] [PubMed]
- Druker, B.J.; Lydon, N.B. Lessons learned from the development of an Abl tyrosine kinase inhibitor for chronic myelogenous leukemia. J. Clin. Investig. 2000, 105, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2018 update on diagnosis, therapy and monitoring. Am. J. Hematol. 2018, 93, 442–459. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Cortes, J.; Kantarjian, H. Estimations of the increasing prevalence and plateau prevalence of chronic myeloid leukemia in the era of tyrosine kinase inhibitor therapy. Cancer 2012, 118, 3123–3127. [Google Scholar] [CrossRef] [PubMed]
- Alves, R.S.; Gonçalves, A.; Jorge, J.; Marques, G.; Ribeiro, A.B.; Tenreiro, R.; Coucelo, M.; Diamond, J.; Pereira, A.; Freitas-Tavares, P.; et al. Influx/efflux transporters in chronic myeloid leukemia: The influence of genetic variants on susceptibility and drug response. HemaSphere 2019, 3, 532. [Google Scholar] [CrossRef]
- Singh, V.K.; Coumar, M.S. Chronic Myeloid Leukemia: Existing Therapeutic Options and Strategies to Overcome Drug Resistance. Mini Rev. Med. Chem. 2019, 19, 333–345. [Google Scholar] [CrossRef]
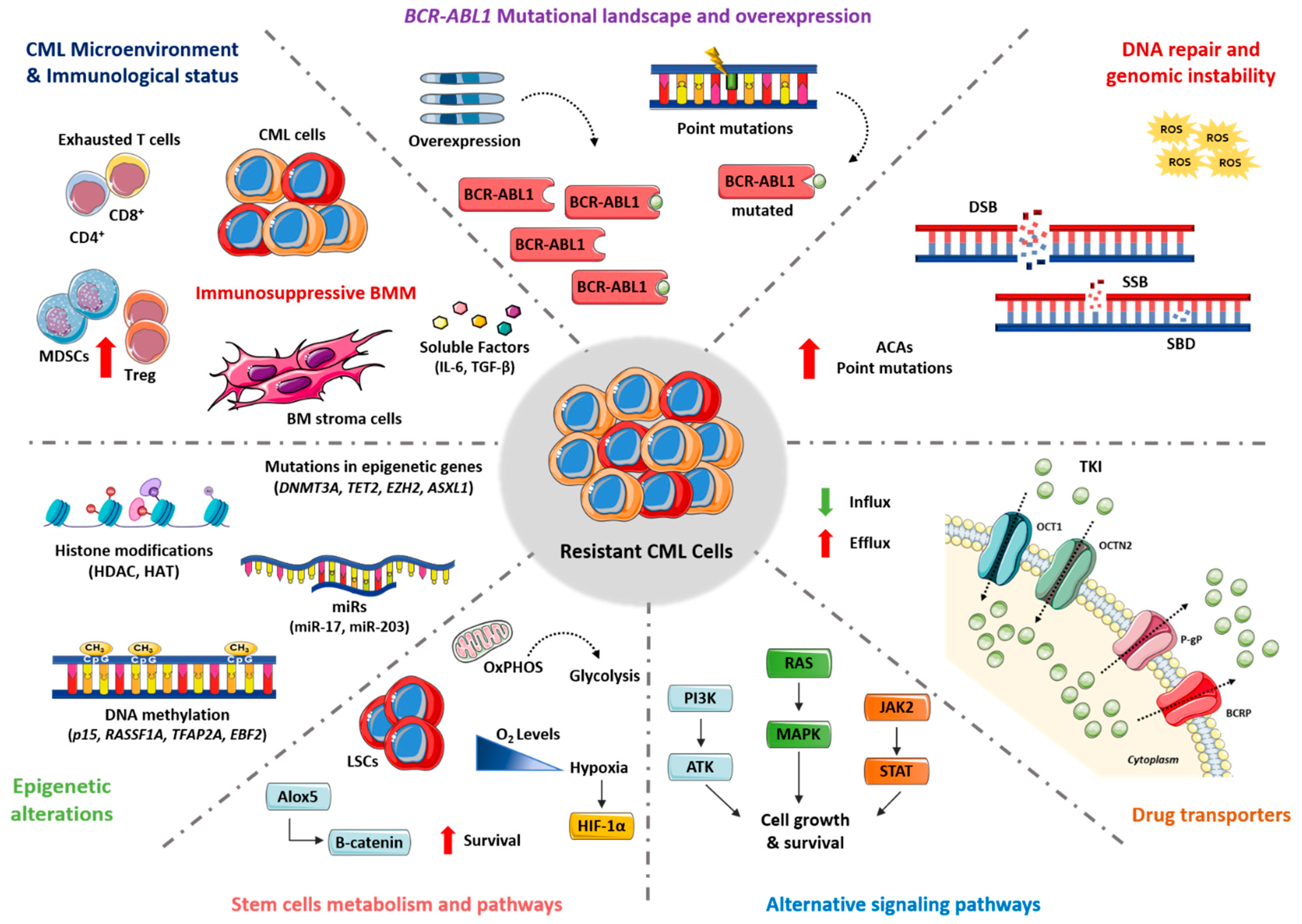
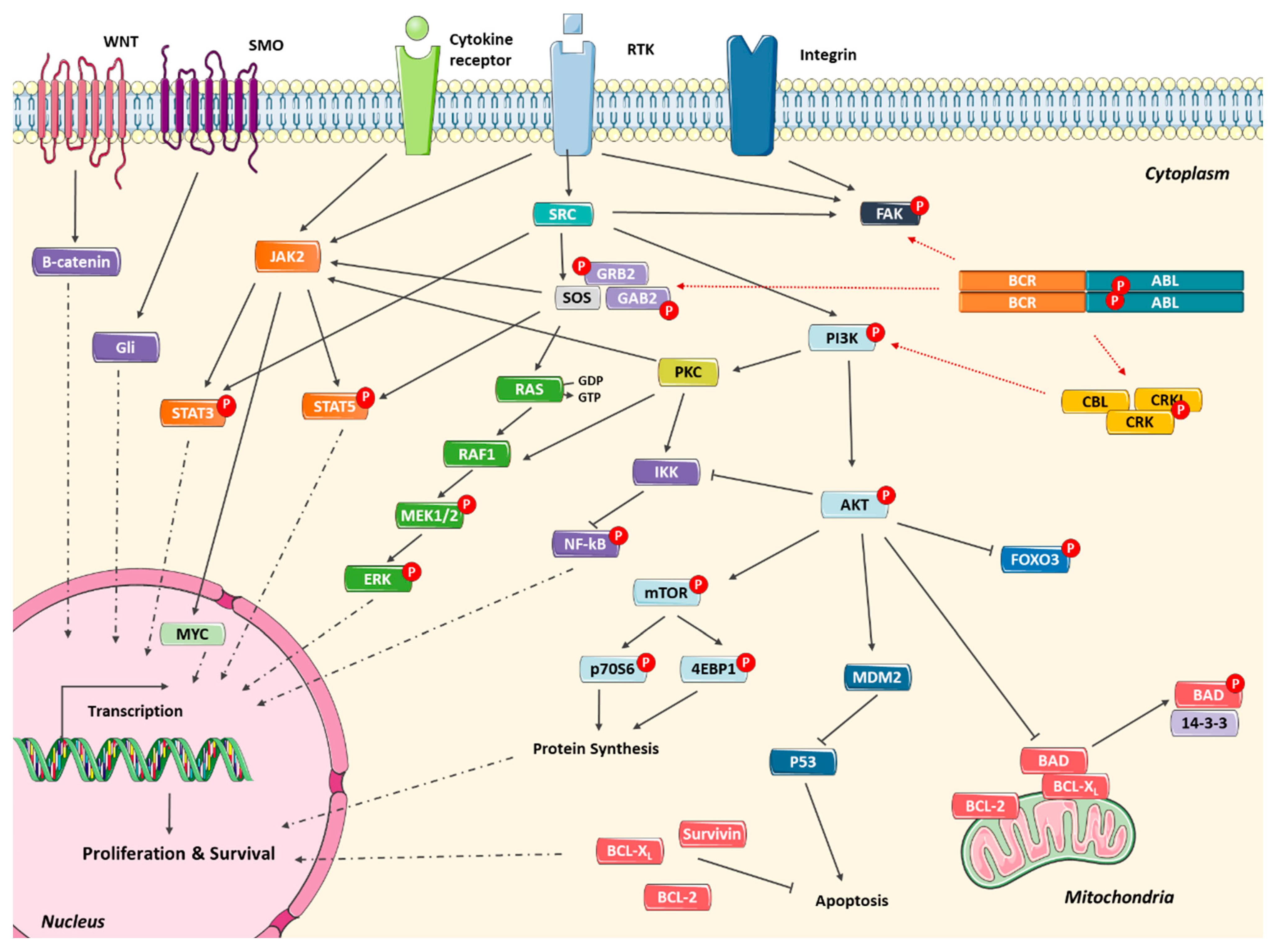
| BCR-ABL1 Mutation | Location $ | Imatinib | Dasatinib | Nilotinib | Bosutinib | Ponatinib | Asciminib |
|---|---|---|---|---|---|---|---|
| Wild-type | Sen | Sen | Sen | Sen | Sen | Sen | |
| M244 | P-loop | Sen | Sen | Sen | Sen | Sen | Sen |
| L248 | P-loop | Int | Int | Sen | Int | Sen | Sen |
| G250 | P-loop | Res | Sen | Int | Res | Sen | Sen |
| Q252 | P-loop | Int | Int | Sen | Sen | Sen | Sen |
| Y253 § | P-loop | Res | Sen | Res | Sen | Sen | Sen |
| E255 § | P-loop | Res | Int | Res | Int | Int | Sen |
| V299 | C-helix | Res | Res | Sen | Res | Sen | Sen |
| T315 ‡, § | Drug contact site | Res | Res | Res | Res | Int | Sen |
| F317 ‡ | Drug contact site | Int | Res | Sen | Int | Sen | Sen |
| A337 | C-loop | Sen | Sen | Sen | Sen | Sen | Int |
| M351 | C-loop | Sen | Sen | Sen | Sen | Sen | Sen |
| M355 | C-loop | Int | Sen | Sen | Sen | Sen | Sen |
| F359 § | C-loop | Int | Sen | Res | Sen | Sen | Sen |
| H396 | A-loop | Int | Sen | Int | Sen | Int | Sen |
| W464 | Myristate pocket | Sen | Sen | Sen | Sen | Sen | Res |
| P465 | Myristate pocket | Sen | Sen | Sen | Sen | Sen | Res |
| V468 | Myristate pocket | Sen | Sen | Sen | Sen | Sen | Res |
| I502 | Myristate pocket | Sen | Sen | Sen | Sen | Sen | Res |
| Drug | Class/Mechanism of Action | NCT Number | Scheme | Phase |
|---|---|---|---|---|
| BCR-ABL1 therapies | ||||
| Asciminib | ABL1 myristoyl pocket inhibitor | NCT02081378 | Mono. and Comb. TKI | 1 |
| NCT03595917 | Plus Dasatinib | 1 | ||
| NCT03906292 | Mono. and Comb. TKI | 2 | ||
| NCT03578367 | Plus Imatinib | 2 | ||
| NCT03106779 | Mono. | 3 | ||
| NCT04971226 | Mono. | 3 | ||
| NCT04948333 | Mono. | 3 | ||
| NCT04877522 | Mono. and Comb. TKI | 3 | ||
| NCT04795427 | Mono. | 2 | ||
| NCT04666259 | Mono. | 3 | ||
| Flumatinib | BCR-ABL1 ATP-binding site inhibitor | NCT04677439 | Mono. | 4 |
| NCT04933526 | Mono. | 4 | ||
| PF-114 | BCR-ABL1 ATP-binding site inhibitor | NCT02885766 | Mono. | 1/2 |
| HQP1351 | BCR-ABL1 ATP-binding site inhibitor | NCT03883100 | Mono. | 2 |
| NCT03883087 | Mono. | 2 | ||
| NCT04126681 | Mono. | 2 | ||
| NCT04260022 | Mono. | 1 | ||
| Vodobatinib | BCR-ABL1 ATP-binding site inhibitor | NCT02629692 | Mono. | 1/2 |
| Non BCR-ABL1 therapies | ||||
| BP1001 | GRB2 antisense oligonucleotide | NCT01159028 | Mono. | 1 |
| NCT02923986 | Plus Dasatinib | 2 | ||
| Tipifarnib | Farnesyltransferase inhibitor | NCT00004009 | Mono. | 1 |
| Lonafarnib | Farnesyltransferase inhibitor | NCT00047502 | Plus Imatinib | 1 |
| Selumetinib | MEK inhibitors | NCT03326310 | Plus Azacitidine | 1 |
| Ruxolitinib | JAK2 inhibitor | NCT03610971 | Comb. TKI | 2 |
| NCT01702064 | Plus Nilotinib | 1 | ||
| Everolimus | mTOR inhibitor | NCT00081874 | Mono. | 1/2 |
| NCT01188889 | Plus Imatinib | 2 | ||
| Sirolimus | mTOR inhibitor | NCT00776373 | Plus Citarabine | 1/2 |
| Venetoclax | BCL-2 inhibitor | NCT02689440 | Plus Dasatinib | 2 |
| NCT04188405 | Plus Ponatinib, Decitabine | 2 | ||
| NCT03576547 | Plus Ponatinib, corticosteroids | 1/2 | ||
| AMG-232 | MDM2 inhibitor | NCT04835584 | Mono. | 1/2 |
| Sonidigib | SHH inhibitor | NCT01456676 | Plus Nilotinib | 1 |
| Pioglitazone | PPAR-γ inhibitor | NCT02852486 | Plus Imatinib | 2 |
| NCT02767063 | Plus TKI | 1/2 | ||
| NCT02889003 | Plus TKI | 2 | ||
| Epigenetic modulators | ||||
| Azacitidine | Hypomethylating agent | NCT03895671 | Plus Ponatinib | 2 |
| NCT01460498 | Plus TKI | 1 | ||
| Panobinostat | Histone Deacetylase Inhibitor | NCT00451035 | Mono. | 1 |
| Immunotherapies | ||||
| Peg-IFNα | Pegylated Interferon alpha | NCT01866553 | Plus Nilotinib | 2 |
| NCT01872442 | Plus Dasatinib | 2 | ||
| NCT03831776 | Plus Bosutinib | 2 | ||
| Nivolumab | Anti-PD-1 antibody | NCT02011945 | Plus Dasatinib | 1 |
| NCT01822509 | Plus Ipilimumab | 1 | ||
| Pembrolizumab | Anti-PD-1 antibody | NCT03516279 | Plus TKI | 2 |
| DC vaccine | Dendritic cells vaccine | NCT02543749 | Mono. | 1/2 |
| CLL1-CD33 cCART | compound CAR (cCAR) T cells | NCT03795779 | Mono. | 1 |
| KDS-1001 | natural killer cell therapy | NCT04808115 | Plus TKI | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alves, R.; Gonçalves, A.C.; Rutella, S.; Almeida, A.M.; De Las Rivas, J.; Trougakos, I.P.; Sarmento Ribeiro, A.B. Resistance to Tyrosine Kinase Inhibitors in Chronic Myeloid Leukemia—From Molecular Mechanisms to Clinical Relevance. Cancers 2021, 13, 4820. https://doi.org/10.3390/cancers13194820
Alves R, Gonçalves AC, Rutella S, Almeida AM, De Las Rivas J, Trougakos IP, Sarmento Ribeiro AB. Resistance to Tyrosine Kinase Inhibitors in Chronic Myeloid Leukemia—From Molecular Mechanisms to Clinical Relevance. Cancers. 2021; 13(19):4820. https://doi.org/10.3390/cancers13194820
Chicago/Turabian StyleAlves, Raquel, Ana Cristina Gonçalves, Sergio Rutella, António M. Almeida, Javier De Las Rivas, Ioannis P. Trougakos, and Ana Bela Sarmento Ribeiro. 2021. "Resistance to Tyrosine Kinase Inhibitors in Chronic Myeloid Leukemia—From Molecular Mechanisms to Clinical Relevance" Cancers 13, no. 19: 4820. https://doi.org/10.3390/cancers13194820
APA StyleAlves, R., Gonçalves, A. C., Rutella, S., Almeida, A. M., De Las Rivas, J., Trougakos, I. P., & Sarmento Ribeiro, A. B. (2021). Resistance to Tyrosine Kinase Inhibitors in Chronic Myeloid Leukemia—From Molecular Mechanisms to Clinical Relevance. Cancers, 13(19), 4820. https://doi.org/10.3390/cancers13194820








