MEIS2 Is an Adrenergic Core Regulatory Transcription Factor Involved in Early Initiation of TH-MYCN-Driven Neuroblastoma Formation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Publicly Available Datasets Used for Analysis
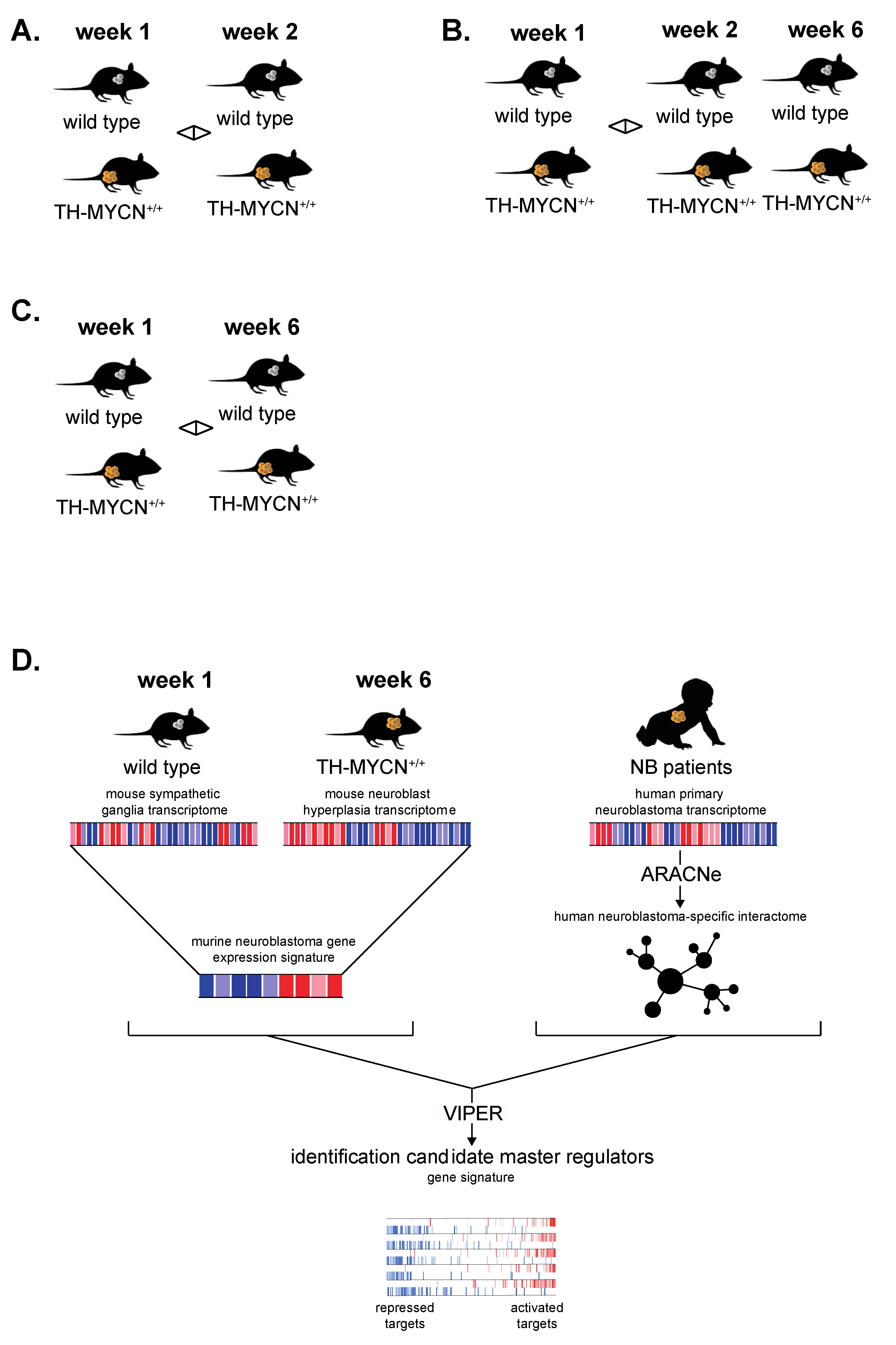
2.2. TH-MYCN Neuroblastoma Progression Model
2.3. RNA Isolation and Gene Expression Analysis of TH-MYCN+/+ and Wild-Type Samples
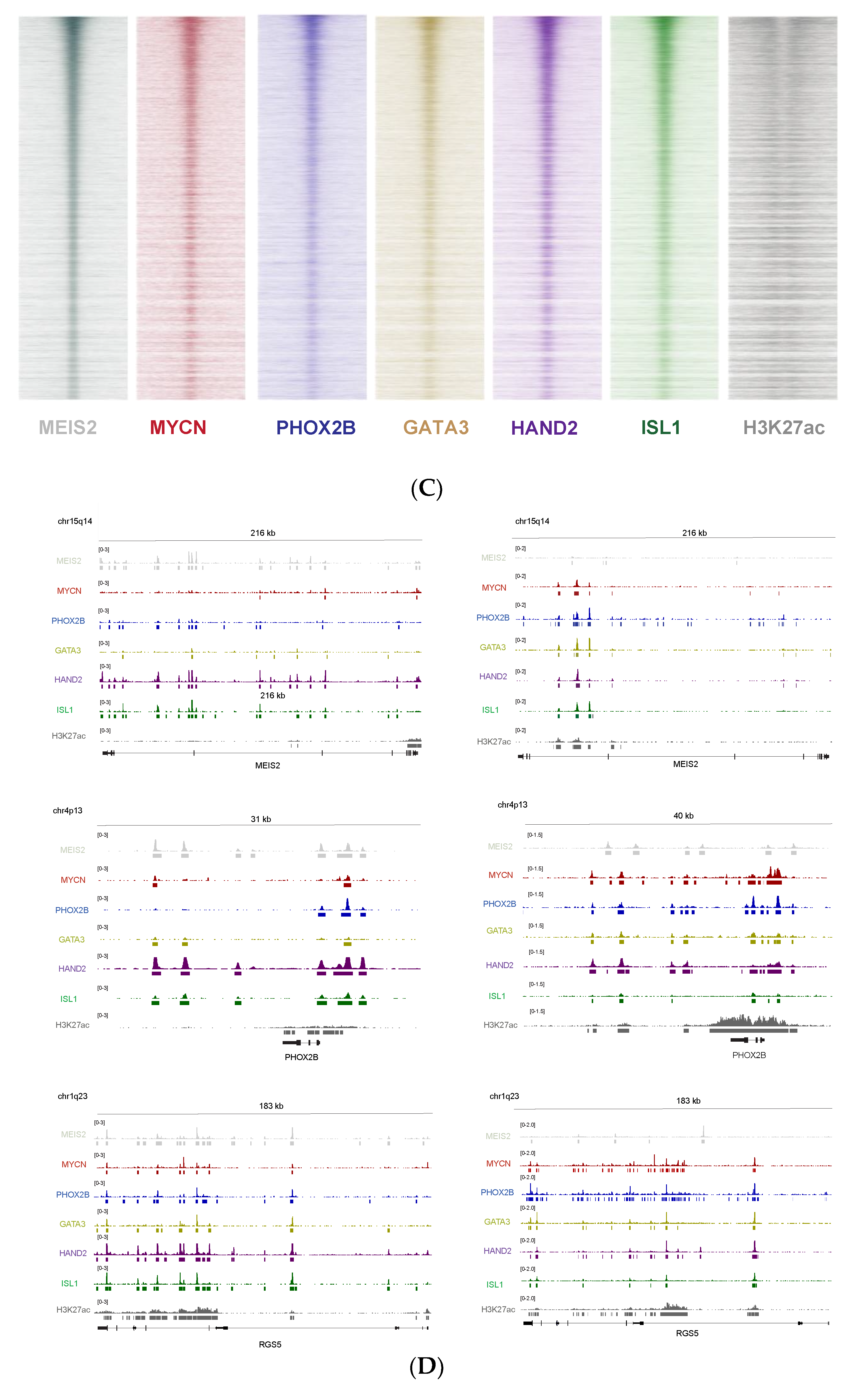
2.4. MEIS2 ChIP-Sequencing
2.5. MEIS2 Pharmacological Inhibition
2.6. Human Neuroblastoma Regulatory Network Assembly and Master Regulator Analysis
2.7. Overall Survival Analysis in Neuroblastoma Tumors
2.8. Statistical Analysis
2.9. Gene Set Enrichment Analysis and Cytoscape Network Visualization
3. Results
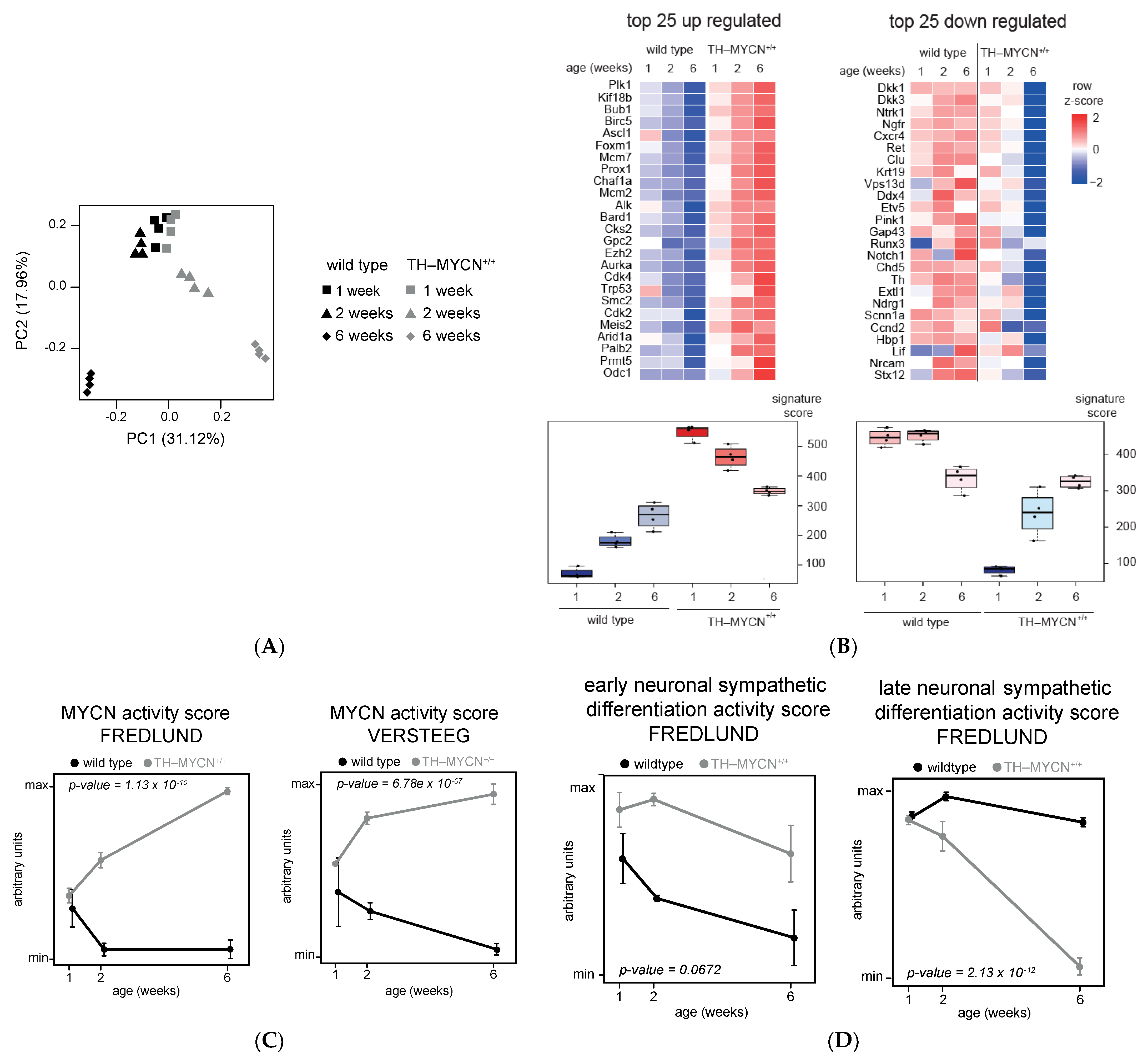
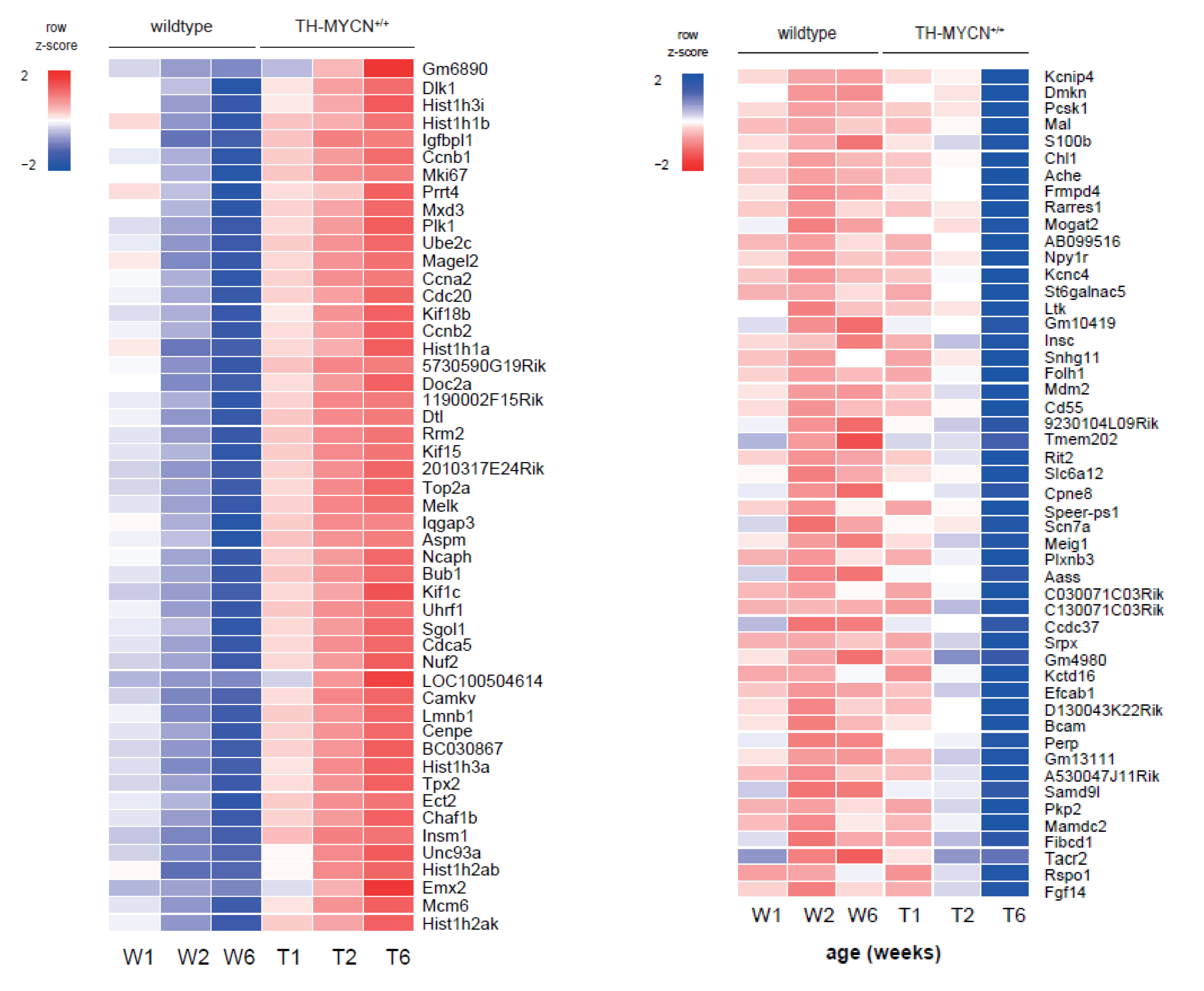
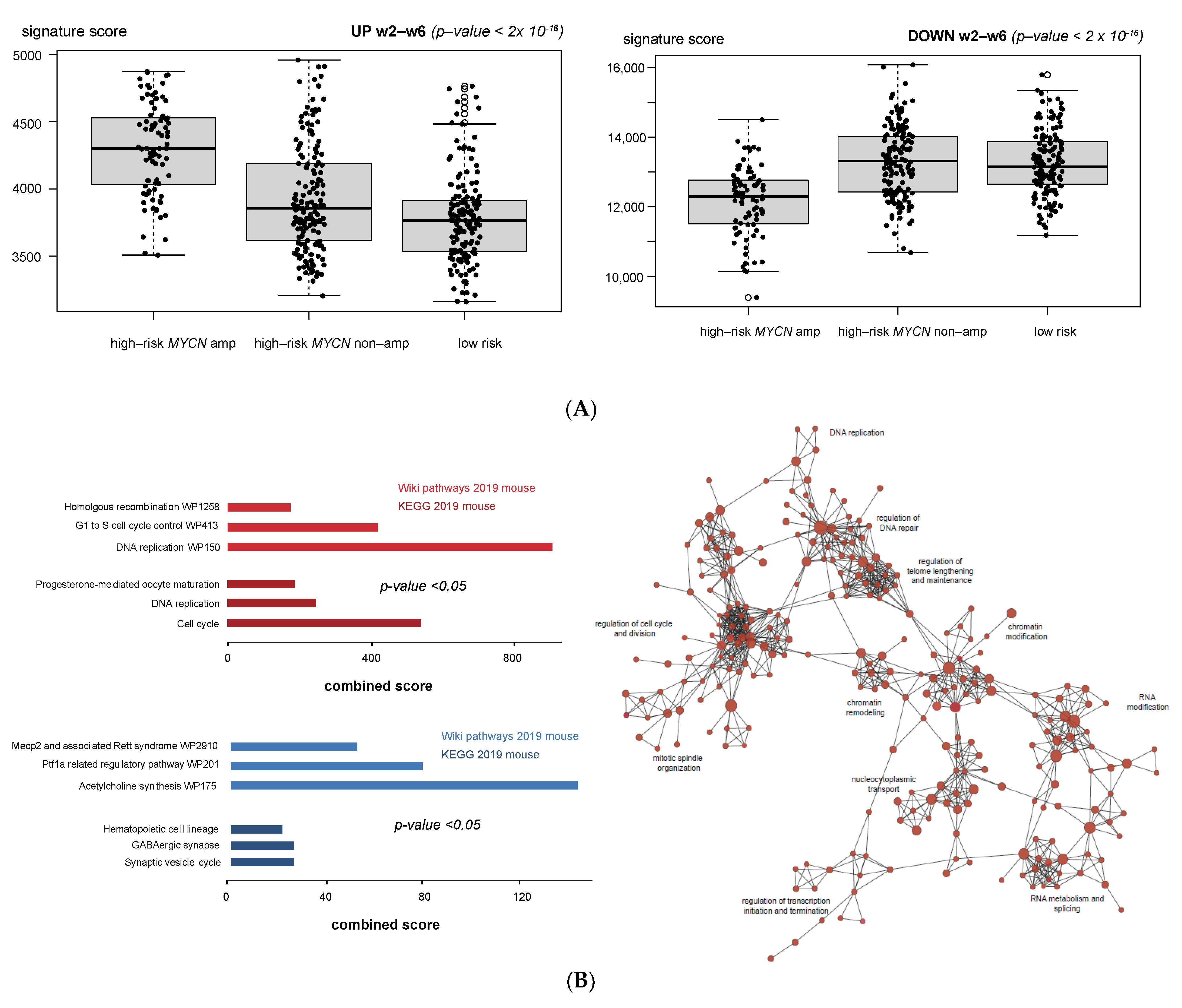
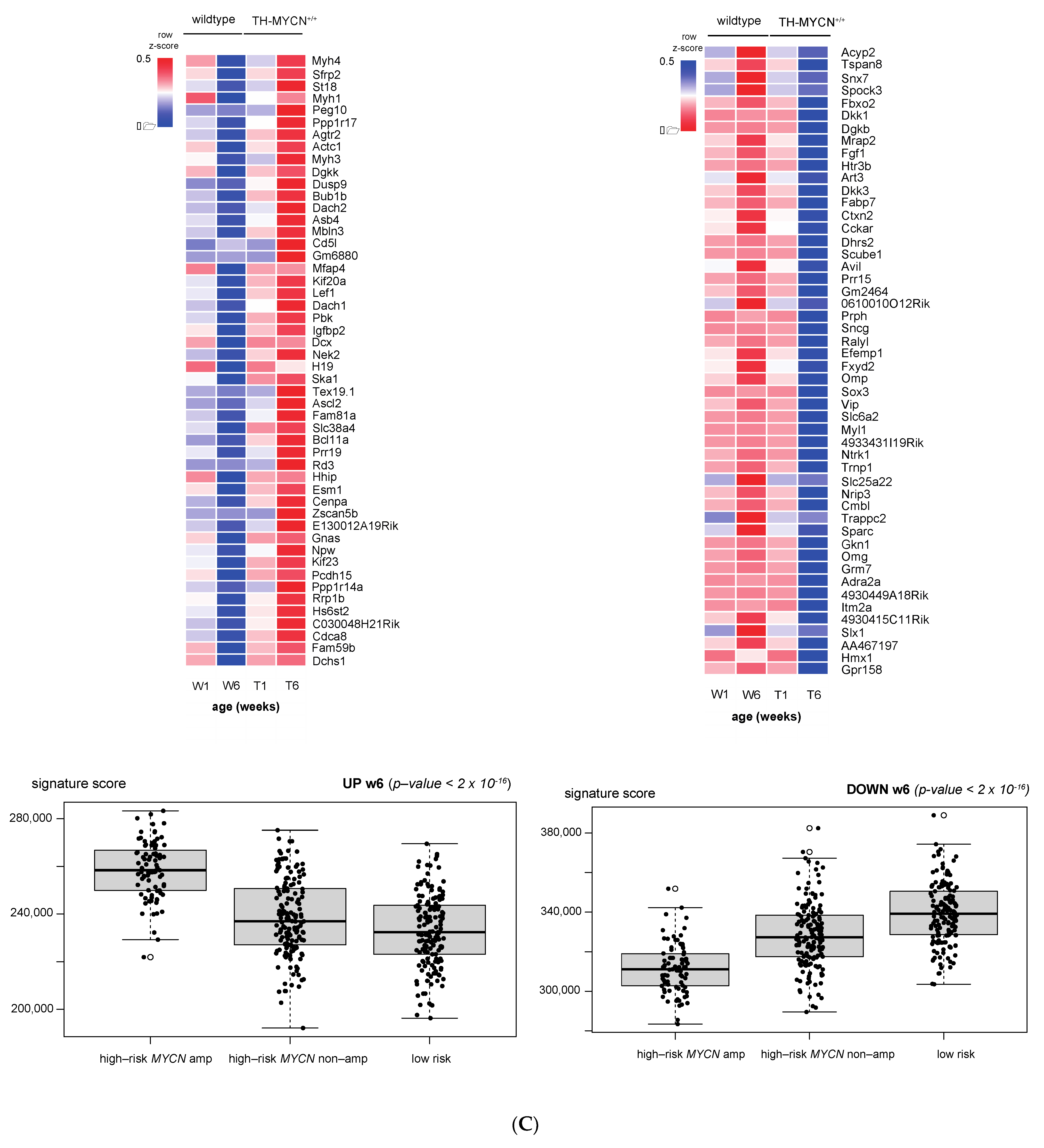
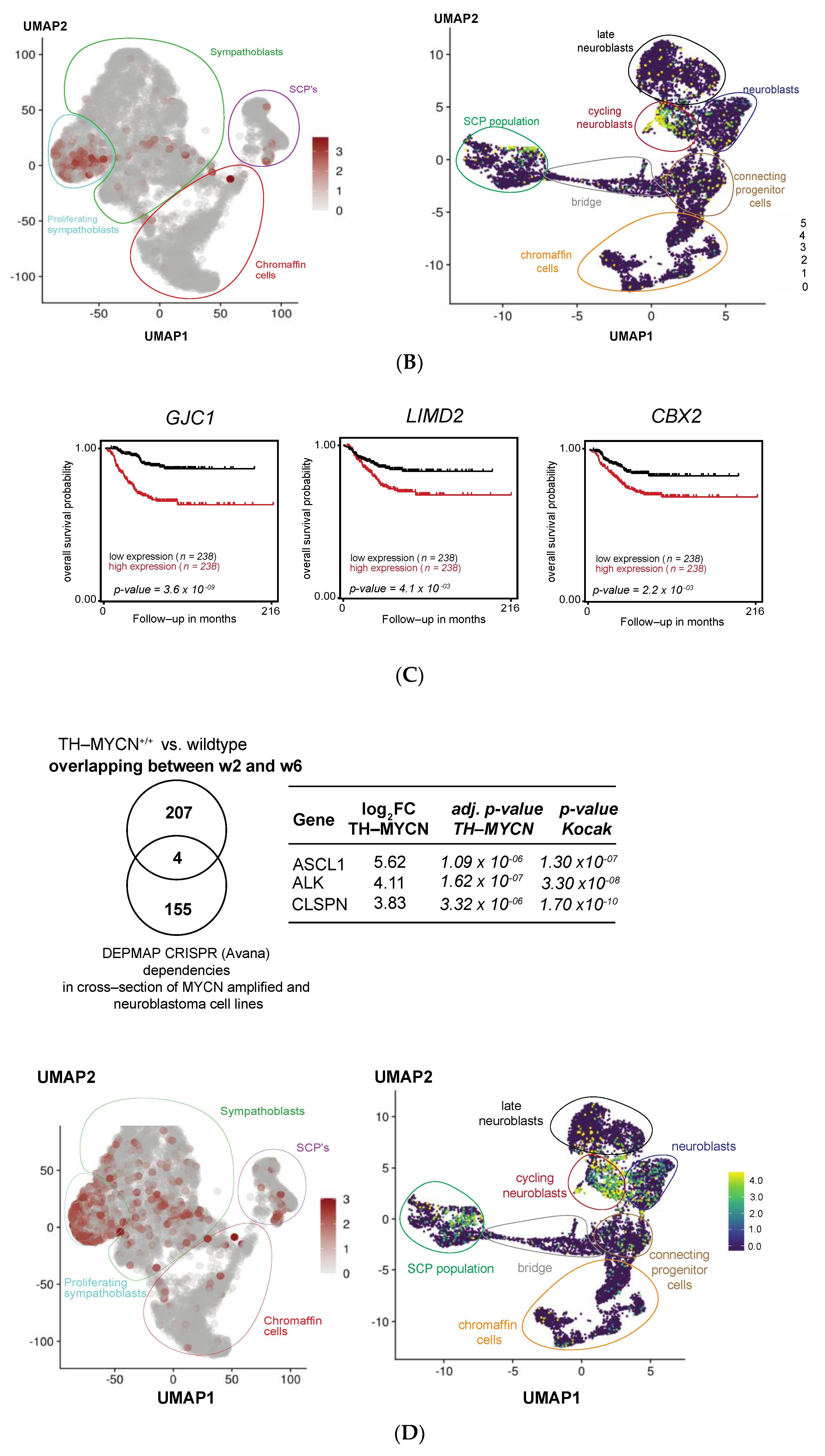
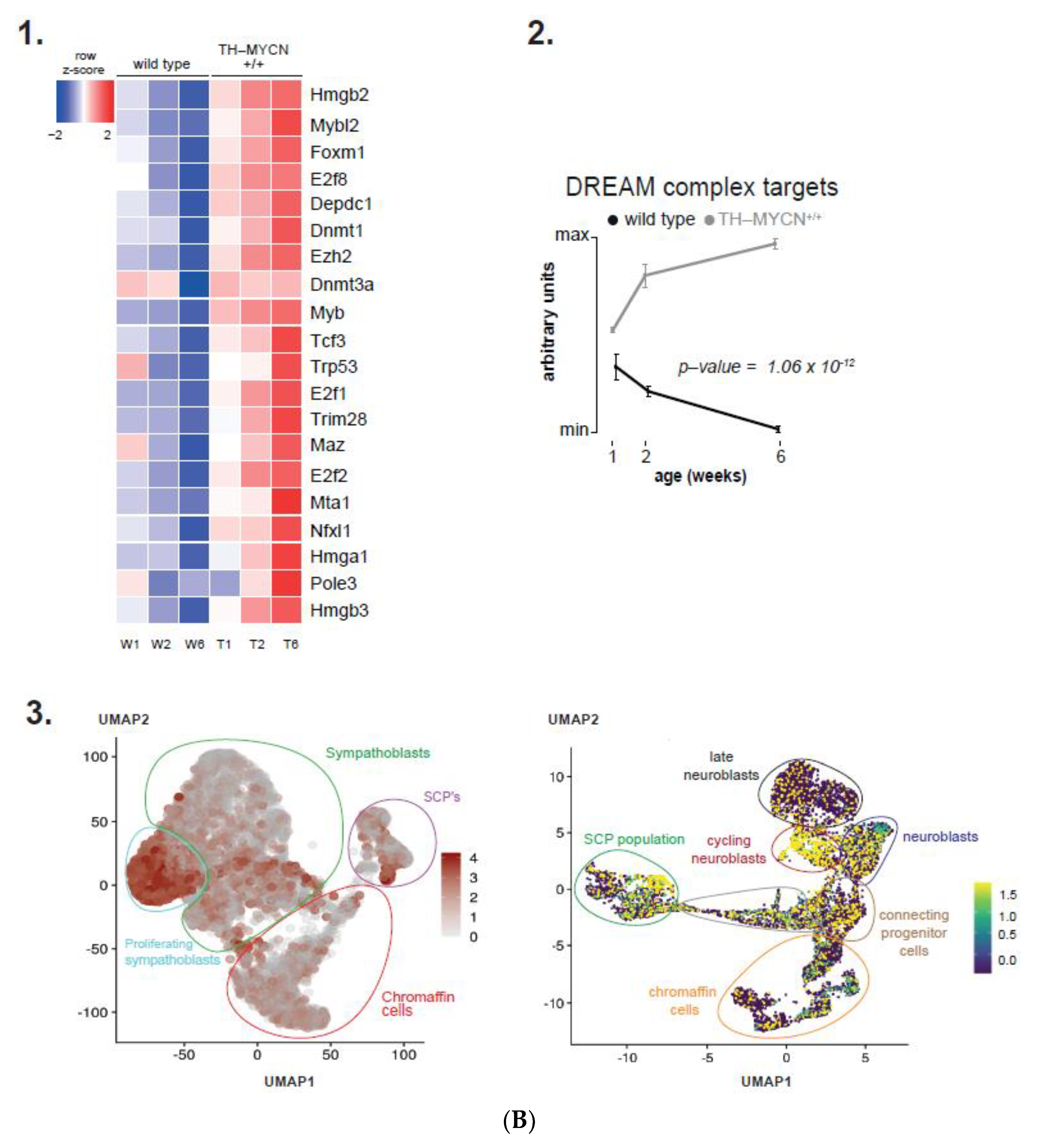
3.1. Validation of Dynamic Gene Regulation during Murine TH-MYCN Tumor Development Using Established Neuroblastoma Gene Signatures
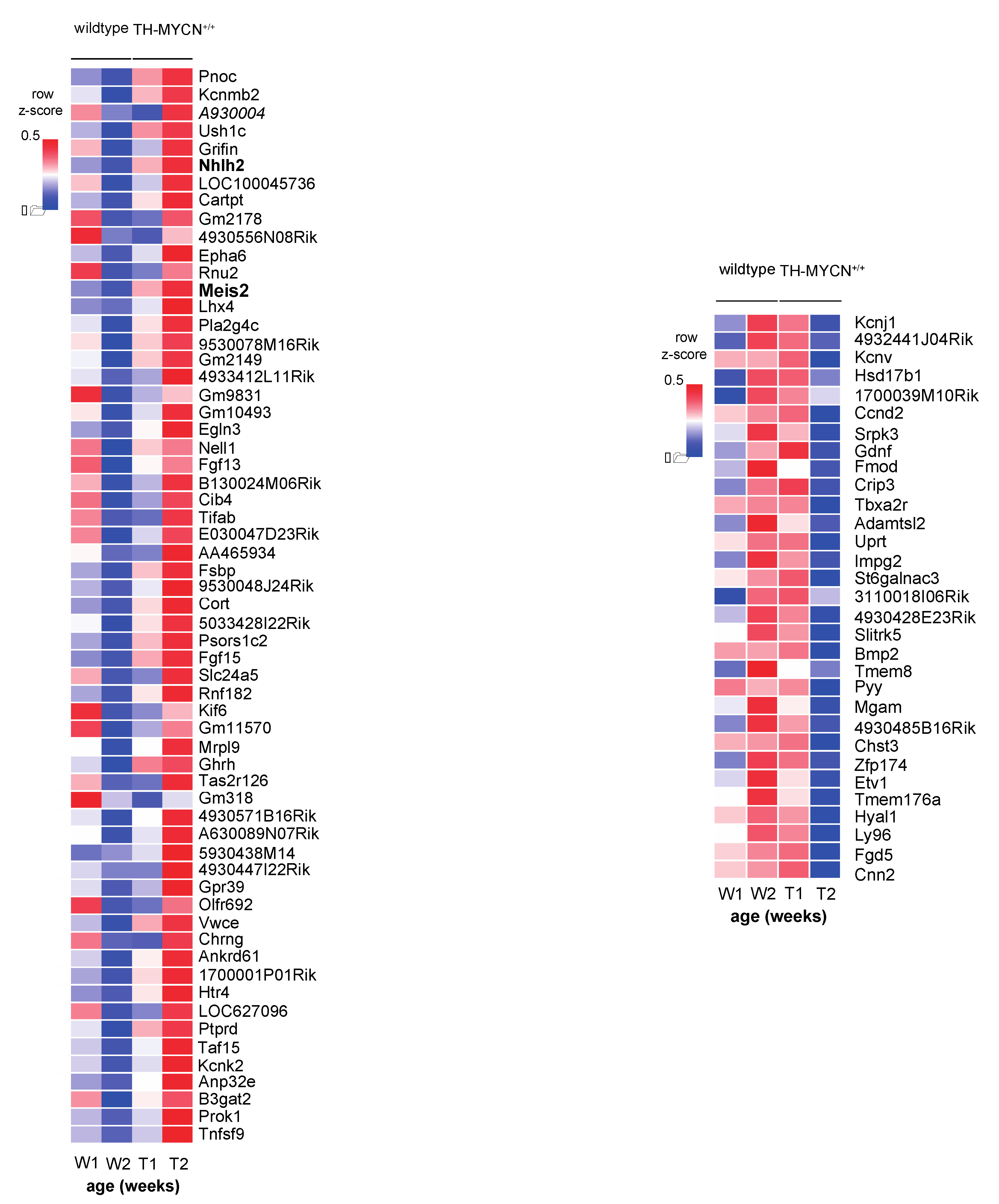
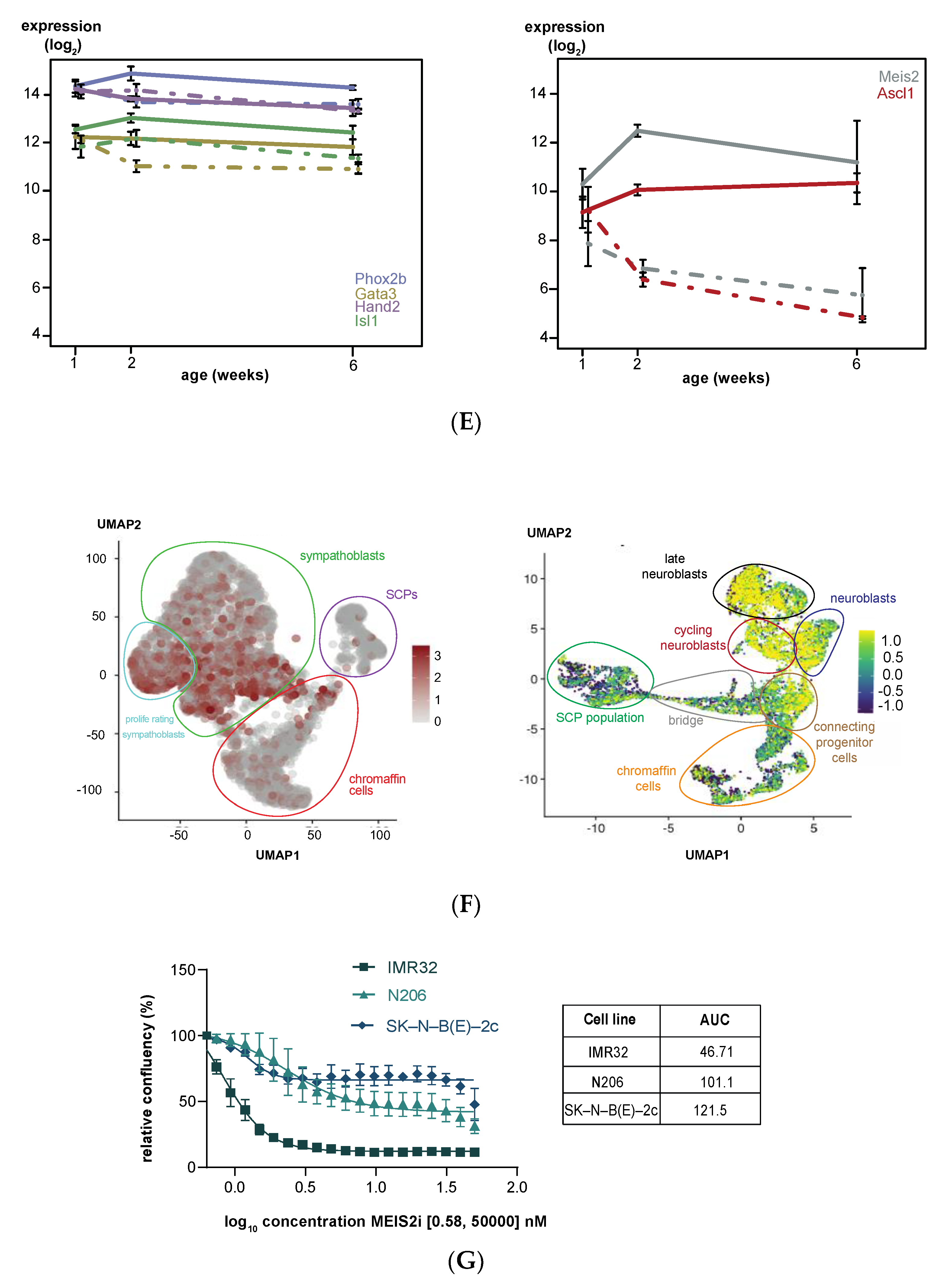
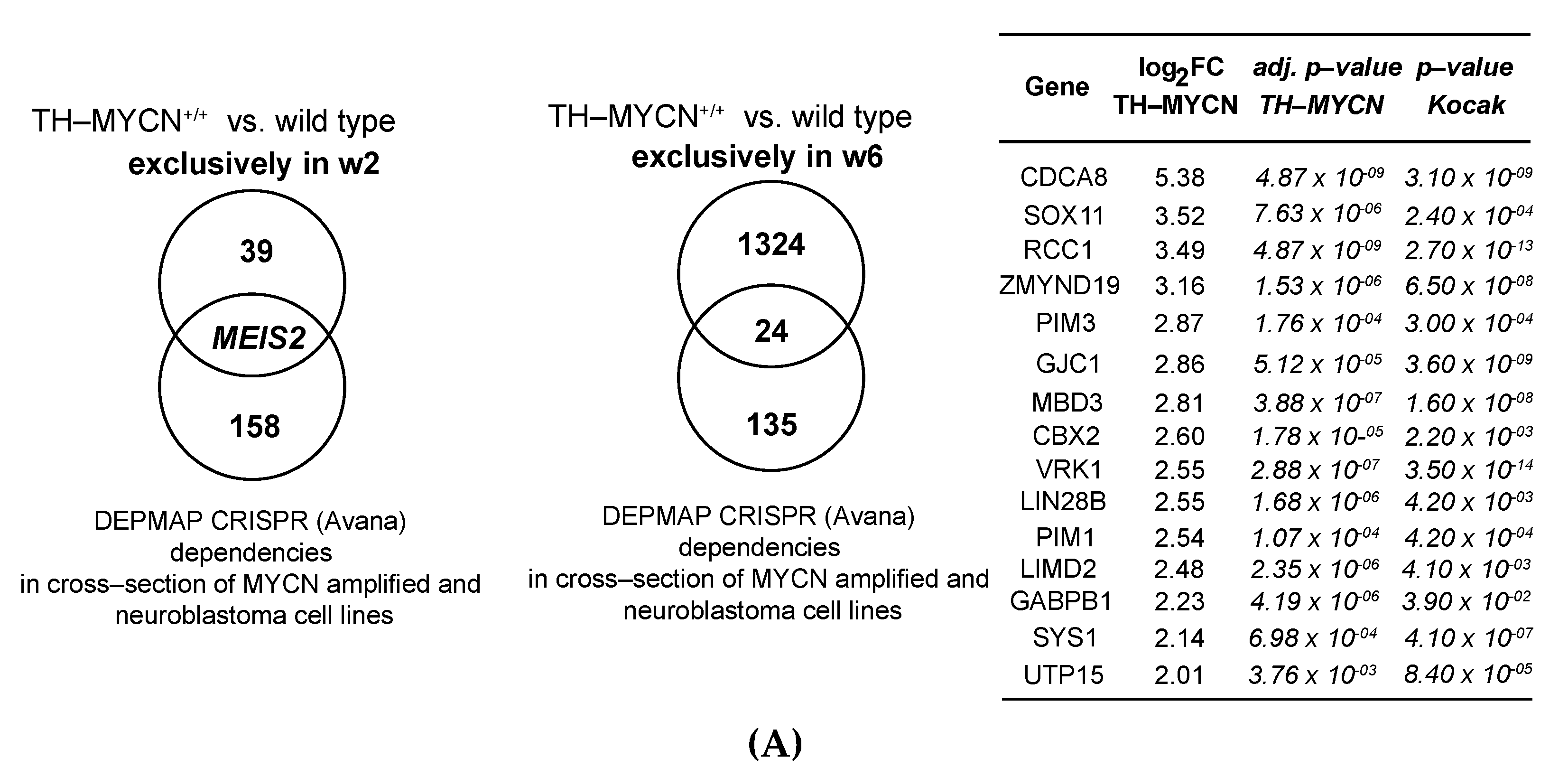
3.2. Upregulated Expression of ASCL1 and MEIS2 as Candidate Early Initiating Events in Murine TH-MYCN Tumor Development
3.3. KEOPS Protein Complex Member LAGE3 Expression Is Strongly Upregulated towards Full-Blown Murine TH-MYCN Tumor Formation
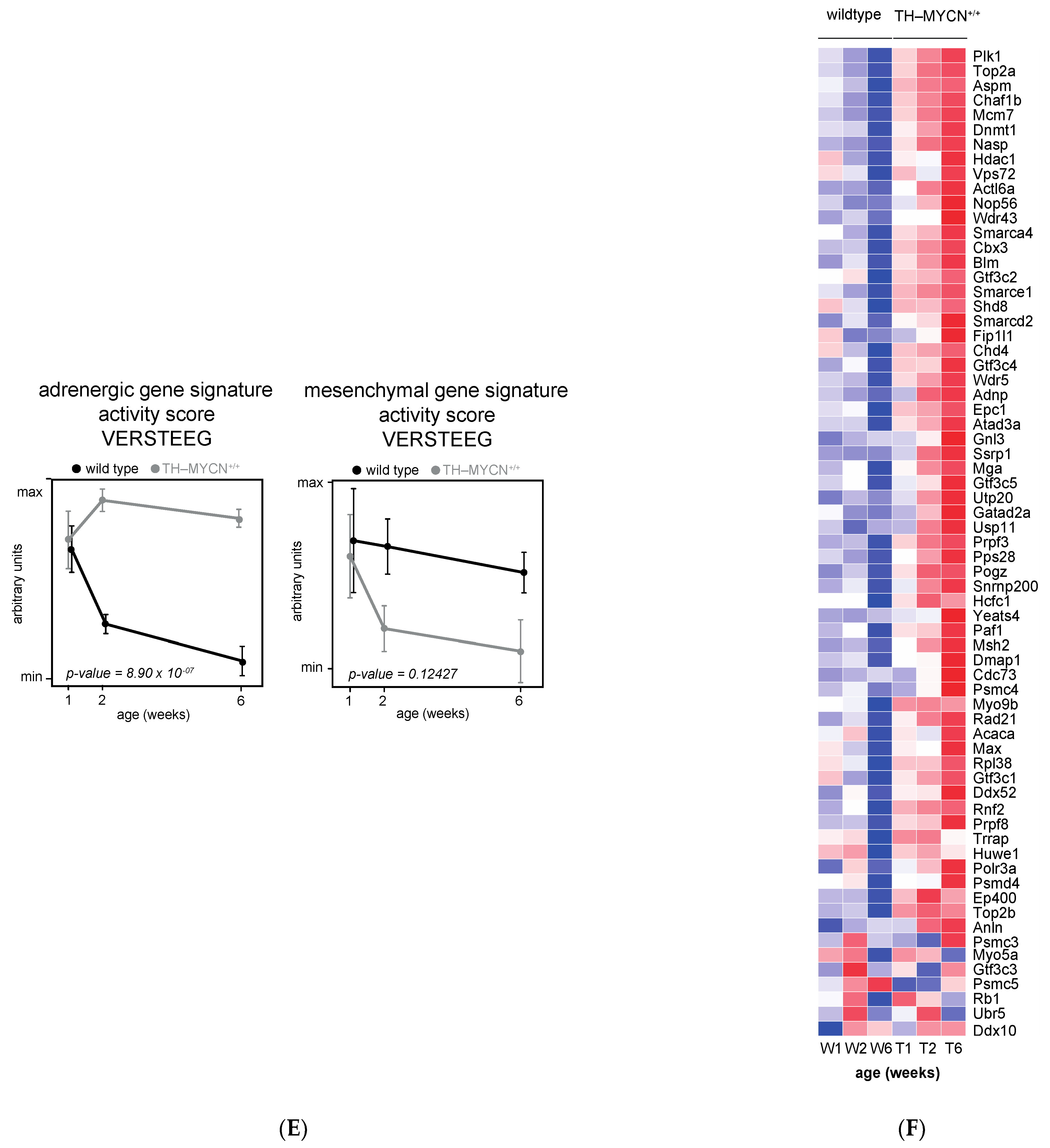
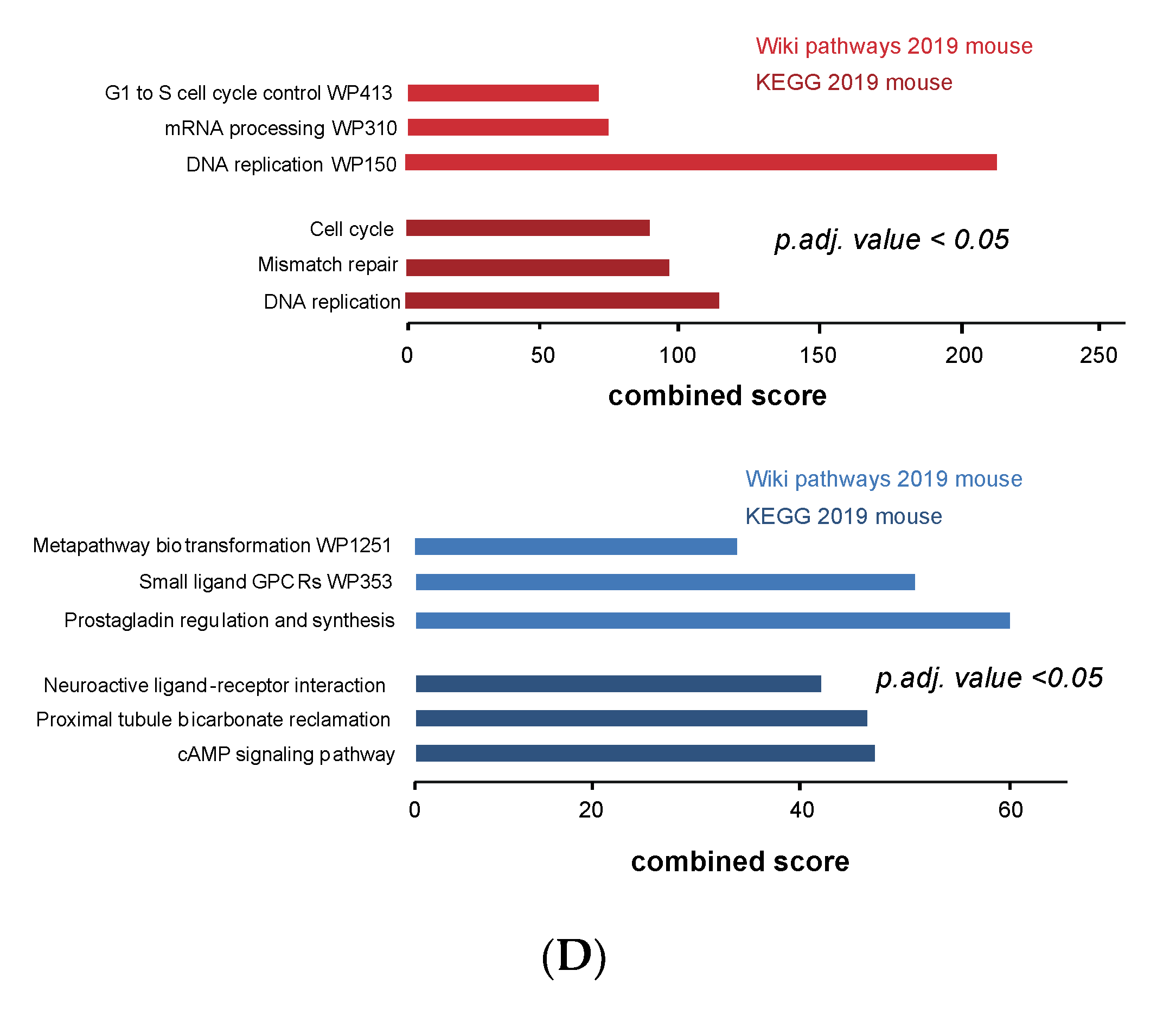
3.4. Cell Cycle and DNA Repair Genes Are Upregulated Late during TH-MYCN-Driven Tumor Formation
3.5. MEIS2 and Other MYCN-Driven Dependency Genes Are Upregulated during Murine TH-MYCN-Driven Neuroblastoma Tumor Formation
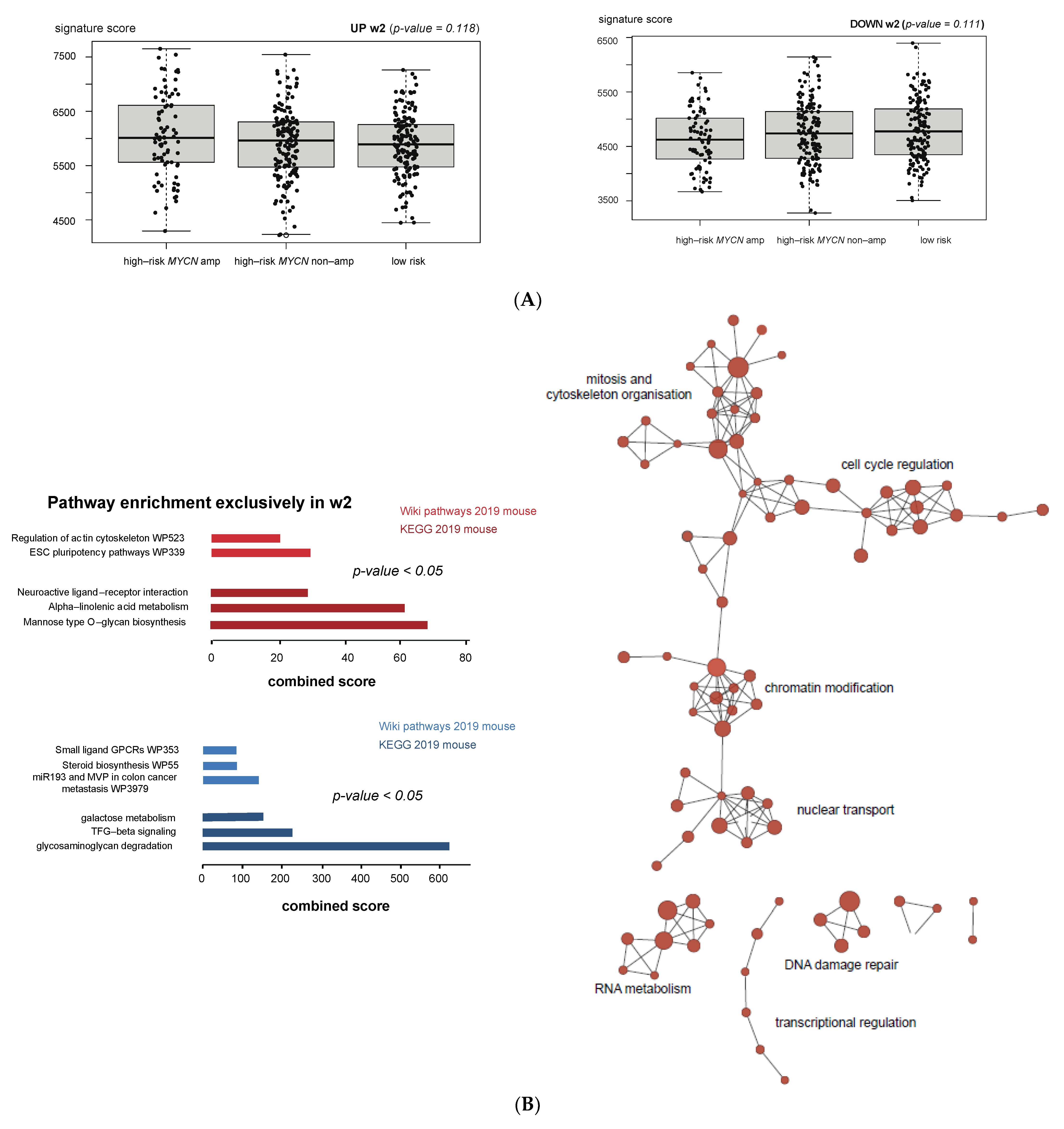
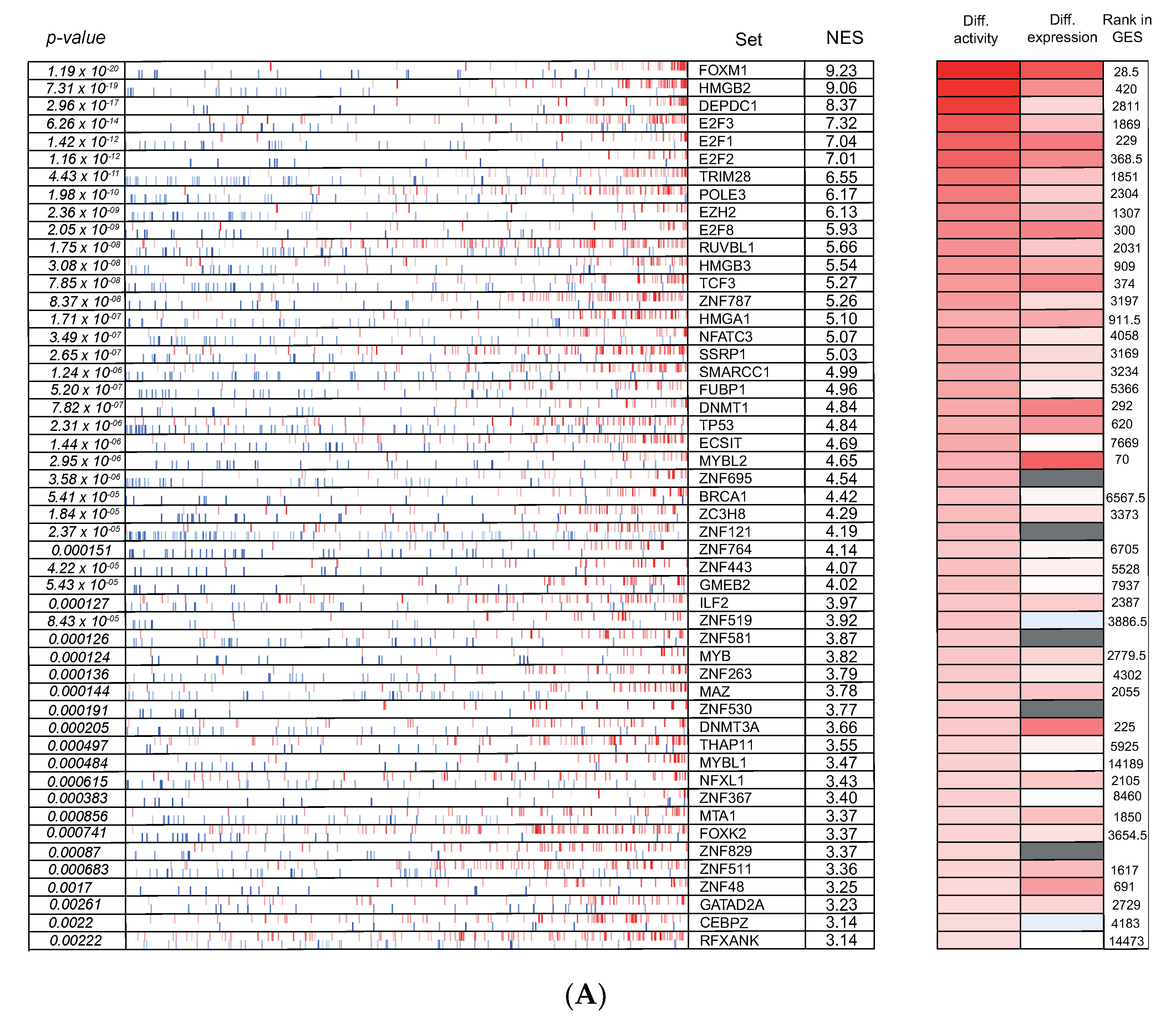
3.6. Cross-Species Master Regulator Analysis in MYCN-Driven Neuroblastoma
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Matthay, K.K.; Maris, J.M.; Schleiermacher, G.; Nakagawara, A.; Mackall, C.L.; Diller, L.; Weiss, W. Neuroblastoma. Nat. Rev. Dis. Prim. 2016, 2, 16078. [Google Scholar] [CrossRef]
- Bown, N.; Cotterill, S.; Lastowska, M.; O’Neill, S.; Pearson, A.D.J.; Plantaz, D.; Meddeb, M.; Danglot, G.; Brinkschmidt, C.; Christiansen, H.; et al. Gain of Chromosome Arm 17q and Adverse Outcome in Patients with Neuroblastoma. N. Engl. J. Med. 1999, 340, 1954–1961. [Google Scholar] [CrossRef]
- Molenaar, J.J.; Domingo-Fernández, R.; Ebus, M.E.; Lindner, S.; Koster, J.; Drabek, K.; Mestdagh, P.; Van Sluis, P.; Valentijn, L.J.; van Nes, J.; et al. LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat. Genet. 2012, 44, 1199–1206. [Google Scholar] [CrossRef] [PubMed]
- Depuydt, P.; Boeva, V.; Hocking, T.D.; Cannoodt, R.; Ambros, I.M.; Ambros, P.F.; Asgharzadeh, S.; Attiyeh, E.F.; Combaret, V.; Defferrari, R.; et al. Genomic Amplifications and Distal 6q Loss: Novel Markers for Poor Survival in High-risk Neuroblastoma Patients. J. Natl. Cancer Inst. 2018, 110, 1084–1093. [Google Scholar] [CrossRef] [PubMed]
- Brady, S.W.; Liu, Y.; Ma, X.; Gout, A.M.; Hagiwara, K.; Zhou, X.; Wang, J.; Macias, M.; Chen, X.; Easton, J.; et al. Pan-neuroblastoma analysis reveals age- and signature-associated driver alterations. Nat. Commun. 2020, 11, 5183. [Google Scholar] [CrossRef] [PubMed]
- Althoff, K.; Beckers, A.; Bell, E.; Nortmeyer, M.; Thor, T.; Sprüssel, A.; Lindner, S.; De Preter, K.; Florin, A.; Heukamp, L.; et al. A Cre-conditional MYCN-driven neuroblastoma mouse model as an improved tool for preclinical studies. Oncogene 2014, 34, 3357–3368. [Google Scholar] [CrossRef]
- Heukamp, L.C.; Thor, T.; Schramm, A.; De Preter, K.; Kumps, C.; De Wilde, B.; Odersky, A.; Peifer, M.; Lindner, S.; Spruessel, A.; et al. Targeted Expression of Mutated ALK Induces Neuroblastoma in Transgenic Mice. Sci. Transl. Med. 2012, 4, 141ra91. [Google Scholar] [CrossRef]
- Zhu, S.; Zhang, X.; Weichert-Leahey, N.; Dong, Z.; Zhang, C.; Lopez, G.; Tao, T.; He, S.; Wood, A.C.; Oldridge, D.; et al. LMO1 Synergizes with MYCN to Promote Neuroblastoma Initiation and Metastasis. Cancer Cell 2017, 32, 310–323.e5. [Google Scholar] [CrossRef]
- Weiss, W.; Aldape, K.; Mohapatra, G.; Feuerstein, B.G.; Bishop, J. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997, 16, 2985–2995. [Google Scholar] [CrossRef]
- Calao, M.; Sekyere, E.O.; Cui, H.J.; Cheung, B.B.; Thomas, W.D.; Keating, J.; Chen, J.; Raif, A.; Jankowski, K.; Davies, N.P.; et al. Direct effects of Bmi1 on p53 protein stability inactivates oncoprotein stress responses in embryonal cancer precursor cells at tumor initiation. Oncogene 2013, 32, 3616–3626. [Google Scholar] [CrossRef]
- Hansford, L.M.; Thomas, W.D.; Keating, J.M.; Burkhart, C.A.; Peaston, A.; Norris, M.; Haber, M.; Armati, P.J.; Weiss, W.; Marshall, G.M. Mechanisms of embryonal tumor initiation: Distinct roles for MycN expression and MYCN amplification. Proc. Natl. Acad. Sci. USA 2004, 101, 12664–12669. [Google Scholar] [CrossRef]
- Alam, G.; Cui, H.; Shi, H.; Yang, L.; Ding, J.; Mao, L.; Maltese, W.A.; Ding, H.-F. MYCN Promotes the Expansion of Phox2B-Positive Neuronal Progenitors to Drive Neuroblastoma Development. Am. J. Pathol. 2009, 175, 856–866. [Google Scholar] [CrossRef] [PubMed]
- Marshall, G.M.; Carter, D.R.; Cheung, B.B.; Liu, T.; Mateos, M.K.; Meyerowitz, J.G.; Weiss, W.A. The prenatal origins of cancer. Nat. Rev. Cancer 2014, 14, 277–289. [Google Scholar] [CrossRef] [PubMed]
- Beckers, A.; Van Peer, G.; Carter, D.R.; Gartlgruber, M.; Herrmann, C.; Agarwal, S.; Helsmoortel, H.H.; Althoff, K.; Molenaar, J.J.; Cheung, B.B.; et al. MYCN-driven regulatory mechanisms controlling LIN28B in neuroblastoma. Cancer Lett. 2015, 366, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Beckers, A.; Van Peer, G.; Carter, D.R.; Mets, E.; Althoff, K.; Cheung, B.B.; Schulte, J.H.; Mestdagh, P.; Vandesompele, J.; Marshall, G.M.; et al. MYCN-targeting miRNAs are predominantly downregulated during MYCN-driven neuroblastoma tumor formation. Oncotarget 2014, 6, 5204–5216. [Google Scholar] [CrossRef]
- Carter, D.R.; Murray, J.; Cheung, B.B.; Gamble, L.; Koach, J.; Tsang, J.; Sutton, S.; Kalla, H.; Syed, S.; Gifford, A.J.; et al. Therapeutic targeting of the MYC signal by inhibition of histone chaperone FACT in neuroblastoma. Sci. Transl. Med. 2015, 7, 312ra176. [Google Scholar] [CrossRef]
- Carter, D.R.; Sutton, S.K.; Pajic, M.; Murray, J.; Sekyere, E.O.; Fletcher, J.; Beckers, A.; De Preter, K.; Speleman, F.; George, R.E.; et al. Glutathione biosynthesis is upregulated at the initiation of MYCN-driven neuroblastoma tumorigenesis. Mol. Oncol. 2016, 10, 866–878. [Google Scholar] [CrossRef]
- Koach, J.; Holien, J.; Massudi, H.; Carter, D.R.; Ciampa, O.C.; Herath, M.; Lim, T.; Seneviratne, J.A.; Milazzo, G.; Murray, J.E.; et al. Drugging MYCN Oncogenic Signaling through the MYCN-PA2G4 Binding Interface. Cancer Res. 2019, 79, 5652–5667. [Google Scholar] [CrossRef]
- Nagy, Z.; Seneviratne, J.A.; Kanikevich, M.; Chang, W.; Mayoh, C.; Venkat, P.; Du, Y.; Jiang, C.; Salib, A.; Koach, J.; et al. An ALYREF-MYCN coactivator complex drives neuroblastoma tumorigenesis through effects on USP3 and MYCN stability. Nat. Commun. 2021, 12, 1881. [Google Scholar] [CrossRef]
- Ooi, C.Y.; Carter, D.R.; Liu, B.; Mayoh, C.; Beckers, A.; Lalwani, A.; Nagy, Z.; De Brouwer, S.; Decaesteker, B.; Hung, T.; et al. Network Modeling of microRNA–mRNA Interactions in Neuroblastoma Tumorigenesis Identifies miR-204 as a Direct Inhibitor of MYCN. Cancer Res. 2018, 78, 3122–3134. [Google Scholar] [CrossRef]
- Lambertz, I.; Kumps, C.; Claeys, S.; Lindner, S.; Beckers, A.; Janssens, E.; Carter, D.R.; Cazes, A.; Cheung, B.B.; De Mariano, M.; et al. Upregulation of MAPK Negative Feedback Regulators and RET in Mutant ALK Neuroblastoma: Implications for Targeted Treatment. Clin. Cancer Res. 2015, 21, 3327–3339. [Google Scholar] [CrossRef]
- R2 Platform. Available online: http://r2.amc.nl (accessed on 22 September 2021).
- Shiny App Hyperplasia Time Series. Available online: https://shiny.dev.cmgg.be/app/01_hyperplasia_time_series (accessed on 22 September 2021).
- Enrichr. Available online: https://maayanlab.cloud/Enrichr/ (accessed on 22 September 2021).
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Sanda, T.; Lawton, L.N.; Barrasa, M.I.; Fan, Z.P.; Kohlhammer, H.; Gutierrez, A.; Ma, W.; Tatarek, J.; Ahn, Y.; Kelliher, M.A.; et al. Core Transcriptional Regulatory Circuit Controlled by the TAL1 Complex in Human T Cell Acute Lymphoblastic Leukemia. Cancer Cell 2012, 22, 209–221. [Google Scholar] [CrossRef]
- Mansour, M.; Abraham, B.; Anders, L.; Berezovskaya, A.; Gutierrez, A.; Durbin, A.; Etchin, J.; Lawton, L.; Sallan, S.E.; Silverman, L.B.; et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 2014, 346, 1373–1377. [Google Scholar] [CrossRef]
- Durbin, A.D.; Zimmerman, M.W.; Dharia, N.V.; Abraham, B.J.; Iniguez, A.B.; Weichert-Leahey, N.; He, S.; Krill-Burger, J.M.; Root, D.E.; Vazquez, F.; et al. Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat. Genet. 2018, 50, 1240–1246. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Turan, R.D.; Albayrak, E.; Uslu, M.; Siyah, P.; Alyazici, L.Y.; Kalkan, B.M.; Aslan, G.S.; Yucel, D.; Aksoz, M.; Tuysuz, E.C.; et al. Development of Small Molecule MEIS Inhibitors that modulate HSC activity. Sci. Rep. 2020, 10, 7994. [Google Scholar] [CrossRef]
- Janky, R.; Verfaillie, A.; Imrichova, H.; Van de Sande, B.; Standaert, L.; Christiaens, V.; Hulselmans, G.; Herten, K.; Sanchez, M.N.; Potier, D.; et al. iRegulon: From a Gene List to a Gene Regulatory Network Using Large Motif and Track Collections. PLoS Comput. Biol. 2014, 10, e1003731. [Google Scholar] [CrossRef]
- Alvarez, M.J.; Shen, M.J.A.Y.; Giorgi, F.; Lachmann, A.; Ding, B.B.; Ye, B.B.D.B.H.; Califano, A. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat. Genet. 2016, 48, 838–847. [Google Scholar] [CrossRef]
- De Brouwer, S.; Mestdagh, P.; Lambertz, I.; Pattyn, F.; De Paepe, A.; Westermann, F.; Schroeder, C.; Schulte, J.H.; Schramm, A.; De Preter, K.; et al. Dickkopf-3 is regulated by the MYCN-induced miR-17-92 cluster in neuroblastoma. Int. J. Cancer 2011, 130, 2591–2598. [Google Scholar] [CrossRef]
- Fujita, T.; Igarashi, J.; Okawa, E.R.; Gotoh, T.; Manne, J.; Kolla, V.; Kim, J.; Zhao, H.; Pawel, B.R.; London, W.B.; et al. CHD5, a Tumor Suppressor Gene Deleted From 1p36.31 in Neuroblastomas. J. Natl. Cancer Inst. 2008, 100, 940–949. [Google Scholar] [CrossRef]
- Fredlund, E.; Ringner, M.; Maris, J.M.; Pahlman, S. High Myc pathway activity and low stage of neuronal differentiation associate with poor outcome in neuroblastoma. Proc. Natl. Acad. Sci. USA 2008, 105, 14094–14099. [Google Scholar] [CrossRef]
- Valentijn, L.J.; Koster, J.; Haneveld, F.; Aissa, R.A.; van Sluis, P.; Broekmans, M.E.C.; Molenaar, J.J.; van Nes, J.; Versteeg, R. Functional MYCN signature predicts outcome of neuroblastoma irrespective of MYCN amplification. Proc. Natl. Acad. Sci. USA 2012, 109, 19190–19195. [Google Scholar] [CrossRef]
- Van Groningen, T.; Koster, J.; Valentijn, L.J.; Zwijnenburg, D.A.; Akogul, N.; Hasselt, N.E.; Broekmans, M.; Haneveld, F.; Nowakowska, N.E.; Bras, J.; et al. Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat. Genet. 2017, 49, 1261–1266. [Google Scholar] [CrossRef]
- van Groningen, T.; Niklasson, C.U.; Chan, A.; Akogul, N.; Westerhout, E.M.; von Stedingk, K.; Hamdi, M.; Valentijn, L.J.; Mohlin, S.; Stroeken, P.; et al. An immature subset of neuroblastoma cells synthesizes retinoic acid and depends on this metabolite. bioRxiv 2021, 444639. [Google Scholar] [CrossRef]
- Baluapuri, A.; Wolf, E.; Eilers, M. Target gene-independent functions of MYC oncoproteins. Nat. Rev. Mol. Cell Biol. 2020, 21, 255–267. [Google Scholar] [CrossRef]
- Xiao, D.; Yue, M.; Su, H.; Ren, P.; Jiang, J.; Li, F.; Hu, Y.; Du, H.-N.; Liu, H.; Qing, G. Polo-like Kinase-1 Regulates Myc Stabilization and Activates a Feedforward Circuit Promoting Tumor Cell Survival. Mol. Cell 2016, 64, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Büchel, G.; Carstensen, A.; Mak, K.-Y.; Roeschert, I.; Leen, E.; Sumara, O.; Hofstetter, J.; Herold, S.; Kalb, J.; Baluapuri, A.; et al. Association with Aurora-A Controls N-MYC-Dependent Promoter Escape and Pause Release of RNA Polymerase II during the Cell Cycle. Cell Rep. 2017, 21, 3483–3497. [Google Scholar] [CrossRef] [PubMed]
- Olsen, R.R.; Otero, J.H.; Lopez, J.G.; Wallace, K.; Finkelstein, D.; Rehg, J.E.; Yin, Z.; Wang, Y.-D.; Freeman, K.W. MYCN induces neuroblastoma in primary neural crest cells. Oncogene 2017, 36, 5075–5082. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, M.; Ozaki, T.; Inuzuka, H.; Tomotsune, D.; Hirato, J.; Okamoto, Y.; Tokita, H.; Ohira, M.; Nakagawara, A. LMO3 Interacts with Neuronal Transcription Factor, HEN2, and Acts as an Oncogene in Neuroblastoma. Cancer Res. 2005, 65, 4587–4597. [Google Scholar] [CrossRef]
- Wang, L.; Tan, T.K.; Durbin, A.D.; Zimmerman, M.W.; Abraham, B.J.; Tan, S.H.; Ngoc, P.C.T.; Weichert-Leahey, N.; Akahane, K.; Lawton, L.N.; et al. ASCL1 is a MYCN- and LMO1-dependent member of the adrenergic neuroblastoma core regulatory circuitry. Nat. Commun. 2019, 10, 5622. [Google Scholar] [CrossRef]
- Geerts, D.; Schilderink, N.; Jorritsma, G.; Versteeg, R. The role of the MEIS homeobox genes in neuroblastoma. Cancer Lett. 2003, 197, 87–92. [Google Scholar] [CrossRef]
- Zha, Y.; Xia, Y.; Ding, J.; Choi, J.-H.; Yang, L.; Dong, Z.; Yan, C.; Huang, S.; Ding, H.-F. MEIS2 is essential for neuroblastoma cell survival and proliferation by transcriptional control of M-phase progression. Cell Death Dis. 2014, 5, e1417. [Google Scholar] [CrossRef]
- Zeid, R.; Lawlor, M.; Poon, E.; Reyes, J.; Fulciniti, M.; Lopez, M.A.; Scott, T.; Nabet, B.; Erb, M.A.; Winter, G.E.; et al. Enhancer invasion shapes MYCN-dependent transcriptional amplification in neuroblastoma. Nat. Genet. 2018, 50, 515–523. [Google Scholar] [CrossRef]
- Kumps, C.; Fieuw, A.; Mestdagh, P.; Menten, B.; Lefever, S.; Pattyn, F.; De Brouwer, S.; Sante, T.; Schulte, J.H.; Schramm, A.; et al. Focal DNA Copy Number Changes in Neuroblastoma Target MYCN Regulated Genes. PLoS ONE 2013, 8, e52321. [Google Scholar] [CrossRef] [PubMed]
- Kameneva, P.; Artemov, A.V.; Kastriti, M.E.; Faure, L.; Olsen, T.K.; Otte, J.; Erickson, A.; Semsch, B.; Andersson, E.R.; Ratz, M.; et al. Single-cell transcriptomics of human embryos identifies multiple sympathoblast lineages with potential implications for neuroblastoma origin. Nat. Genet. 2021, 53, 694–706. [Google Scholar] [CrossRef] [PubMed]
- Jansky, S.; Sharma, A.K.; Körber, V.; Quintero, A.; Toprak, U.H.; Wecht, E.M.; Gartlgruber, M.; Greco, A.; Chomsky, E.; Grünewald, T.G.P.; et al. Single-cell transcriptomic analyses provide insights into the developmental origins of neuroblastoma. Nat. Genet. 2021, 53, 683–693. [Google Scholar] [CrossRef]
- Perrochia, L.; Guetta, D.; Hecker, A.; Forterre, P.; Basta, T. Functional assignment of KEOPS/EKC complex subunits in the biosynthesis of the universal t 6 A tRNA modification. Nucleic Acids Res. 2013, 41, 9484–9499. [Google Scholar] [CrossRef] [PubMed]
- He, M.-H.; Liu, J.-C.; Lu, Y.-S.; Wu, Z.-J.; Liu, Y.-Y.; Wu, Z.; Peng, J.; Zhou, J.-Q. KEOPS complex promotes homologous recombination via DNA resection. Nucleic Acids Res. 2019, 47, 5684–5697. [Google Scholar] [CrossRef] [PubMed]
- Downey, M.; Houlsworth, R.; Maringele, L.; Rollie, A.; Brehme, M.; Galicia, S.; Guillard, S.; Partington, M.; Zubko, M.K.; Krogan, N.J.; et al. A Genome-Wide Screen Identifies the Evolutionarily Conserved KEOPS Complex as a Telomere Regulator. Cell 2006, 124, 1155–1168. [Google Scholar] [CrossRef]
- El Faitwri, T.; Huber, K. Expression pattern of delta-like 1 homolog in developing sympathetic neurons and chromaffin cells. Gene Expr. Patterns 2018, 30, 49–54. [Google Scholar] [CrossRef]
- Ackermann, S.; Goeser, F.; Schulte, J.H.; Schramm, A.; Ehemann, V.; Hero, B.; Eggert, A.; Berthold, F.; Fischer, M. Polo-Like Kinase 1 is a Therapeutic Target in High-Risk Neuroblastoma. Clin. Cancer Res. 2011, 17, 731–741. [Google Scholar] [CrossRef]
- Chen, D.; Cox, J.; Annam, J.; Weingart, M.; Essien, G.; Rathi, K.S.; Rokita, J.L.; Khurana, P.; Cuya, S.M.; Bosse, K.R.; et al. LIN28B-PDZ Binding Kinase Signaling Promotes Neuroblastoma Metastasis. Neoplasia 2020, 22, 231–241. [Google Scholar] [CrossRef]
- Sultan, I.; Tbakhi, A. BCL11A gene over-expression in high risk neuroblastoma. Cancer Genet. 2020, 244, 30–31. [Google Scholar] [CrossRef]
- Niu, J.; Li, X.-M.; Wang, X.; Liang, C.; Zhang, Y.-D.; Li, H.-Y.; Liu, F.-Y.; Sun, H.; Xie, S.-Q.; Fang, D. DKK1 inhibits breast cancer cell migration and invasion through suppression of β-catenin/MMP7 signaling pathway. Cancer Cell Int. 2019, 19, 168. [Google Scholar] [CrossRef]
- Cecchi, C.; Liguri, G.; Fiorillo, C.; Bogani, F.; Gambassi, M.; Giannoni, E.; Cirri, P.; Baglioni, S.; Ramponi, G. Acylphosphatase overexpression triggers SH-SY5Y differentiation towards neuronal phenotype. Cell. Mol. Life Sci. 2004, 61, 1775–1784. [Google Scholar] [CrossRef]
- Muñoz-Llancao, P.; De Gregorio, C.; Heras, M.L.; Meinohl, C.; Noorman, K.; Boddeke, E.; Cheng, X.; Lezoualc’H, F.; Schmidt, M.; Gonzalez-Billault, C. Microtubule-regulating proteins and cAMP-dependent signaling in neuroblastoma differentiation. Cytoskeleton 2017, 74, 143–158. [Google Scholar] [CrossRef] [PubMed]
- Meyers, R.M.; Bryan, J.G.; McFarland, J.M.; Weir, B.A.; Sizemore, A.E.; Xu, H.; Dharia, N.V.; Montgomery, P.G.; Cowley, G.S.; Pantel, S.; et al. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat. Genet. 2017, 49, 1779–1784. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Wang, X.; Su, D.; Su, X.; Lin, L.; Li, S.; Wu, Q.; Liu, S.; Zhang, P.; Zhu, X.; et al. CBX2 Inhibits Neurite Development by Regulating Neuron-Specific Genes Expression. Front. Mol. Neurosci. 2018, 11, 46. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, Z.; Xu, L.; Li, Y.; Li, Y.; Zhang, X.; Wang, Y.; Liu, D. LIMD2 targeted by miR-34a promotes the proliferation and invasion of non-small cell lung cancer cells. Mol. Med. Rep. 2018, 18, 4760–4766. [Google Scholar] [CrossRef]
- Cole, K.A.; Attiyeh, E.F.; Mosse, Y.P.; Laquaglia, M.J.; Diskin, S.J.; Brodeur, G.M.; Maris, J.M. A Functional Screen Identifies miR-34a as a Candidate Neuroblastoma Tumor Suppressor Gene. Mol. Cancer Res. 2008, 6, 735–742. [Google Scholar] [CrossRef]
- Mozos, A.; Royo, C.; Hartmann, E.; De Jong, D.; Baró, C.; Valera, A.; Fu, K.; Weisenburger, D.D.; Delabie, J.; Chuang, S.-S.; et al. SOX11 expression is highly specific for mantle cell lymphoma and identifies the cyclin D1-negative subtype. Haematologica 2009, 94, 1555–1562. [Google Scholar] [CrossRef] [PubMed]
- Decaesteker, B.; Louwagie, A.; Loontiens, S.; De Vloed, F.; Roels, J.; Vanhauwaert, S.; De Brouwer, S.; Sanders, E.; Denecker, G.; D’haene, E.; et al. SOX11 is a lineage-dependency factor and master epigenetic regulator in neuroblastoma. bioRxiv 2020, 261131. [Google Scholar] [CrossRef]
- Bianco, J.N.; Bergoglio, V.; Lin, Y.-L.; Pillaire, M.-J.; Schmitz, A.-L.; Gilhodes, J.; Lusque, A.; Mazières, J.; Lacroix-Triki, M.; Roumeliotis, T.I.; et al. Overexpression of Claspin and Timeless protects cancer cells from replication stress in a checkpoint-independent manner. Nat. Commun. 2019, 10, 910. [Google Scholar] [CrossRef] [PubMed]
- Dharia, N.V.; Kugener, G.; Guenther, L.M.; Malone, C.F.; Durbin, A.D.; Hong, A.L.; Howard, T.P.; Bandopadhayay, P.; Wechsler, C.S.; Fung, I.; et al. A first-generation pediatric cancer dependency map. Nat. Genet. 2021, 53, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Rajbhandari, P.; Lopez, G.; Capdevila, C.; Salvatori, B.; Yu, J.; Rodriguez-Barrueco, R.; Martinez, D.; Yarmarkovich, M.; Weichert-Leahey, N.; Abraham, B.J.; et al. Cross-Cohort Analysis Identifies a TEAD4–MYCN Positive Feedback Loop as the Core Regulatory Element of High-Risk Neuroblastoma. Cancer Discov. 2018, 8, 582–599. [Google Scholar] [CrossRef]
- Margolin, A.A.; Wang, K.; Lim, W.K.; Kustagi, M.; Nemenman, I.; Califano, A. Reverse engineering cellular networks. Nat. Protoc. 2006, 1, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Sadasivam, S.; DeCaprio, J.A. The DREAM complex: Master coordinator of cell cycle-dependent gene expression. Nat. Rev. Cancer 2013, 13, 585–595. [Google Scholar] [CrossRef]
- Engeland, K. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ. 2018, 25, 114–132. [Google Scholar] [CrossRef]
- Wapinski, O.L.; Vierbuchen, T.; Qu, K.; Lee, Q.Y.; Chanda, S.; Fuentes, D.; Giresi, P.G.; Ng, Y.H.; Marro, S.; Neff, N.F.; et al. Hierarchical Mechanisms for Direct Reprogramming of Fibroblasts to Neurons. Cell 2013, 155, 621–635. [Google Scholar] [CrossRef]
- Srinivasan, M.; Mehta, P.; Yu, Y.; Prugar, E.; Koonin, E.V.; Karzai, A.W.; Sternglanz, R. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J. 2010, 30, 873–881. [Google Scholar] [CrossRef]
- Bianchi, A.; Shore, D. The KEOPS Complex: A Rosetta Stone for Telomere Regulation? Cell 2006, 124, 1125–1128. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, A.; Sheltzer, J.M. Discovering and validating cancer genetic dependencies: Approaches and pitfalls. Nat. Rev. Genet. 2020, 21, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Murphy, D.M.; Buckley, P.G.; Bryan, K.; Watters, K.M.; Koster, J.; van Sluis, P.; Molenaar, J.; Versteeg, R.; Stallings, R.L. Dissection of the oncogenic MYCN transcriptional network reveals a large set of clinically relevant cell cycle genes as drivers of neuroblastoma tumorigenesis. Mol. Carcinog. 2011, 50, 403–411. [Google Scholar] [CrossRef]
- Trigg, R.M.; Lee, L.C.; Prokoph, N.; Jahangiri, L.; Reynolds, C.P.; Burke, G.A.A.; Probst, N.A.; Han, M.; Matthews, J.; Lim, H.K.; et al. The targetable kinase PIM1 drives ALK inhibitor resistance in high-risk neuroblastoma independent of MYCN status. Nat. Commun. 2019, 10, 5428. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Wyn, J.; Zimmerman, M.W.; Weichert-Leahey, N.; Nunes, C.; Cheung, B.B.; Abraham, B.J.; Beckers, A.; Volders, P.-J.; Decaesteker, B.; Carter, D.R.; et al. MEIS2 Is an Adrenergic Core Regulatory Transcription Factor Involved in Early Initiation of TH-MYCN-Driven Neuroblastoma Formation. Cancers 2021, 13, 4783. https://doi.org/10.3390/cancers13194783
De Wyn J, Zimmerman MW, Weichert-Leahey N, Nunes C, Cheung BB, Abraham BJ, Beckers A, Volders P-J, Decaesteker B, Carter DR, et al. MEIS2 Is an Adrenergic Core Regulatory Transcription Factor Involved in Early Initiation of TH-MYCN-Driven Neuroblastoma Formation. Cancers. 2021; 13(19):4783. https://doi.org/10.3390/cancers13194783
Chicago/Turabian StyleDe Wyn, Jolien, Mark W. Zimmerman, Nina Weichert-Leahey, Carolina Nunes, Belamy B. Cheung, Brian J. Abraham, Anneleen Beckers, Pieter-Jan Volders, Bieke Decaesteker, Daniel R. Carter, and et al. 2021. "MEIS2 Is an Adrenergic Core Regulatory Transcription Factor Involved in Early Initiation of TH-MYCN-Driven Neuroblastoma Formation" Cancers 13, no. 19: 4783. https://doi.org/10.3390/cancers13194783
APA StyleDe Wyn, J., Zimmerman, M. W., Weichert-Leahey, N., Nunes, C., Cheung, B. B., Abraham, B. J., Beckers, A., Volders, P.-J., Decaesteker, B., Carter, D. R., Look, A. T., De Preter, K., Van Loocke, W., Marshall, G. M., Durbin, A. D., Speleman, F., & Durinck, K. (2021). MEIS2 Is an Adrenergic Core Regulatory Transcription Factor Involved in Early Initiation of TH-MYCN-Driven Neuroblastoma Formation. Cancers, 13(19), 4783. https://doi.org/10.3390/cancers13194783







