Microbe-Mediated Activation of Toll-like Receptor 2 Drives PDL1 Expression in HNSCC
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. PDL1 Induction
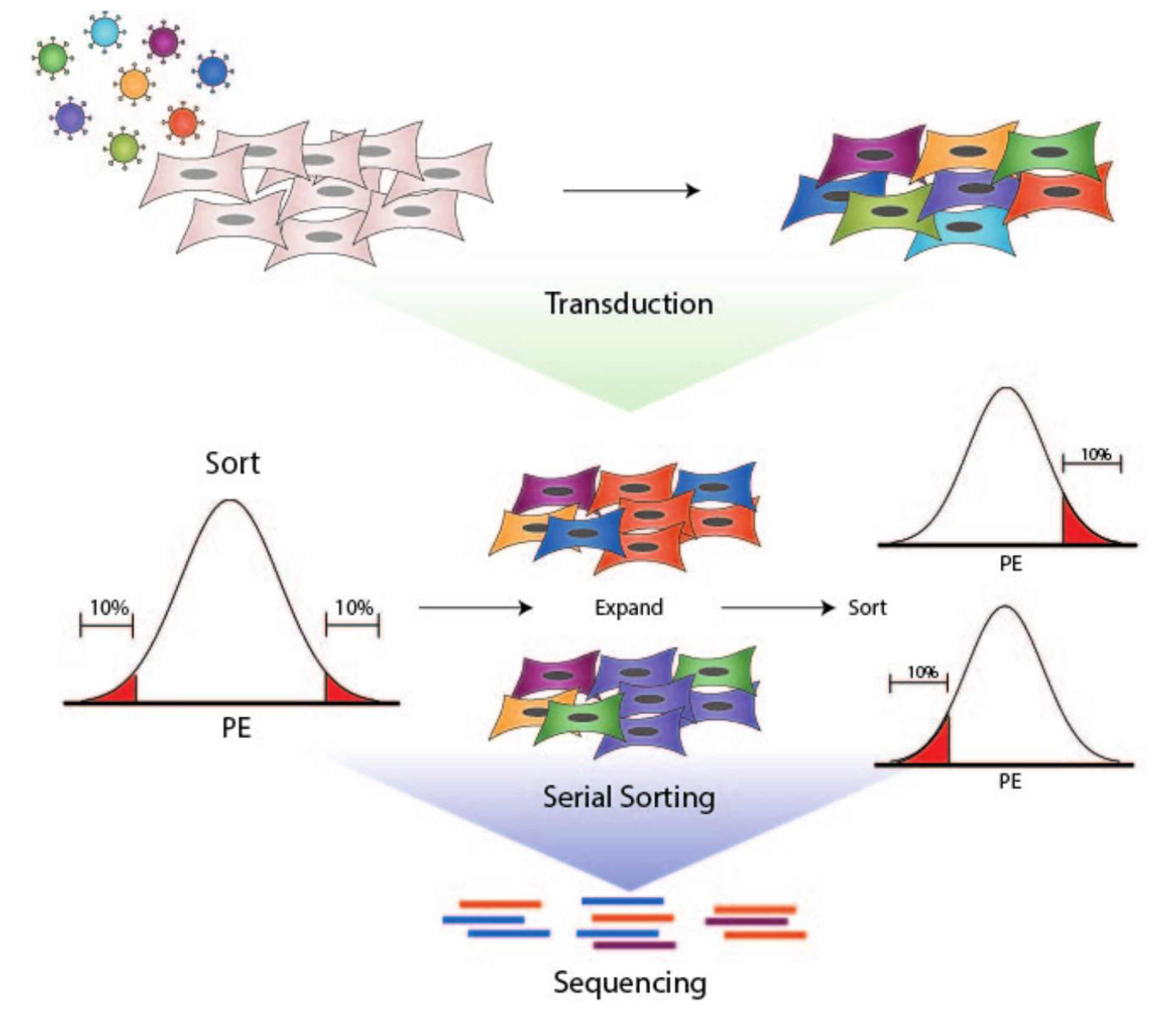
2.3. Transduction
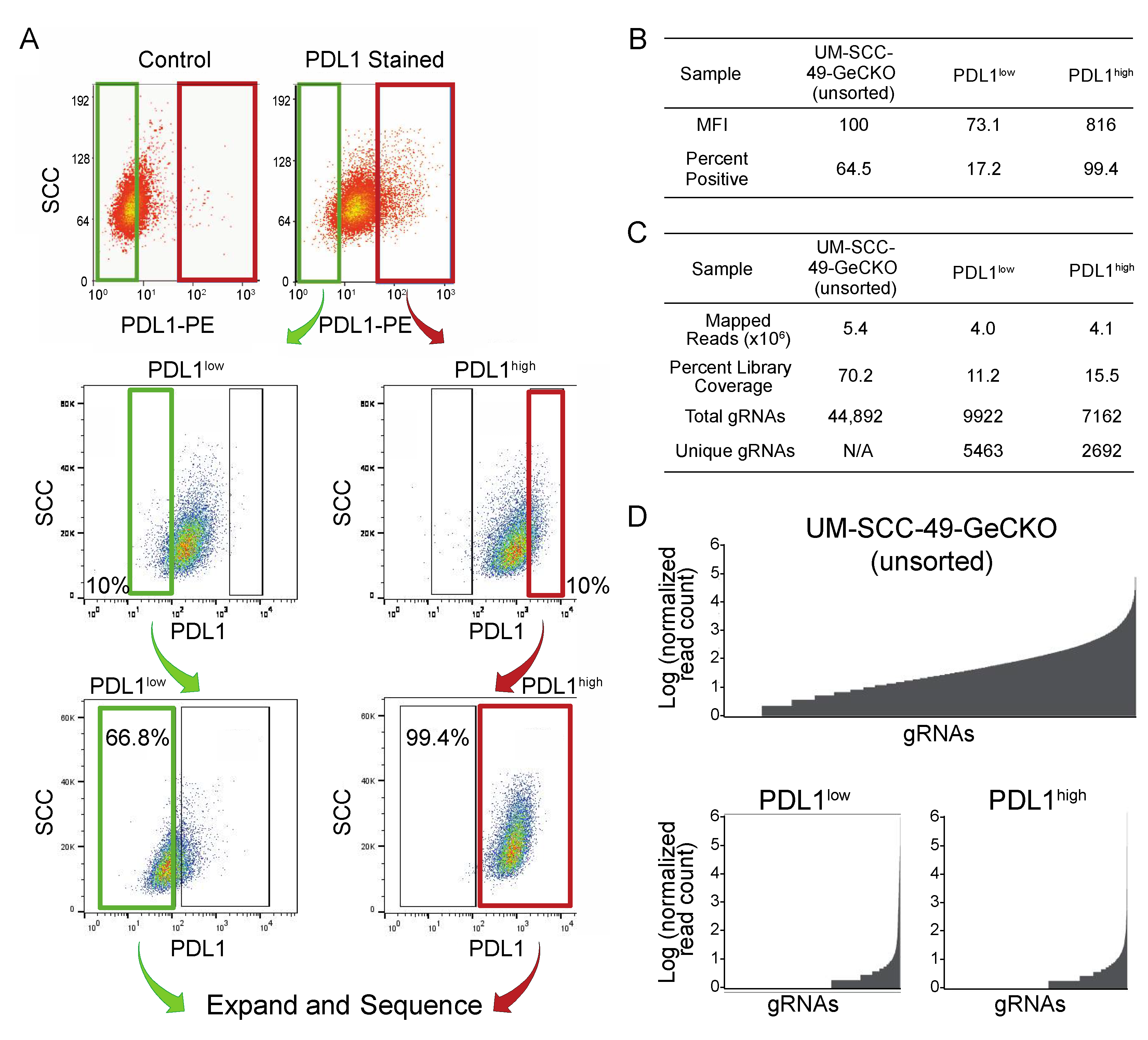
2.4. Fluorescence-Activated Cell Sorting
2.5. GeCKO Library Preparation
2.6. Analysis of CRISPR Libraries
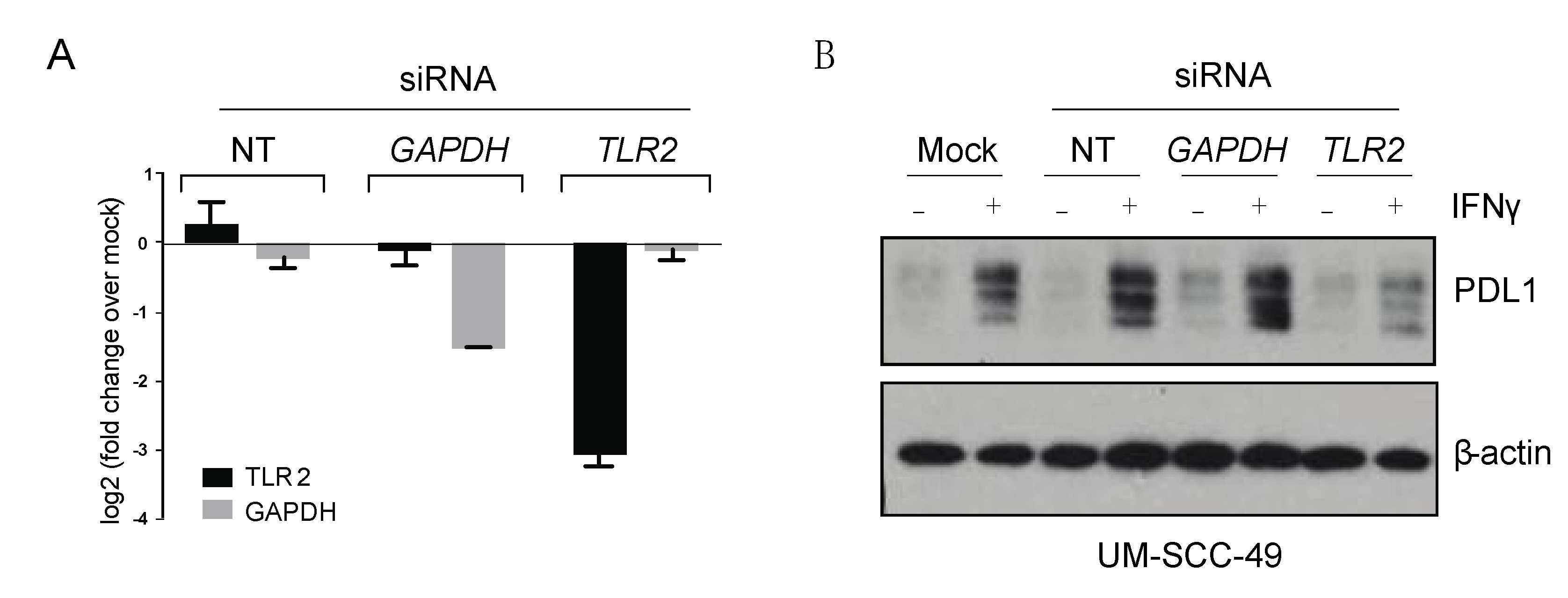
2.7. SiRNA Transfection
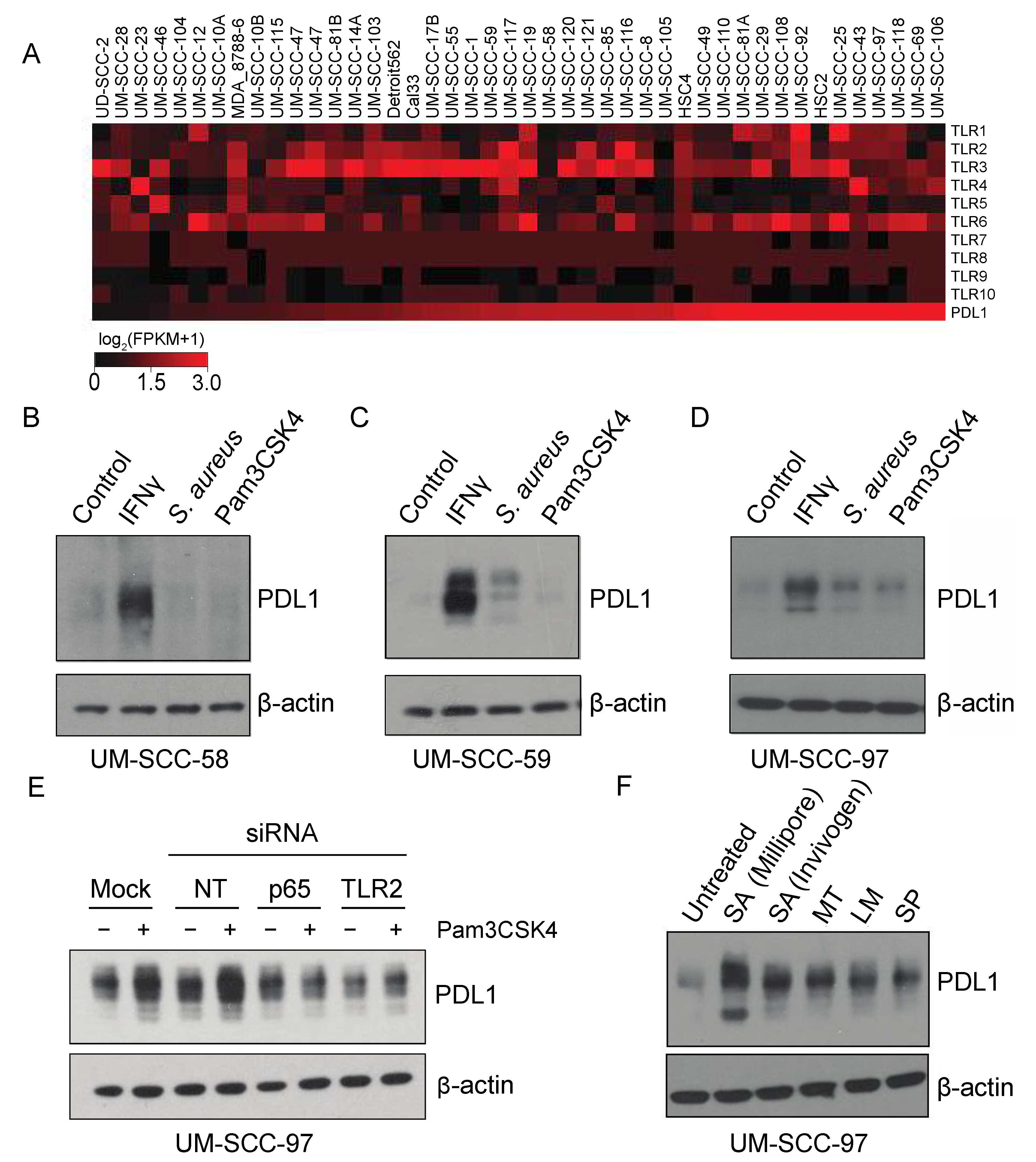
2.8. Western Blotting
2.9. qPCR
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef]
- Bauml, J.M.; Aggarwal, C.; Cohen, R.B. Immunotherapy for head and neck cancer: Where are we now and where are we going? Ann. Transl. Med. 2019, 7, S75. [Google Scholar] [CrossRef] [PubMed]
- Cohen, E.E.W.; Soulières, D.; Le Tourneau, C.; Dinis, J.; Licitra, L.; Ahn, M.J.; Soria, A.; Machiels, J.P.; Mach, N.; Mehra, R.; et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): A randomised, open-label, phase 3 study. Lancet 2019, 393, 156–167. [Google Scholar] [CrossRef]
- Ferris, R.L.; Licitra, L.; Fayette, J.; Even, C.; Blumenschein, G.; Harrington, K.J.; Guigay, J.; Vokes, E.E.; Saba, N.F.; Haddad, R.; et al. Nivolumab in Patients with Recurrent or Metastatic Squamous Cell Carcinoma of the Head and Neck: Efficacy and Safety in CheckMate 141 by Prior Cetuximab Use. Clin. Cancer Res. 2019, 25, 5221–5230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burtness, B.; Harrington, K.J.; Greil, R.; Soulières, D.; Tahara, M.; de Castro, G.; Psyrri, A.; Basté, N.; Neupane, P.; Bratland, Å.; et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): A randomised, open-label, phase 3 study. Lancet 2019, 394, 1915–1928. [Google Scholar] [CrossRef]
- Sun, C.; Mezzadra, R.; Schumacher, T.N. Regulation and Function of the PD-L1 Checkpoint. Immunity 2018, 48, 434–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Concha-Benavente, F.; Srivastava, R.M.; Trivedi, S.; Lei, Y.; Chandran, U.; Seethala, R.R.; Freeman, G.J.; Ferris, R.L. Identification of the Cell-Intrinsic and -Extrinsic Pathways Downstream of EGFR and IFNγ That Induce PD-L1 Expression in Head and Neck Cancer. Cancer Res. 2016, 76, 1031–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuster, A.; Erasimus, H.; Fritah, S.; Nazarov, P.V.; van Dyck, E.; Niclou, S.P.; Golebiewska, A. RNAi/CRISPR Screens: From a Pool to a Valid Hit. Trends Biotechnol. 2019, 37, 38–55. [Google Scholar] [CrossRef]
- Smith, I.; Greenside, P.G.; Natoli, T.; Lahr, D.L.; Wadden, D.; Tirosh, I.; Narayan, R.; Root, D.E.; Golub, T.R.; Subramanian, A.; et al. Evaluation of RNAi and CRISPR technologies by large-scale gene expression profiling in the Connectivity Map. PLoS Biol. 2017, 15, e2003213. [Google Scholar] [CrossRef] [Green Version]
- Brenner, J.C.; Graham, M.P.; Kumar, B.; Saunders, L.M.; Kupfer, R.; Lyons, R.H.; Bradford, C.R.; Carey, T.E. Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head Neck 2010, 32, 417–426. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, M.L.; Kulkarni, A.; Birkeland, A.C.; Michmerhuizen, N.L.; Foltin, S.K.; Mann, J.E.; Hoesli, R.C.; Devenport, S.N.; Jewell, B.M.; Shuman, A.G.; et al. The genomic landscape of UM-SCC oral cavity squamous cell carcinoma cell lines. Oral Oncol. 2018, 87, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Benakanakere, M.R.; Li, Q.; Eskan, M.A.; Singh, A.V.; Zhao, J.; Galicia, J.C.; Stathopoulou, P.; Knudsen, T.B.; Kinane, D.F. Modulation of TLR2 protein expression by miR-105 in human oral keratinocytes. J. Biol. Chem. 2009, 284, 23107–23115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawasaki, T.; Kawai, T. Toll-like receptor signaling pathways. Front. Immunol 2014, 5, 461. [Google Scholar] [CrossRef] [Green Version]
- Fitzgerald, K.A.; Kagan, J.C. Toll-like Receptors and the Control of Immunity. Cell 2020, 180, 1044–1066. [Google Scholar] [CrossRef] [PubMed]
- Motshwene, P.G.; Moncrieffe, M.C.; Grossmann, J.G.; Kao, C.; Ayaluru, M.; Sandercock, A.M.; Robinson, C.V.; Latz, E.; Gay, N.J. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J. Biol. Chem. 2009, 284, 25404–25411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.C.; Lo, Y.C.; Wu, H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature 2010, 465, 885–890. [Google Scholar] [CrossRef] [Green Version]
- Lyford-Pike, S.; Peng, S.; Young, G.D.; Taube, J.M.; Westra, W.H.; Akpeng, B.; Bruno, T.C.; Richmon, J.D.; Wang, H.; Bishop, J.A.; et al. Evidence for a role of the PD-1:PD-L1 pathway in immune resistance of HPV-associated head and neck squamous cell carcinoma. Cancer Res. 2013, 73, 1733–1741. [Google Scholar] [CrossRef] [Green Version]
- Cheng, K.; Gao, M.; Godfroy, J.I.; Brown, P.N.; Kastelowitz, N.; Yin, H. Specific activation of the TLR1-TLR2 heterodimer by small-molecule agonists. Sci. Adv. 2015, 1, e1400139. [Google Scholar] [CrossRef] [Green Version]
- Kumar, H.; Kawai, T.; Akira, S.; Akira, S. Pathogen recognition by the innate immune system. Int. Immunol. 2011, 30, 16–34. [Google Scholar] [CrossRef]
- Wang, J.; Roderiquez, G.; Norcross, M.A. Control of adaptive immune responses by Staphylococcus aureus through IL-10, PD-L1, and TLR2. Sci. Rep. 2012, 2, 606. [Google Scholar] [CrossRef] [Green Version]
- Chattopadhyay, I.; Verma, M.; Panda, M. Role of Oral Microbiome Signatures in Diagnosis and Prognosis of Oral Cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033819867354. [Google Scholar] [CrossRef] [Green Version]
- Madonna, S.; Scarponi, C.; Sestito, R.; Sestito, R.; Pallotta, S.; Pallotta, S.; Cavani, A.; Cavani, A.; Albanesi, C.; Albanesi, C. The IFN-gamma-dependent suppressor of cytokine signaling 1 promoter activity is positively regulated by IFN regulatory factor-1 and Sp1 but repressed by growth factor independence-1b and Krüppel-like factor-4, and it is dysregulated in psoriatic keratinocytes. J. Immunol. 2010, 185, 2467–2481. [Google Scholar] [PubMed] [Green Version]
- Libermann, T.A.; Baltimore, D. Activation of interleukin-6 gene expression through the NF-kappa B transcription factor. Mol. Cell. Biol. 1990, 10, 2327–2334. [Google Scholar] [CrossRef]
- Ozinsky, A.; Underhill, D.M.; Fontenot, J.D.; Hajjar, A.M.; Smith, K.D.; Wilson, C.B.; Schroeder, L.; Aderem, A. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc. Natl. Acad. Sci. USA 2000, 97, 13766–13771. [Google Scholar] [CrossRef] [Green Version]
- Bhatt, K.; Salgame, P. Host innate immune response to Mycobacterium tuberculosis. J. Clin. Immunol. 2007, 27, 347–362. [Google Scholar] [CrossRef] [PubMed]
- Lembo, A.; Kalis, C.; Kirschning, C.J.; Mitolo, V.; Jirillo, E.; Wagner, H.; Galanos, C.; Freudenberg, M.A. Differential contribution of Toll-like receptors 4 and 2 to the cytokine response to Salmonella enterica serovar Typhimurium and Staphylococcus aureus in mice. Infect. Immun. 2003, 71, 6058–6062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshimura, A.; Lien, E.; Ingalls, R.R.; Tuomanen, E.; Dziarski, R.; Golenbock, D. Cutting edge: Recognition of Gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J. Immunol. 1999, 163, 1–5. [Google Scholar] [PubMed]
- Flo, T.H.; Halaas, O.; Lien, E.; Ryan, L.; Teti, G.; Golenbock, D.T.; Sundan, A.; Espevik, T.; Espevik, T. Human toll-like receptor 2 mediates monocyte activation by Listeria monocytogenes, but not by group B streptococci or lipopolysaccharide. J. Immunol. 2000, 164, 2064–2069. [Google Scholar] [CrossRef] [Green Version]
- Javaid, N.; Choi, S. Toll-like Receptors from the Perspective of Cancer Treatment. Cancers 2020, 12, 297. [Google Scholar] [CrossRef] [Green Version]
- Fessler, J.; Matson, V.; Gajewski, T.F. Exploring the emerging role of the microbiome in cancer immunotherapy. J. Immunother. Cancer 2019, 7, 108. [Google Scholar] [CrossRef]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillère, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef] [Green Version]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Preston, R.; White, J.R.; Godoy-Vitorino, F.; Rodríguez-Hilario, A.; Navarro, K.; González, H.; Michailidi, C.; Jedlicka, A.; Canapp, S.; Bondy, J.; et al. High-resolution microbiome profiling uncovers. Oncotarget 2017, 8, 110931–110948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makkawi, H.; Hoch, S.; Burns, E.; Hosur, K.; Hajishengallis, G.; Kirschning, C.J.; Nussbaum, G. Stimulates TLR2-PI3K Signaling to Escape Immune Clearance and Induce Bone Resorption Independently of MyD88. Front. Cell. Infect. Microbiol. 2017, 7, 359. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.P.; Wang, K.; Zhang, Z.J.; Tong, Y.N.; Han, D.; Hu, C.Y.; Li, Q.; Xiang, Y.; Mao, X.H.; Tang, B. TLR2/TLR4 activation induces Tregs and suppresses intestinal inflammation caused by Fusobacterium nucleatum in vivo. PLoS ONE 2017, 12, e0186179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braunstein, M.A.-O.; Kucharczyk, J.; Adams, S. Targeting Toll-Like Receptors for Cancer Therapy. Target. Oncol. 2018, 13, 583–598. [Google Scholar] [CrossRef]
- Guo, H.; Chen, Y.; Hu, X.; Qian, G.; Ge, S.; Zhang, J. The regulation of Toll-like receptor 2 by miR-143 suppresses the invasion and migration of a subset of human colorectal carcinoma cells. Mol. Cancer 2013, 12, 77. [Google Scholar] [CrossRef] [Green Version]
- West, A.C.; Tang, K.; Tye, H.; Yu, L.; Deng, N.; Najdovska, M.; Lin, S.J.; Balic, J.J.; Okochi-Takada, E.; McGuirk, P.; et al. Identification of a TLR2-regulated gene signature associated with tumor cell growth in gastric cancer. Oncogene 2017, 36, 5134–5144. [Google Scholar] [CrossRef]
- Tye, H.; Kennedy, C.L.; Najdovska, M.; McLeod, L.; McCormack, W.; Hughes, N.; Dev, A.; Sievert, W.; Ooi, C.H.; Ishikawa, T.O.; et al. STAT3-driven upregulation of TLR2 promotes gastric tumorigenesis independent of tumor inflammation. Cancer Cell 2012, 22, 466–478. [Google Scholar] [CrossRef] [Green Version]
- Cheng, K.; Wang, X.; Zhang, S.; Yin, H. Discovery of small-molecule inhibitors of the TLR1/TLR2 complex. Angew. Chem. Int. Ed. Engl. 2012, 51, 12246–12249. [Google Scholar] [CrossRef] [PubMed]
- Mistry, P.; Laird, M.H.; Schwarz, R.S.; Greene, S.; Dyson, T.; Snyder, G.A.; Xiao, T.S.; Chauhan, J.; Fletcher, S.; Toshchakov, V.Y.; et al. Inhibition of TLR2 signaling by small molecule inhibitors targeting a pocket within the TLR2 TIR domain. Proc. Natl. Acad. Sci. USA 2015, 112, 5455–5460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reilly, M.; Miller, R.M.; Thomson, M.H.; Patris, V.; Ryle, P.; McLoughlin, L.; Mutch, P.; Gilboy, P.; Miller, C.; Broekema, M.; et al. Randomized, double-blind, placebo-controlled, dose-escalating phase I, healthy subjects study of intravenous OPN-305, a humanized anti-TLR2 antibody. Clin. Pharm. 2013, 94, 593–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Luo, F.; Cai, Y.; Liu, N.; Wang, L.; Xu, D.; Chu, Y. TLR1/TLR2 agonist induces tumor regression by reciprocal modulation of effector and regulatory T cells. J. Immunol. 2011, 186, 1963–1969. [Google Scholar] [CrossRef] [Green Version]
- Chua, B.Y.; Olson, M.R.; Bedoui, S.; Sekiya, T.; Wong, C.Y.; Turner, S.J.; Jackson, D.C. The use of a TLR2 agonist-based adjuvant for enhancing effector and memory CD8 T-cell responses. Immunol. Cell Biol. 2014, 92, 377–383. [Google Scholar] [CrossRef]
- Miyauchi, S.; Kim, S.S.; Pang, J.; Gold, K.A.; Gutkind, J.S.; Califano, J.A.; Mell, L.K.; Cohen, E.E.W.; Sharabi, A.B. Immune Modulation of Head and Neck Squamous Cell Carcinoma and the Tumor Microenvironment by Conventional Therapeutics. Clin. Cancer Res. 2019, 25, 4211–4223. [Google Scholar] [CrossRef] [Green Version]
- Rajasekaran, N.; Chester, C.; Yonezawa, A.; Zhao, X.; Kohrt, H.E. Enhancement of antibody-dependent cell mediated cytotoxicity: A new era in cancer treatment. Immunotargets 2015, 4, 91–100. [Google Scholar] [CrossRef] [Green Version]

| Rank | gRNA Target | PDL1low/Control | PDL1high/Control |
|---|---|---|---|
| 1 | RNF130 | 130.4702 | 0.61437 |
| 2 | UIMC1 | 97.62247 | 0.533461 |
| 3 | KLHL30 | 97.47681 | 0 |
| 4 | C1orf116 | 93.29673 | 0.604938 |
| 5 | SLC2A6 | 93.00492 | 0.467358 |
| 6 | TLR2 | 80.91354 | 0.363622 |
| 7 | KIDINS220 | 74.24762 | 0.376101 |
| 8 | COMP | 73.34039 | 0.374752 |
| 9 | STAB1 | 48.71035 | 0.181629 |
| 10 | NFYB | 45.65152 | 0.163268 |
| 11 | DISP2 | 38.79179 | 0.872197 |
| 12 | SLC25A25 | 36.74726 | 0.14868 |
| 13 | C7orf55 | 36.6708 | 0.194452 |
| 14 | SSMEM1 | 34.35596 | 0.153269 |
| 15 | TSPAN31 | 30.65758 | 0.121086 |
| 16 | UNKL | 27.33073 | 0.127668 |
| 17 | ITSN1 | 14.07346 | 0.076536 |
| 18 | HIRA | 13.86975 | 0.066925 |
| 19 | ZDHHC24 | 11.48397 | 5.910346 |
| 20 | NODAL | 11.19729 | 0.06114 |
| 21 | TTL | 7.297645 | 0.027327 |
| 22 | C1orf35 | 7.269804 | 0.038215 |
| 23 | ULBP2 | 6.721698 | 0.028772 |
| 24 | AXDND1 | 6.5365 | 0.040082 |
| 25 | ZFP64 | 6.110201 | 0.042713 |
| 26 | MAGEA6 | 5.625484 | 0.045438 |
| 27 | PCDHGB6 | 5.256831 | 0.039738 |
| 28 | FAM168B | 4.806231 | 0.031167 |
| 29 | FABP6 | 4.648937 | 0.03478 |
| 30 | USP16 | 4.350071 | 0.028961 |
| 31 | VPS25 | 4.180224 | 0.033329 |
| 32 | SPATA31A2 | 4.175848 | 0.035267 |
| 33 | NPR1 | 3.975242 | 0.033517 |
| 34 | CKMT1A | 3.801816 | 0.020674 |
| 35 | CCDC117 | 3.520316 | 0.02719 |
| 36 | KIF13B | 3.131082 | 0.022894 |
| 37 | FZR1 | 2.847764 | 0.022893 |
| 38 | ZNRF3 | 2.696549 | 0.02445 |
| 39 | STRN3 | 2.642702 | 0.026301 |
| 40 | BLCAP | 2.598926 | 0.020274 |
| 41 | GMNC | 2.586674 | 0.020701 |
| 42 | KDM6A | 2.56034 | 0.026508 |
| 43 | OR5T2 | 2.554679 | 0.028434 |
| 44 | CLNS1A | 2.489466 | 0.021168 |
| 45 | RAD50 | 2.088789 | 0.0218049 |
| 46 | NARR | 2.070283 | 0.020284 |
| 47 | HINT1 | 2.068037 | 0.018824 |
| 48 | IRG1 | 2.043546 | 0.0089151 |
| 49 | SLC9A6 | 2.043088 | 0 |
| 50 | TTC34 | 1.554888 | 0.022822 |
| Rank | gRNA Target | PDL1high/Control | PDL1low/Control |
|---|---|---|---|
| 1 | IZUMO3 | 223.6348 | 1.0669 |
| 2 | hsa-mir-105-1 | 167.3409 | 1.178067 |
| 3 | GLS | 111.713 | 0.854239 |
| 4 | FAT2 | 104.4253 | 0.590414 |
| 5 | SCGB3A1 | 50.81308 | 0.337907 |
| 6 | RAPGEF3 | 37.84282 | 0.242991 |
| 7 | AP2A1 | 31.35016 | 0.214096 |
| 8 | TRIM49 | 29.33133 | 0.181142 |
| 9 | hsa-mir-105-2 | 9.937754 | 0.060283 |
| 10 | FIGLA | 6.562241 | 0.021861 |
| 11 | POGLUT1 | 6.558918 | 0.06121 |
| 12 | ZDHHC24 | 5.910346 | 11.48397 |
| 13 | KCNIP2 | 5.604062 | 0.0485690 |
| 14 | ZNF37A | 4.446165 | 0.038013 |
| 15 | TTC5 | 3.348804 | 0 |
| 16 | SLC35F3 | 2.39854 | 0 |
| 17 | FAM71C | 2.325857 | 0 |
| 18 | CXorf21 | 2.179963 | 0.014468 |
| 19 | RBBP8 | 2.111634 | 0.04188 |
| 20 | CCDC129 | 1.992549 | 0 |
| 21 | SLC5A8 | 1.902974 | 0 |
| 22 | CHST2 | 1.864696 | 0.068597 |
| 23 | P2RY2 | 1.836203 | 0 |
| 24 | SEMA6C | 1.834026 | 0.015726 |
| 25 | SPHK1 | 1.797793 | 0 |
| 26 | KLHL30 | 1.794423 | 0.007607 |
| 27 | hsa-mir-212 | 1.77299 | 0 |
| 28 | CARNS1 | 1.744393 | 0 |
| 29 | BRF2 | 1.744393 | 0 |
| 30 | LMNB2 | 1.744393 | 0 |
| 31 | MCL1 | 1.744393 | 0 |
| 32 | GNA13 | 1.744393 | 0 |
| 33 | NF2 | 1.722029 | 0.714116 |
| Gene Set | Gene | Pearson’s R | Adjusted p-Value |
|---|---|---|---|
| gRNAs enriched in PDL1low | RNF130 | 0.082815 | NS |
| UIMC1 | 0.136287 | 0.03342197 | |
| KLHL30 | 0.046933 | NS | |
| C1orf116 | 0.033465 | NS | |
| SLC2A6 | 0.143694 | 0.01757946 | |
| TLR2 | 0.324183 | 0.01798496 | |
| KIDINS220 | 0.143437 | NS | |
| COMP | −0.004867 | 1.13 × 10−11 | |
| STAB1 | 0.29879 | 0.00021754 | |
| NFYB | −0.187014 | NS | |
| gRNAs enriched in PDL1high | GLS | 0.137618 | 0.02984585 |
| FAT2 | 0.242119 | 1.56 × 10−7 | |
| SCGB3A1 | −0.22681 | 1.42 × 10−6 | |
| RAPGEF3 | −0.023542 | NS | |
| AP2A1 | −0.159187 | 0.00414437 | |
| TRIM49 | −0.093222 | NS | |
| TLR and IFNγ pathway genes | STAT1 | 0.60212877 | 1.15 × 10−55 |
| JAK2 | 0.69457561 | 3.05 × 10−81 | |
| TLR2 | 0.32418282 | 7.46 × 10−14 | |
| TLR1 | 0.30202603 | 6.12 × 10−12 | |
| TLR3 | 0.44556113 | 1.71 × 10−27 | |
| TLR4 | 0.39553731 | 3.55 × 10−21 | |
| TLR5 | 0.08243978 | NS | |
| TLR6 | 0.28567334 | 1.25 × 10−10 | |
| TLR7 | 0.41385687 | 2.29 × 10−23 | |
| TLR8 | 0.53208367 | 3.15 × 10−41 | |
| TLR9 | 0.17934421 | 0.00051316 | |
| TLR10 | 0.27146896 | 1.48 × 10−9 | |
| MYD88 | 0.42093739 | 2.99 × 10−24 | |
| RELA | −0.0036575 | NS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mann, J.E.; Ludwig, M.L.; Kulkarni, A.; Scheftz, E.B.; Murray, I.R.; Zhai, J.; Gensterblum-Miller, E.; Jiang, H.; Brenner, J.C. Microbe-Mediated Activation of Toll-like Receptor 2 Drives PDL1 Expression in HNSCC. Cancers 2021, 13, 4782. https://doi.org/10.3390/cancers13194782
Mann JE, Ludwig ML, Kulkarni A, Scheftz EB, Murray IR, Zhai J, Gensterblum-Miller E, Jiang H, Brenner JC. Microbe-Mediated Activation of Toll-like Receptor 2 Drives PDL1 Expression in HNSCC. Cancers. 2021; 13(19):4782. https://doi.org/10.3390/cancers13194782
Chicago/Turabian StyleMann, Jacqueline E, Megan L Ludwig, Aditi Kulkarni, Erin B Scheftz, Isabel R Murray, Jingyi Zhai, Elizabeth Gensterblum-Miller, Hui Jiang, and J Chad Brenner. 2021. "Microbe-Mediated Activation of Toll-like Receptor 2 Drives PDL1 Expression in HNSCC" Cancers 13, no. 19: 4782. https://doi.org/10.3390/cancers13194782
APA StyleMann, J. E., Ludwig, M. L., Kulkarni, A., Scheftz, E. B., Murray, I. R., Zhai, J., Gensterblum-Miller, E., Jiang, H., & Brenner, J. C. (2021). Microbe-Mediated Activation of Toll-like Receptor 2 Drives PDL1 Expression in HNSCC. Cancers, 13(19), 4782. https://doi.org/10.3390/cancers13194782








