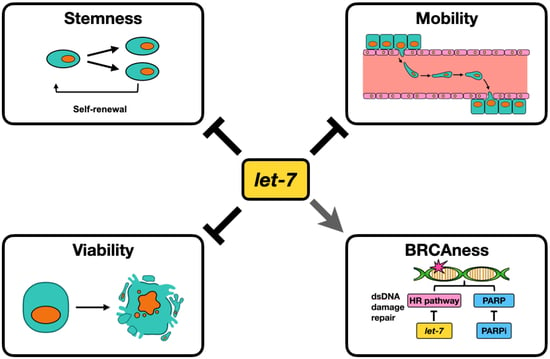
Let-7i Reduces Aggressive Phenotype and Induces BRCAness in Ovarian Cancer Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Let-7 Over-Expression in PD HGSOC Samples
2.2. Let-7 Represses Self-Renewal
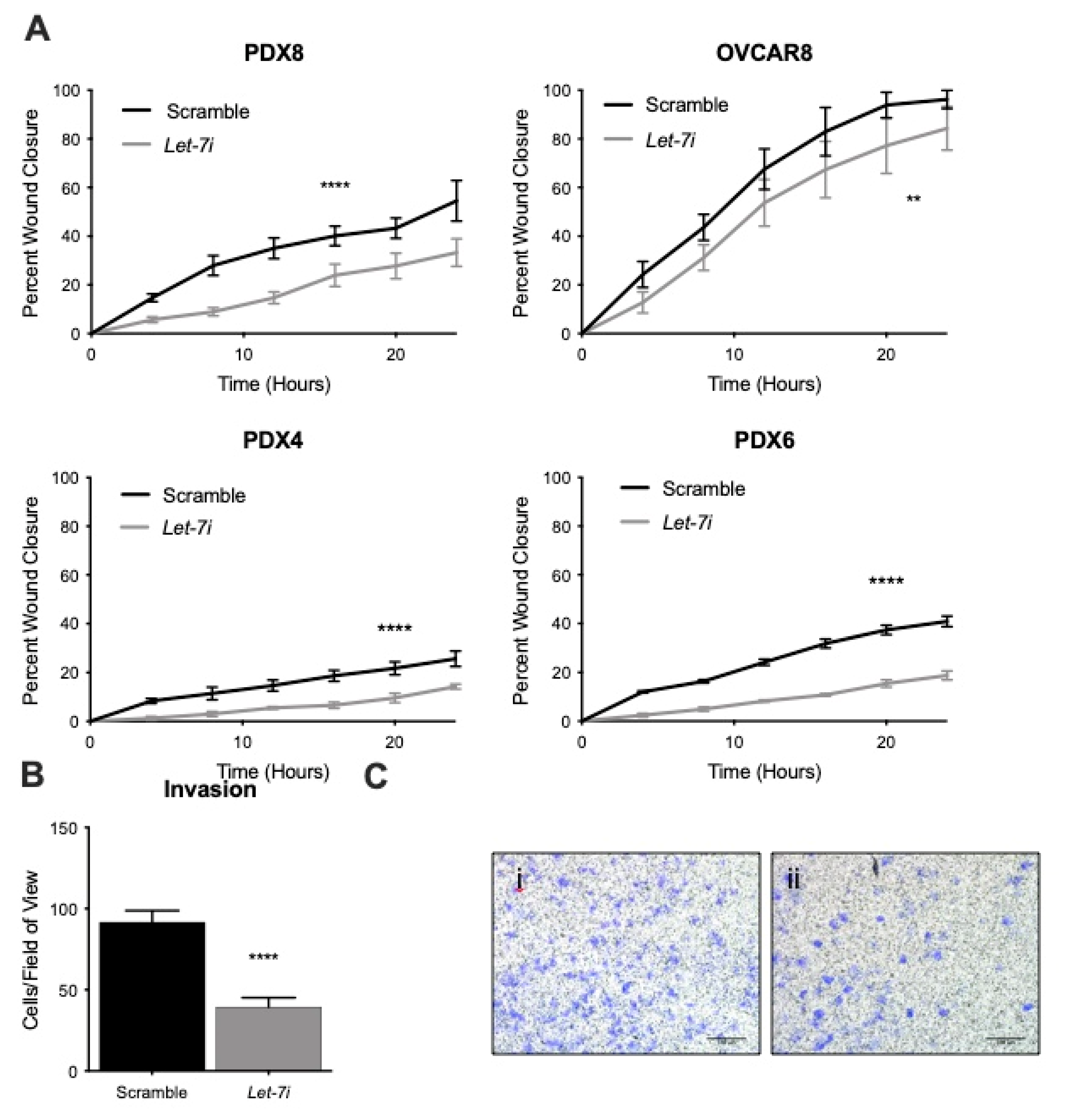
2.3. Let-7 Represses Migration and Invasion of HGSOC
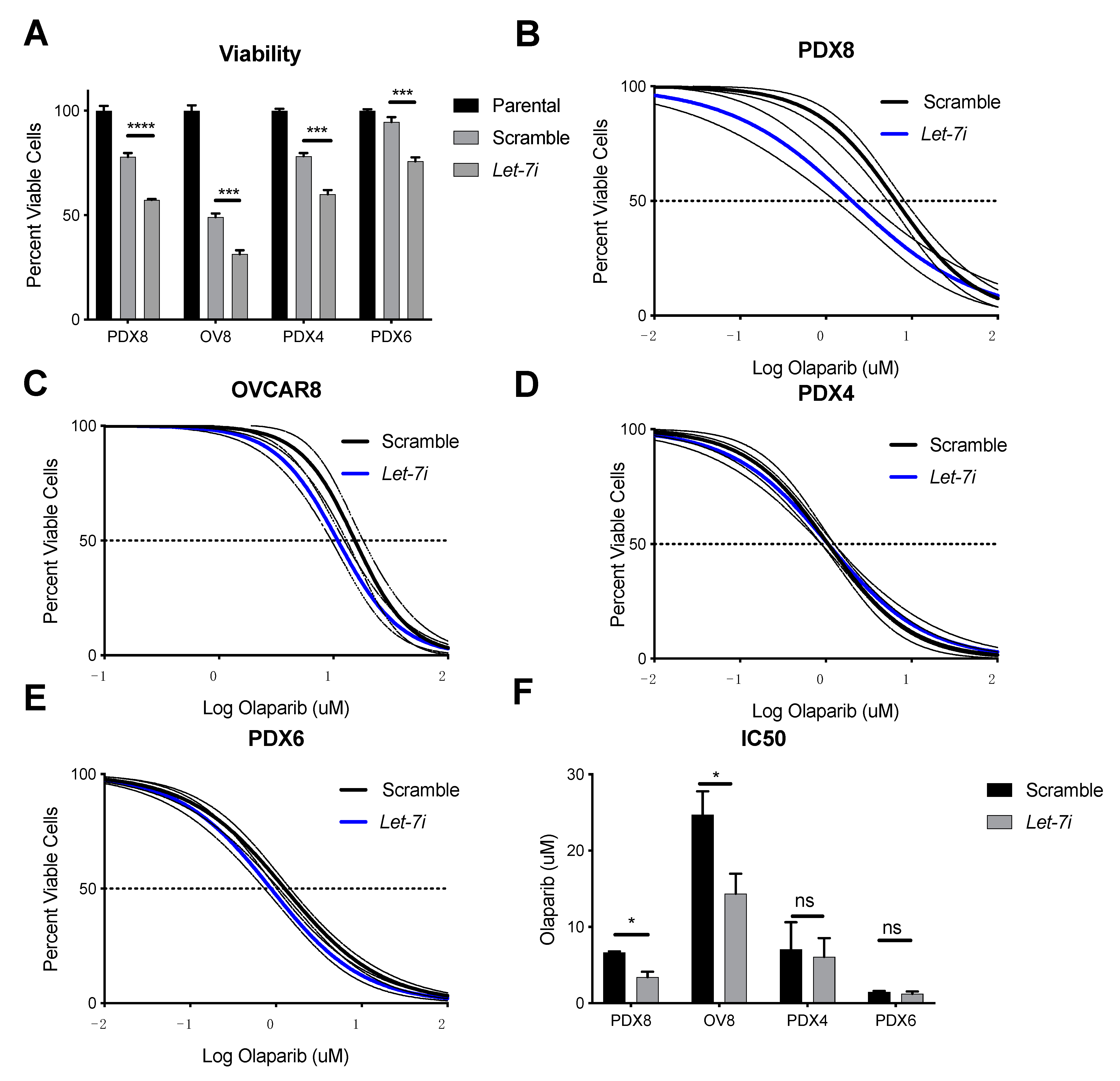
2.4. Let-7i Enhances HGSOC Sensitivity to PARP Inhibitors
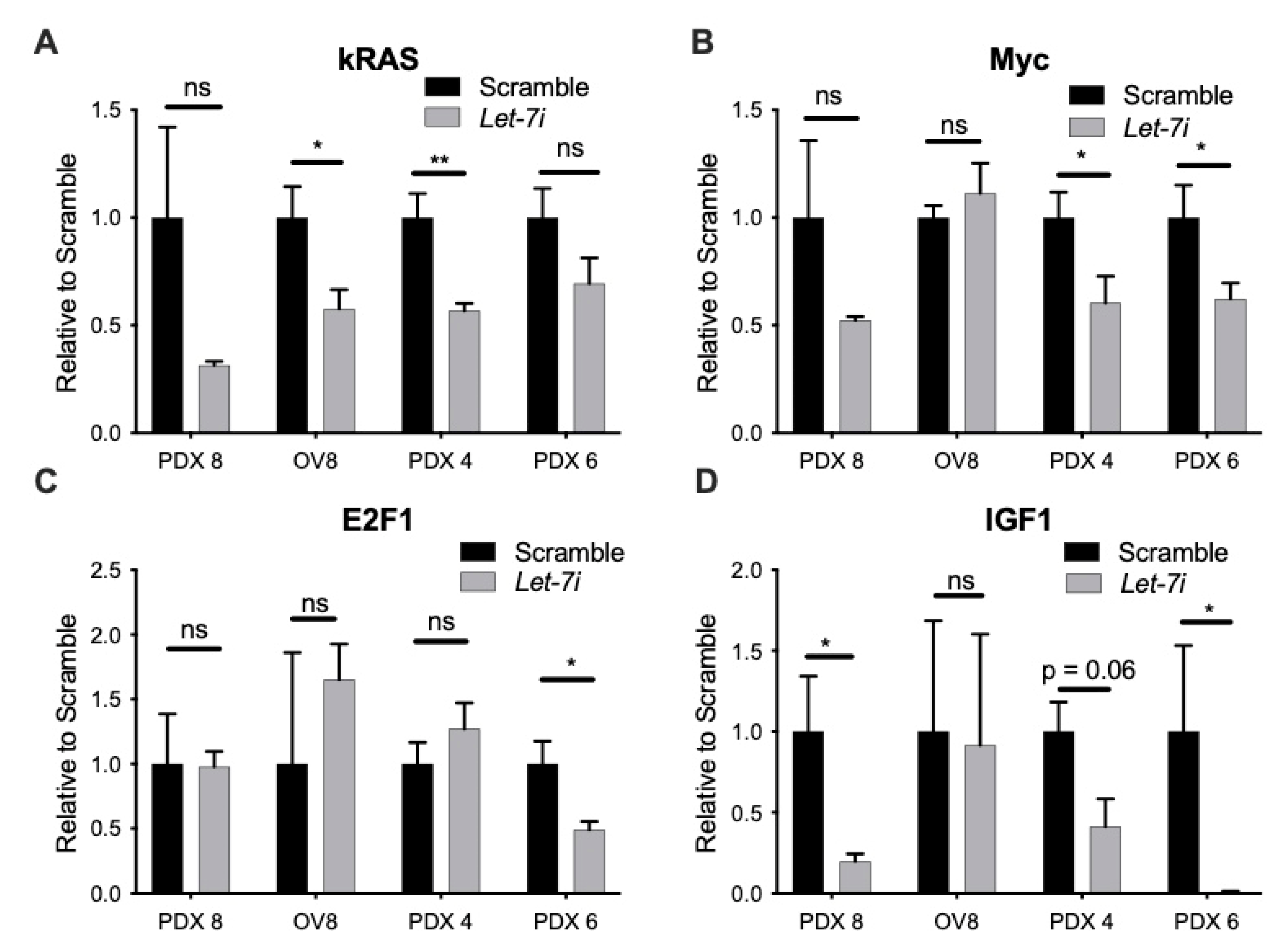
2.5. Let-7i Represses Factors Involved in HR Repair
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. MicroRNA Let-7 Overexpression
- Scramble S: 5′-mCmArUmArUmUrGmCrGmCrGmUrAmUrAmGrUmCrGC.
- Scramble AS: 5′-/5Phos/rGrCrGrArCrUrArUrArCrGrCrGrCrArArUrArUmGmGrU-3′.
- Let-7i-5P S: 5′-mCmArGmCrAmCrAmArAmCrUmArCmUrAmCrCmUrCA-3′.
- Let-7i-5P AS: 5′-/5Phos/rUrGrArGrGrUrArGrUrArGrUrUrUrGrUrGrCrUmGmUrU-3′.
4.3. Real-Time Quantitative Reverse-Transcription PCR (RT-qPCR)
4.4. Spheroid Formation Assay
4.5. Scratch Assay (Wound Healing Cell Migration Assay)
4.6. Western Blot
4.7. Invasion Assay
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- SEER Cancer Stat Facts: Ovarian Cancer; National Cancer Institute: Bethesda, MD, USA. Available online: https://seer.cancer.gov/statfacts/html/ovary.html (accessed on 27 June 2021).
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, N.S.; Miller, A.; Rungruang, B.; Richard, S.D.; Rodriguez, N.; Bookman, M.A.; Hamilton, C.A.; Krivak, T.C.; Maxwell, G.L. Does Aggressive Surgery Improve Outcomes? Interaction Between Preoperative Disease Burden and Complex Surgery in Patients with Advanced-Stage Ovarian Cancer: An Analysis of GOG 182. J. Clin. Oncol. 2015, 33, 937–943. [Google Scholar] [CrossRef] [PubMed]
- Hoppenot, C.; Eckert, M.; Tienda, S.M.; Lengyel, E. Who are the long-term survivors of high grade serous ovarian cancer? Gynecol. Oncol. 2018, 148, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Beck, B.; Blanpain, C. Unravelling cancer stem cell potential. Nat. Rev. Cancer 2013, 13, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, M.; Friedrich, D.; Kraft, C.; Rogmans, C. Multimodal Treatment of Primary Advanced Ovarian Cancer. Anticancer. Res. 2021, 41, 3253–3260. [Google Scholar] [CrossRef]
- Cortez, A.J.; Tudrej, P.; Kujawa, K.; Lisowska, K.M. Advances in ovarian cancer therapy. Cancer Chemother. Pharmacol. 2017, 81, 17–38. [Google Scholar] [CrossRef]
- Vergote, I.; Coens, C.; Nankivell, M.; Kristensen, G.B.; Parmar, M.K.B.; Ehlen, T.; Jayson, G.; Johnson, N.; Swart, A.M.; Verheijen, R.; et al. Neoadjuvant chemotherapy versus debulking surgery in advanced tubo-ovarian cancers: Pooled analysis of individual patient data from the EORTC 55971 and CHORUS trials. Lancet Oncol. 2018, 19, 1680–1687. [Google Scholar] [CrossRef]
- Kuroki, L.; Guntupalli, S.R. Treatment of epithelial ovarian cancer. BMJ 2020, 371, m3773. [Google Scholar] [CrossRef]
- Byrum, A.; Vindigni, A.; Mosammaparast, N. Defining and Modulating ‘BRCAness’. Trends Cell Biol. 2019, 29, 740–751. [Google Scholar] [CrossRef]
- Gupta, S.; Nag, S.; Aggarwal, S.; Rauthan, A.; Warrier, N. Maintenance therapy for recurrent epithelial ovarian cancer: Current therapies and future perspectives—A review. J. Ovarian Res. 2019, 12, 1–15. [Google Scholar] [CrossRef]
- Coleman, R.L.; Oza, A.; Lorusso, D.; Aghajanian, C.; Oaknin, A.; Dean, A.; Colombo, N.; Weberpals, J.I.; Clamp, A.; Scambia, G.; et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017, 390, 1949–1961. [Google Scholar] [CrossRef]
- Moore, K.N.; Secord, A.A.; Geller, M.A.; Miller, D.; Cloven, N.; Fleming, G.F.; Hendrickson, A.E.W.; Azodi, M.; DiSilvestro, P.; Oza, A.; et al. Niraparib monotherapy for late-line treatment of ovarian cancer (QUADRA): A multicentre, open-label, single-arm, phase 2 trial. Lancet Oncol. 2019, 20, 636–648. [Google Scholar] [CrossRef]
- Martín, A.G.; Pothuri, B.; Vergote, I.; Christensen, R.D.; Graybill, W.; Mirza, M.R.; McCormick, C.; Lorusso, D.; Hoskins, P.; Freyer, G.; et al. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2391–2402. [Google Scholar] [CrossRef] [PubMed]
- Mirza, M.R.; Monk, B.J.; Herrstedt, J.; Oza, A.; Mahner, S.; Redondo, A.; Fabbro, M.; Ledermann, J.A.; Lorusso, D.; Vergote, I.; et al. Niraparib Maintenance Therapy in Platinum-Sensitive, Recurrent Ovarian Cancer. N. Engl. J. Med. 2016, 375, 2154–2164. [Google Scholar] [CrossRef]
- Poveda, A.; Floquet, A.; Ledermann, J.A.; Asher, R.; Penson, R.T.; Oza, A.M.; Korach, J.; Huzarski, T.; Pignata, S.; Friedlander, M.; et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): A final analysis of a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2021, 22, 620–631. [Google Scholar] [CrossRef]
- Falzone, L.; Scandurra, G.; Lombardo, V.; Gattuso, G.; Lavoro, A.; Distefano, A.B.; Scibilia, G.; Scollo, P. A multidisciplinary approach remains the best strategy to improve and strengthen the management of ovarian cancer (Review). Int. J. Oncol. 2021, 59, 1–14. [Google Scholar] [CrossRef]
- Jiang, X.; Li, W.; Li, X.; Bai, H.; Zhang, Z. Current status and future prospects of PARP inhibitor clinical trials in ovarian cancer. Cancer Manag. Res. 2019, ume 11, 4371–4390. [Google Scholar] [CrossRef]
- An, D.; Banerjee, S.; Lee, J.-M. Recent advancements of antiangiogenic combination therapies in ovarian cancer. Cancer Treat. Rev. 2021, 98, 102224. [Google Scholar] [CrossRef] [PubMed]
- Siomi, H.; Siomi, M.C. Posttranscriptional Regulation of MicroRNA Biogenesis in Animals. Mol. Cell 2010, 38, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Kong, W.; He, L.; Zhao, J.-J.; O’Donnell, J.D.; Wang, J.; Wenham, R.M.; Coppola, D.; Kruk, P.A.; Nicosia, S.V.; et al. MicroRNA Expression Profiling in Human Ovarian Cancer: miR-214 Induces Cell Survival and Cisplatin Resistance by Targeting PTEN. Cancer Res. 2008, 68, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Slack, F.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef]
- Hayes, G.D.; Ruvkun, G. Misexpression of the Caenorhabditis elegans miRNA let-7 Is Sufficient to Drive Developmental Programs. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 21–27. [Google Scholar] [CrossRef]
- Unternaehrer, J.J.; Zhao, R.; Kim, K.; Cesana, M.; Powers, J.T.; Ratanasirintrawoot, S.; Onder, T.; Shibue, T.; Weinberg, R.A.; Daley, G.Q. The Epithelial-Mesenchymal Transition Factor SNAIL Paradoxically Enhances Reprogramming. Stem Cell Rep. 2014, 3, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Worringer, K.; Rand, T.A.; Hayashi, Y.; Sami, S.; Takahashi, K.; Tanabe, K.; Narita, M.; Srivastava, D.; Yamanaka, S. The let-7/LIN-41 Pathway Regulates Reprogramming to Human Induced Pluripotent Stem Cells by Controlling Expression of Prodifferentiation Genes. Cell Stem Cell 2013, 14, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Melton, C.; Judson, R.L.; Blelloch, R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 2010, 463, 621–626. [Google Scholar] [CrossRef]
- Xiao, M.; Cai, J.; Cai, L.; Jia, J.; Xie, L.; Zhu, Y.; Huang, B.; Jin, D.; Wang, Z. Let-7e sensitizes epithelial ovarian cancer to cisplatin through repressing DNA double strand break repair. J. Ovarian Res. 2017, 10, 1–13. [Google Scholar] [CrossRef]
- Song, H.; Zhang, Y.; Liu, N.; Zhang, D.; Wan, C.; Zhao, S.; Kong, Y.; Yuan, L. Let-7b inhibits the malignant behavior of glioma cells and glioma stem-like cells via downregulation of E2F2. J. Physiol. Biochem. 2016, 72, 733–744. [Google Scholar] [CrossRef]
- Sun, X.; Xu, C.; Tang, S.-C.; Wang, J.; Wang, H.; Wang, P.; Du, N.; Qin, S.; Li, G.; Xu, S.; et al. Let-7c blocks estrogen-activated Wnt signaling in induction of self-renewal of breast cancer stem cells. Cancer Gene Ther. 2016, 23, 83–89. [Google Scholar] [CrossRef]
- Lee, H.; Han, S.; Kwon, C.S.; Lee, D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell 2015, 7, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.-H.; Lan, H.-Y.; Huang, C.-H.; Tai, S.-K.; Tzeng, C.-H.; Kao, S.-Y.; Wu, K.-J.; Hung, M.-C.; Yang, M.-H. RAC1 activation mediates Twist1-induced cancer cell migration. Nature 2012, 14, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Shyh-Chang, N.; Daley, G.Q. Lin28: Primal Regulator of Growth and Metabolism in Stem Cells. Cell Stem Cell 2013, 12, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chirshev, E.; Hojo, N.; Suzuki, T.; Bertucci, A.; Pierce, M.; Perry, C.; Wang, R.; Zink, J.; Glackin, C.; et al. The Epithelial–Mesenchymal Transcription Factor SNAI1 Represses Transcription of the Tumor Suppressor miRNA let-7 in Cancer. Cancers 2021, 13, 1469. [Google Scholar] [CrossRef] [PubMed]
- Chirshev, E.; Hojo, N.; Bertucci, A.; Sanderman, L.; Nguyen, A.; Wang, H.; Suzuki, T.; Brito, E.; Martinez, S.; Castañón, C.; et al. Epithelial/mesenchymal heterogeneity of high-grade serous ovarian carcinoma samples correlates with miRNA let-7 levels and predicts tumor growth and metastasis. Mol. Oncol. 2020, 14, 2796–2813. [Google Scholar] [CrossRef]
- Zhou, J.; Ng, S.-B.; Chng, W.-J. LIN28/LIN28B: An emerging oncogenic driver in cancer stem cells. Int. J. Biochem. Cell Biol. 2013, 45, 973–978. [Google Scholar] [CrossRef]
- Büssing, I.; Slack, F.; Grosshans, H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008, 14, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Parisi, S.; Passaro, F.; Russo, L.; Musto, A.; Navarra, A.; Romano, S.; Petrosino, G.; Russo, T. Lin28 is induced in primed embryonic stem cells and regulates let-7-independent events. FASEB J. 2016, 31, 1046–1058. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wang, H.; Yan, M.; Yang, X.; Shen, R.; Ni, X.; Chen, X.; Yang, P.; Chen, M.; Lu, X.; et al. High LIN28A and PLK4 co-expression is associated with poor prognosis in epithelial ovarian cancer. Mol. Med. Rep. 2018, 18, 5327–5336. [Google Scholar] [CrossRef] [PubMed]
- Albino, D.; Civenni, G.; Dallavalle, C.; Roos, M.; Jahns, H.; Curti, L.; Rossi, S.; Pinton, S.; D’Ambrosio, G.; Sessa, F.; et al. Activation of the Lin28/let-7 Axis by Loss of ESE3/EHF Promotes a Tumorigenic and Stem-like Phenotype in Prostate Cancer. Cancer Res. 2016, 76, 3629–3643. [Google Scholar] [CrossRef]
- Navarra, A.; Musto, A.; Gargiulo, A.; Petrosino, G.; Pierantoni, G.M.; Fusco, A.; Russo, T.; Parisi, S. Hmga2 is necessary for Otx2-dependent exit of embryonic stem cells from the pluripotent ground state. BMC Biol. 2016, 14, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Chirshev, E.; Oberg, K.; Ioffe, Y.J.; Unternaehrer, J.J. Let-7as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin. Transl. Med. 2019, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bao, W.; Liu, Y.; Wang, S.; Xu, S.; Li, X.; Li, Y.; Wu, S. miR-98-5p contributes to cisplatin resistance in epithelial ovarian cancer by suppressing miR-152 biogenesis via targeting Dicer1. Cell Death Dis. 2018, 9, 1–17. [Google Scholar] [CrossRef]
- Brueckner, B.; Stresemann, C.; Kuner, R.; Mund, C.; Musch, T.; Meister, M.; Sültmann, H.; Lyko, F. The Human let-7a-3 Locus Contains an Epigenetically Regulated MicroRNA Gene with Oncogenic Function. Cancer Res. 2007, 67, 1419–1423. [Google Scholar] [CrossRef]
- Shi, W.; Zhang, Z.; Yang, B.; Guo, H.; Jing, L.; Liu, T.; Luo, Y.; Liu, H.; Li, Y.; Gao, Y. Overexpression of microRNA let-7 correlates with disease progression and poor prognosis in hepatocellular carcinoma. Medicine 2017, 96, e7764. [Google Scholar] [CrossRef] [PubMed]
- Wielgos, M.E.; Rajbhandari, R.; Cooper, T.S.; Wei, S.; Nozell, S.; Yang, E.S. Let-7 Status Is Crucial for PARP1 Expression in HER2-Overexpressing Breast Tumors. Mol. Cancer Res. 2016, 15, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Hong, X.; Hu, J.; Lu, Q. Targeted regulation of MiR-98 on E2F1 increases chemosensitivity of leukemia cells K562/A02. OncoTargets Ther. 2017, ume 10, 3233–3239. [Google Scholar] [CrossRef]
- Shen, G.; Wu, R.; Liu, B.; Dong, W.; Tu, Z.; Yang, J.; Xu, Z.; Pan, T. Upstream and downstream mechanisms for the promoting effects of IGF-1 on differentiation of spermatogonia to primary spermatocytes. Life Sci. 2014, 101, 49–55. [Google Scholar] [CrossRef]
- Yu, F.; Yao, H.; Zhu, P.; Zhang, X.; Pan, Q.; Gong, C.; Huang, Y.; Hu, X.; Su, F.; Lieberman, J.; et al. let-7 Regulates Self Renewal and Tumorigenicity of Breast Cancer Cells. Cell 2007, 131, 1109–1123. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Clarke, I.D.; Terasaki, M.; Bonn, V.E.; Hawkins, C.; Squire, J.; Dirks, P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003, 63, 5821–5828. [Google Scholar]
- Zhang, S.; Balch, C.; Chan, M.; Lai, H.-C.; Matei, D.; Schilder, J.M.; Yan, P.S.; Huang, T.H.-M.; Nephew, K.P. Identification and Characterization of Ovarian Cancer-Initiating Cells from Primary Human Tumors. Cancer Res. 2008, 68, 4311–4320. [Google Scholar] [CrossRef]
- Hagedorn, E.J.; Sherwood, D.R. Cell invasion through basement membrane: The anchor cell breaches the barrier. Curr. Opin. Cell Biol. 2011, 23, 589–596. [Google Scholar] [CrossRef][Green Version]
- Dallas, N.A.; Xia, L.; Fan, F.; Gray, M.J.; Gaur, P.; Van Buren, G.; Samuel, S.; Kim, M.P.; Lim, S.J.; Ellis, L.M. Chemoresistant Colorectal Cancer Cells, the Cancer Stem Cell Phenotype, and Increased Sensitivity to Insulin-like Growth Factor-I Receptor Inhibition. Cancer Res. 2009, 69, 1951–1957. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.-H.; Kim, K.P. E2F1 facilitates DNA break repair by localizing to break sites and enhancing the expression of homologous recombination factors. Exp. Mol. Med. 2019, 51, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Luoto, K.R.; Meng, A.X.; Wasylishen, A.; Zhao, H.; Coackley, C.L.; Penn, L.; Bristow, R. Tumor Cell Kill by c-MYC Depletion: Role of MYC-Regulated Genes that Control DNA Double-Strand Break Repair. Cancer Res. 2010, 70, 8748–8759. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhang, Z.; Zhao, L.; Li, L.; Zuo, W.; Han, W.Z.A.L. High expression of RAD51 promotes DNA damage repair and survival in KRAS-mutant lung cancer cells. BMB Rep. 2019, 52, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Chintapalli, J.; Sodagum, L.; Baskin, S.; Malhotra, A.; Reiss, K.; Meggs, L.G. Activated IGF-1R inhibits hyperglycemia-induced DNA damage and promotes DNA repair by homologous recombination. Am. J. Physiol. Physiol. 2005, 289, F1144–F1152. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007, 21, 1025–1030. [Google Scholar] [CrossRef] [PubMed]
- Madison, B.B.; Jeganathan, A.N.; Mizuno, R.; Winslow, M.M.; Castells, A.; Cuatrecasas, M.; Rustgi, A.K. Let-7 Represses Carcinogenesis and a Stem Cell Phenotype in the Intestine via Regulation of Hmga2. PLoS Genet. 2015, 11, e1005408. [Google Scholar] [CrossRef]
- Bapat, S.A.; Mali, A.M.; Koppikar, C.B.; Kurrey, N. Stem and Progenitor-Like Cells Contribute to the Aggressive Behavior of Human Epithelial Ovarian Cancer. Cancer Res. 2005, 65, 3025–3029. [Google Scholar] [CrossRef]
- Park, H.; Hwang, S.; Jeong, J.-Y.; Jung, S.G.; Choi, M.C.; Joo, W.D.; Song, S.H.; Lee, C.; An, H.J. Integrative analysis of transcription factors and microRNAs in ovarian cancer cell spheroids. J. Ovarian Res. 2020, 13, 1–10. [Google Scholar] [CrossRef]
- Gao, Q.; Yang, Z.; Xu, S.; Li, X.; Yang, X.; Jin, P.; Liu, Y.; Zhou, X.; Zhang, T.; Gong, C.; et al. Heterotypic CAF-tumor spheroids promote early peritoneal metastasis of ovarian cancer. J. Exp. Med. 2019, 216, 688–703. [Google Scholar] [CrossRef]
- Zong, X.; Wang, W.; Ozes, A.; Fang, F.; Sandusky, G.E.; Nephew, K.P. EZH2-Mediated Downregulation of the Tumor Suppressor DAB2IP Maintains Ovarian Cancer Stem Cells. Cancer Res. 2020, 80, 4371–4385. [Google Scholar] [CrossRef] [PubMed]
- Ogishima, J.; Taguchi, A.; Kawata, A.; Kawana, K.; Yoshida, M.; Yoshimatsu, Y.; Sato, M.; Nakamura, H.; Kawata, Y.; Nishijima, A.; et al. The oncogene KRAS promotes cancer cell dissemination by stabilizing spheroid formation via the MEK pathway. BMC Cancer 2018, 18, 1201. [Google Scholar] [CrossRef]
- Chaffer, C.L.; Weinberg, R.A. A Perspective on Cancer Cell Metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef] [PubMed]
- Dudjak, L.A. Cancer metastasis. Semin. Oncol. Nurs. 1992, 8, 40–50. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, X.; Chen, D.; Ma, C. Let-7a suppresses glioma cell proliferation and invasion through TGF-β/Smad3 signaling pathway by targeting HMGA2. Tumor Biol. 2015, 37, 8107–8119. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, Y.; Huang, L.; Xiao, Q.; Chen, X.; Zhong, J.; Chen, Y.; Yang, D.; Han, Z.; Shu, Y.; et al. Let-7a suppresses macrophage infiltrations and malignant phenotype of Ewing sarcoma via STAT3/NF-κB positive regulatory circuit. Cancer Lett. 2016, 374, 192–201. [Google Scholar] [CrossRef]
- Tang, R.; Yang, C.; Ma, X.; Wang, Y.; Luo, D.; Huang, C.; Xu, Z.; Liu, P.; Yang, L. MiR-let-7a inhibits cell proliferation, migration, and invasion by down-regulating PKM2 in gastric cancer. Oncotarget 2016, 7, 5972–5984. [Google Scholar] [CrossRef]
- Xu, X.; Bao, Z.; Liu, Y.; Ji, J.; Liu, N. MicroRNA-98 Attenuates Cell Migration and Invasion in Glioma by Directly Targeting Pre-B Cell Leukemia Homeobox 3. Cell. Mol. Neurobiol. 2017, 37, 1359–1371. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Han, H.; Chen, J.; Zhang, Z.; Li, S.; Fang, F.; Zheng, Q.; Ma, Y.; Zhang, J.; Wu, N.; et al. MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 2014, 342, 43–51. [Google Scholar] [CrossRef]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS Is Regulated by the let-7 MicroRNA Family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [CrossRef]
- Hira, M.T.; Razzaque, M.A.; Angione, C.; Scrivens, J.; Sawan, S.; Sarker, M. Integrated multi-omics analysis of ovarian cancer using variational autoencoders. Sci. Rep. 2021, 11, 1–16. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, T.; Zhang, Z.; Payne, S.; Zhang, B.; McDermott, J.; Zhou, J.-Y.; Petyuk, V.A.; Chen, L.; Ray, D.; et al. Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell 2016, 166, 755–765. [Google Scholar] [CrossRef]
- Matulonis, U.; Shapira-Frommer, R.; Santin, A.; Lisyanskaya, A.; Pignata, S.; Vergote, I.; Raspagliesi, F.; Sonke, G.; Birrer, M.; Provencher, D.; et al. Antitumor activity and safety of pembrolizumab in patients with advanced recurrent ovarian cancer: Results from the phase II KEYNOTE-100 study. Ann. Oncol. 2019, 30, 1080–1087. [Google Scholar] [CrossRef] [PubMed]
- Marabelle, A.; Fakih, M.; Lopez, J.; Shah, M.; Shapira-Frommer, R.; Nakagawa, K.; Chung, H.C.; Kindler, H.L.; Lopez-Martin, J.A.; Miller, W.H.; et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: Prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020, 21, 1353–1365. [Google Scholar] [CrossRef]
- Khella, C.; Mehta, G.; Mehta, R.; Gatza, M. Recent Advances in Integrative Multi-Omics Research in Breast and Ovarian Cancer. J. Pers. Med. 2021, 11, 149. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Bankhead, A.; Ljungman, M.; Neamati, N. Multi-omics profiling reveals key signaling pathways in ovarian cancer controlled by STAT3. Theranostics 2019, 9, 5478–5496. [Google Scholar] [CrossRef]
- Liu, X.; Ory, V.; Chapman, S.; Yuan, H.; Albanese, C.; Kallakury, B.; Timofeeva, O.A.; Nealon, C.; Dakic, A.; Simic, V.; et al. ROCK Inhibitor and Feeder Cells Induce the Conditional Reprogramming of Epithelial Cells. Am. J. Pathol. 2012, 180, 599–607. [Google Scholar] [CrossRef]
- Edelstein, A.D.; Amodaj, N.; Hoover, K.H.; Vale, R.D.; Stuurman, N. Computer Control of Microscopes Using µManager. Curr. Protoc. Mol. Biol. 2010, 92, 14.20.1–14.20.17. [Google Scholar] [CrossRef]
- Moutasim, K.A.; Nystrom, M.L.; Thomas, G.J. Cell Migration and Invasion Assays. J. Vis. Exp. 2011, 731, 333–343. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chirshev, E.; Suzuki, T.; Wang, H.; Nguyen, A.; Hojo, N.; Sanderman, L.; Mirshahidi, S.; Ioffe, Y.J.; Unternaehrer, J.J. Let-7i Reduces Aggressive Phenotype and Induces BRCAness in Ovarian Cancer Cells. Cancers 2021, 13, 4617. https://doi.org/10.3390/cancers13184617
Chirshev E, Suzuki T, Wang H, Nguyen A, Hojo N, Sanderman L, Mirshahidi S, Ioffe YJ, Unternaehrer JJ. Let-7i Reduces Aggressive Phenotype and Induces BRCAness in Ovarian Cancer Cells. Cancers. 2021; 13(18):4617. https://doi.org/10.3390/cancers13184617
Chicago/Turabian StyleChirshev, Evgeny, Tise Suzuki, Hanmin Wang, Anthony Nguyen, Nozomi Hojo, Linda Sanderman, Saied Mirshahidi, Yevgeniya J. Ioffe, and Juli J. Unternaehrer. 2021. "Let-7i Reduces Aggressive Phenotype and Induces BRCAness in Ovarian Cancer Cells" Cancers 13, no. 18: 4617. https://doi.org/10.3390/cancers13184617
APA StyleChirshev, E., Suzuki, T., Wang, H., Nguyen, A., Hojo, N., Sanderman, L., Mirshahidi, S., Ioffe, Y. J., & Unternaehrer, J. J. (2021). Let-7i Reduces Aggressive Phenotype and Induces BRCAness in Ovarian Cancer Cells. Cancers, 13(18), 4617. https://doi.org/10.3390/cancers13184617