Hepatic Cancer Stem Cells: Molecular Mechanisms, Therapeutic Implications, and Circulating Biomarkers
Abstract
Simple Summary
Abstract
1. Introduction
2. Molecular Pathways Regulating CSCs in HCC
2.1. Beta-Catenin Pathway
2.2. The NOTCH Pathway
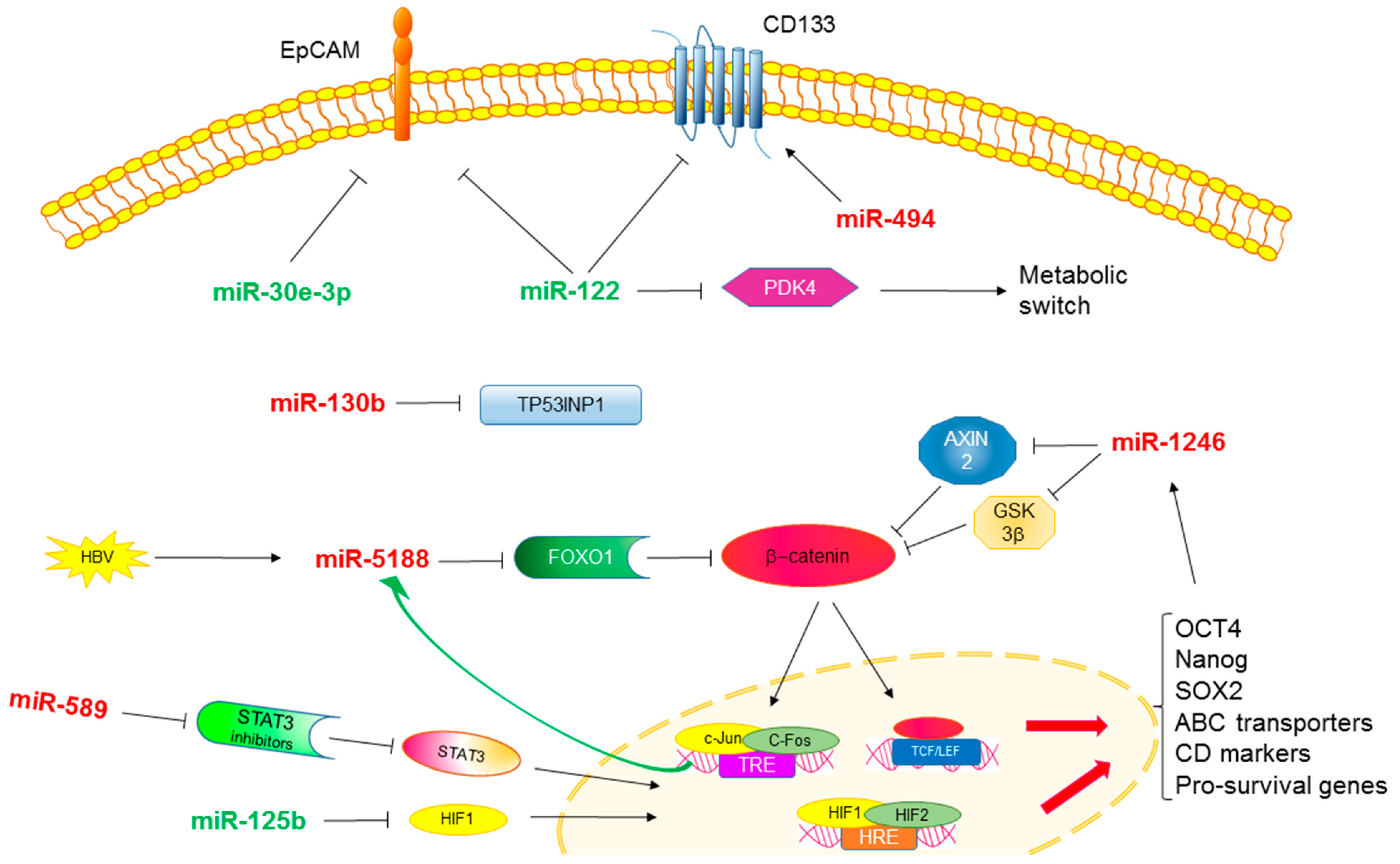
3. MicroRNAs and Stemness in HCC
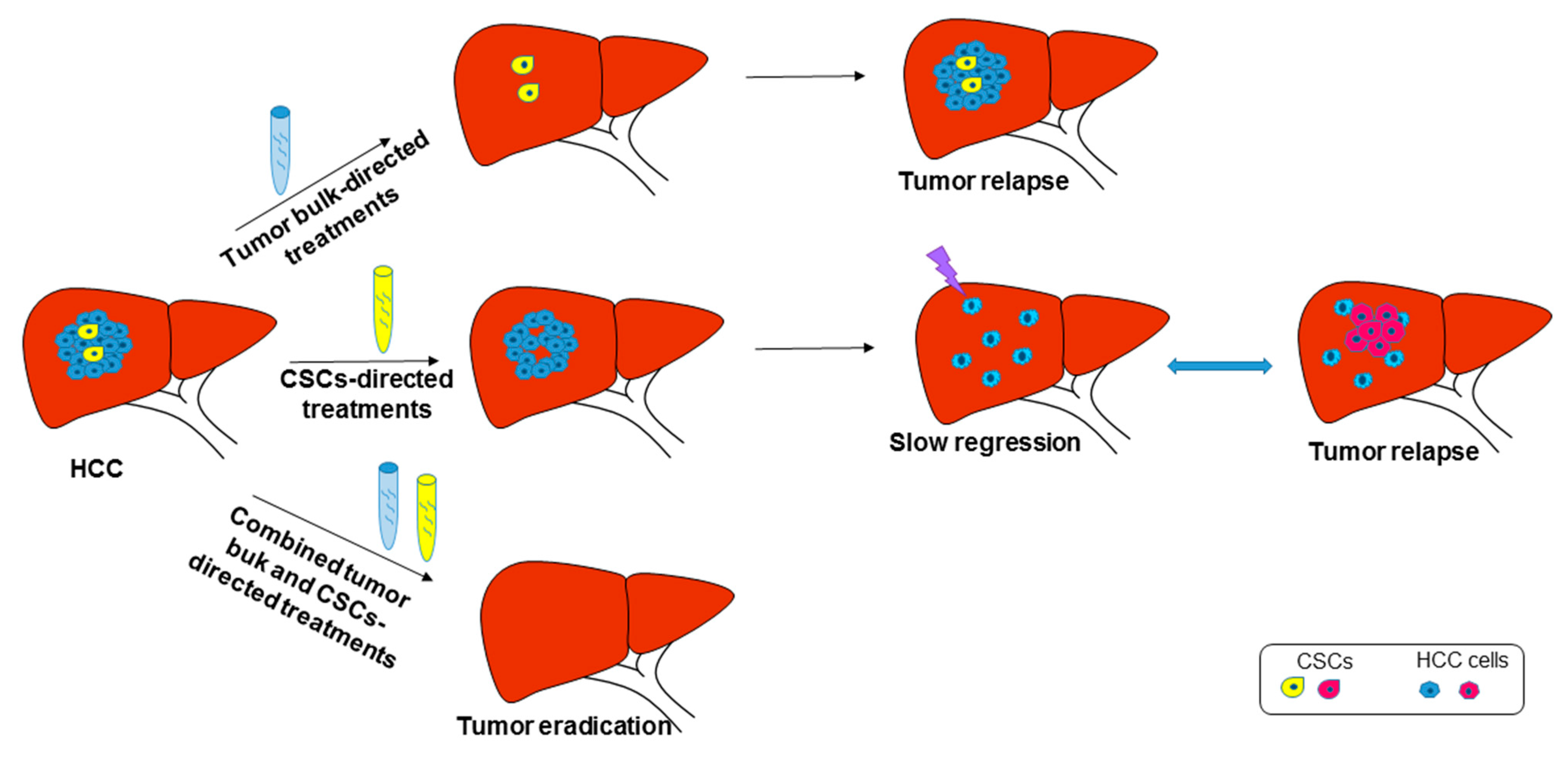
4. CSC Targeting and Therapeutic Implications
5. Circulating Cancer Stem Cells
6. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepathol. 2018, 69, 182–236. [Google Scholar]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus Bevacizumab in unresectable hepatocellular carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef] [PubMed]
- Rimassa, L.; Pressiani, T.; Merle, P. Systemic treatment options in hepatocellular carcinoma. Liver Cancer 2019, 8, 427–446. [Google Scholar] [CrossRef] [PubMed]
- Xue, R.; Li, R.; Guo, H.; Guo, L.; Su, Z.; Ni, X.; Qi, L.; Zhang, T.; Li, Q.; Zhang, Z.; et al. Variable intra-tumor genomic heterogeneity of multiple lesions in patients with hepatocellular carcinoma. Gastroenterology 2016, 150, 998–1008. [Google Scholar] [CrossRef] [PubMed]
- Feo, F.; Pascale, R.M. Multifocal hepatocellular carcinoma: Intrahepatic metastasis or multicentric carcinogenesis? Ann. Transl. Med. 2015, 3, 4. [Google Scholar]
- Zhang, Q.; Lou, Y.; Yang, J.; Wang, J.; Feng, J.; Zhao, Y.; Wang, L.; Huang, X.; Fu, Q.; Ye, M.; et al. Integrated multiomic analysis reveals comprehensive tumour heterogeneity and novel immunophenotypic classification in hepatocellular carcinomas. Gut 2019, 68, 2019–2031. [Google Scholar] [CrossRef] [PubMed]
- Shackleton, M.; Quintana, E.; Fearon, E.R.; Morrison, S.J. Heterogeneity in cancer: Cancer stem cells versus clonal evolution. Cell 2009, 138, 822–829. [Google Scholar] [CrossRef] [PubMed]
- Dagogo-Jack, I.; Shaw, A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Gramantieri, L.; Giovannini, C.; Lanzi, A.; Chieco, P.; Ravaioli, M.; Venturi, A.; Grazi, G.L.; Bolondi, L. Aberrant Notch3 and Notch4 expression in human hepatocellular carcinoma. Liver Int. 2007, 27, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Perugorria, M.J.; Olaizola, P.; Labiano, I.; Esparza-Baquer, A.; Marzioni, M.; Marin, J.J.G.; Bujanda, L.; Banales, J.M. Wnt-beta-catenin signalling in liver development, health and disease. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 121–136. [Google Scholar] [CrossRef]
- Pez, F.; Lopez, A.; Kim, M.; Wands, J.R.; Caron de Fromentel, C.; Merle, P. Wnt signaling and hepatocarcinogenesis: Molecular targets for the development of innovative anticancer drugs. J. Hepatol. 2013, 59, 1107–1117. [Google Scholar] [CrossRef]
- Zucman-Rossi, J.; Villanueva, A.; Nault, J.C.; Llovet, J.M. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 2015, 149, 1226–1239.e4. [Google Scholar] [CrossRef]
- Bengochea, A.; de Souza, M.M.; Lefrancois, L.; Le Roux, E.; Galy, O.; Chemin, I.; Kim, M.; Wands, J.R.; Trepo, C.; Hainaut, P.; et al. Common dysregulation of Wnt/Frizzled receptor elements in human hepatocellular carcinoma. Br. J. Cancer 2008, 99, 143–150. [Google Scholar] [CrossRef]
- Laurent-Puig, P.; Legoix, P.; Bluteau, O.; Belghiti, J.; Franco, D.; Binot, F.; Monges, G.; Thomas, G.; Bioulac-Sage, P.; Zucman-Rossi, J. Genetic alterations associated with hepatocellular carcinomas define distinct pathways of hepatocarcinogenesis. Gastroenterology 2001, 120, 1763–1773. [Google Scholar] [CrossRef] [PubMed]
- Hoshida, Y.; Nijman, S.M.; Kobayashi, M.; Chan, J.A.; Brunet, J.P.; Chiang, D.Y.; Villanueva, A.; Newell, P.; Ikeda, K.; Hashimoto, M.; et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009, 69, 7385–7392. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Yan, H.X.; Chen, L.; Liu, Q.; He, Y.Q.; Yu, L.X.; Zhang, S.H.; Huang, D.D.; Tang, L.; Kong, X.N.; et al. Wnt/beta-catenin signaling contributes to activation of normal and tumorigenic liver progenitor cells. Cancer Res. 2008, 68, 4287–4295. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.Y.; Hsu, C.C.; Chan, T.S.; Yen, C.J.; Chen, W.Y.; Pan, H.W.; Tsai, K.K. Dishevelled 1-regulated superpotent cancer stem cells mediate wnt heterogeneity and tumor progression in hepatocellular carcinoma. Stem Cell Rep. 2020, 14, 462–477. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.L.; Bamodu, O.A.; Ong, J.R.; Lee, W.H.; Yeh, C.T.; Tsai, J.T. Targeting the epigenetic non-coding RNA MALAT1/Wnt signaling axis as a therapeutic approach to suppress stemness and metastasis in hepatocellular carcinoma. Cells 2020, 9, 1020. [Google Scholar] [CrossRef]
- Ma, S.; Chan, K.W.; Lee, T.K.; Tang, K.H.; Wo, J.Y.; Zheng, B.J.; Guan, X.Y. Aldehyde dehydrogenase discriminates the CD133 liver cancer stem cell populations. Mol. Cancer Res. 2008, 6, 1146–1153. [Google Scholar] [CrossRef]
- Slmile, M.M.; Feo, C.F.; Calvisi, D.F.; Pascale, R.M.; Feo, F. GNMT: A multifaceted suppressor of hepatocarcinogenesis. Hepatoma Res. 2021, 7, 35. [Google Scholar]
- Martinez-Chantar, M.L.; Lu, S.C.; Mato, J.M.; Luka, Z.; Wagner, C.; French, B.A.; Franch, S.W. The role of stem cells/progenitor cells in liver carcinogenesis in glycine N-methyltransferase deficient mice. Exp. Mol. Pathol. 2010, 88, 234–237. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, B.; Cao, Y.; Meng, G.; Qian, L.; Xu, T.; Yan, C.; Luo, O.; Wang, S.; Wei, J.; Ding, Y.; et al. Targeting glutaminase 1 attenuates stemness properties in hepatocellular carcinoma by increasing reactive oxygen species and suppressing Wnt/beta-catenin pathway. EBioMedicine 2019, 39, 239–254. [Google Scholar] [CrossRef]
- Soth, M.J.; Le, K.; Di Francesco, M.E.; Hamilton, M.M.; Liu, G.; Burke, J.P.; Carroll, C.L.; Kovacs, J.J.; Bardehangen, J.P.; Bristow, C.A.; et al. Discovery of IPN60090, a Clinical Stage Selective Glutaminase-1 (GLS-1) inhibitor with excellent pharmacokinetic and physicochemical properties. J. Med. Chem. 2020, 63, 12957–12977. [Google Scholar] [CrossRef] [PubMed]
- Fornari, F.; Gramantieri, L.; Callegari, E.; Shankaraiah, R.C.; Piscaglia, F.; Negrini, M.; Giovannini, C. MicroRNAs in animal, models of HCC. Cancers 2019, 11, 1906. [Google Scholar] [CrossRef] [PubMed]
- Toh, T.B.; Lim, J.J.; Hooi, L.; Rashid, M.; Chow, E.K. Targeting Jak/Stat pathway as a therapeutic strategy against SP/CD44+ tumorigenic cells in Akt/beta-catenin-driven hepatocellular carcinoma. J. Hepatol. 2020, 72, 104–118. [Google Scholar] [CrossRef]
- Ishizawa, K.; Rasheed, Z.A.; Karisch, R.; Wang, Q.; Kowalski, J.; Susky, E.; Pereira, K.; Karamboulas, C.; Moghal, N.; Rajeshkumar, N.V.; et al. Tumor-initiating cells are rare in many human tumors. Cell Stem Cell 2010, 7, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Jarriault, S.; Brou, C.; Logeat, F.; Schroeter, E.H.; Kopan, R.; Israel, A. Signalling downstream of activated mammalian Notch. Nature 1995, 377, 355–358. [Google Scholar] [CrossRef]
- Kopan, R.; Ilagan, M.X. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef]
- Majumder, S.; Crabtree, J.S.; Golde, T.E.; Minter, L.M.; Osborne, B.A.; Miele, L. Targeting Notch in oncology: The path forward. Nat. Rev. Drug Discov. 2021, 20, 125–144. [Google Scholar] [CrossRef] [PubMed]
- Tanimizu, N.; Miyajima, A. Notch signaling controls hepatoblast differentiation by altering the expression of liver-enriched transcription factors. J. Cell Sci. 2004, 117, 3165–3174. [Google Scholar] [CrossRef]
- Wang, T.; You, N.; Tao, K.; Wang, X.; Zhao, G.; Xia, N.; Li, N.; Tang, L.; Liu, W.; Dou, K.; et al. Notch is the key factor in the process of fetal liver stem/progenitor cells differentiation into hepatocytes. Dev. Growth Diff. 2012, 54, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Liu, W.H.; Liang, H.Y.; Yan, H.T.; Lin, N.; Li, D.Y.; Wang, T.; Tang, L.J. Differentiation-inducing therapeutic effect of Notch inhibition in reversing malignant transformation of liver normal stem cells via MET. Oncotarget 2018, 9, 18885–18895. [Google Scholar] [CrossRef]
- Giovannini, C.; Gramantieri, L.; Minguzzi, M.; Fornari, F.; Chieco, P.; Grazi, G.L.; Bolondi, L. CDKN1C/P57 is regulated by the Notch target gene Hes1 and induces senescence in human hepatocellular carcinoma. Am. J. Pathol. 2012, 181, 413–422. [Google Scholar] [CrossRef] [PubMed]
- Razumilava, N.; Gores, G.J. Notch-driven carcinogenesis: The merging of hepatocellular cancer and cholangiocarcinoma into a common molecular liver cancer subtype. J. Hepatol. 2013, 58, 1244–1245. [Google Scholar] [CrossRef] [PubMed]
- Giovannini, C.; Baglioni, M.; Baron Toaldo, M.; Ventrucci, C.; D’Adamo, S.; Cipone, M.; Chieco, P.; Gramantieri, L.; Bolondi, L. Notch3 inhibition enhances sorafenib cytotoxic efficacy by promoting GSK3b phosphorylation and p21 down-regulation in hepatocellular carcinoma. Oncotarget 2013, 4, 1618–1631. [Google Scholar] [CrossRef]
- Giovannini, C.; Gramantieri, L.; Chieco, P.; Minguzzi, M.; Lago, F.; Pianetti, S.; Ramazzoti, E.; Marcu, K.B.; Bolondi, L. Selective ablation of Notch3 in HCC enhances doxorubicin’s death promoting effect by a p53 dependent mechanism. J. Hepatol. 2009, 50, 969–979. [Google Scholar] [CrossRef]
- Giovannini, C.; Salzano, A.M.; Baglioni, M.; Vitale, M.; Scaloni, A.; Zambrano, N.; Giannone, F.A.; Vasuri, F.; D’Errico, A.; Svegliati Baroni, G.; et al. Brivanib in combination with Notch3 silencing shows potent activity in tumour models. Br. J. Cancer 2019, 120, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lu, C.; Fang, T.; Wang, Y.; Hu, W.; Qiao, J.; Liu, B.; Liu, J.; Chen, N.; Li, M.; et al. Notch3 functions as a regulator of cell self-renewal by interacting with the beta-catenin pathway in hepatocellular carcinoma. Oncotarget 2015, 6, 3669–3679. [Google Scholar] [CrossRef]
- Wang, R.; Sun, Q.; Wang, P.; Liu, M.; Xiong, S.; Luo, J.; Huang, H.; Du, Q.; Galler, D.A.; Cheng, B. Notch and Wnt/beta-catenin signaling pathway play important roles in activating liver cancer stem cells. Oncotarget 2016, 7, 5754–5768. [Google Scholar] [CrossRef]
- Giovannini, C.; Minguzzi, M.; Genovese, F.; Baglioni, M.; Gualandi, A.; Ravaioli, M.; Milazzo, M.; Tavolari, S.; Bolondi, L.; Gramantieri, L. Molecular and proteomic insight into Notch1 characterization in hepatocellular carcinoma. Oncotarget 2016, 7, 39609–39626. [Google Scholar] [CrossRef]
- Piguet, A.C.; Saar, B.; Hlushchuk, R.; St-Pierre, M.V.; McSheehy, P.M.; Radojevic, V.; Afthinos, M.; Terracciano, L.; Djonov, V.; Dufour, J.F. Everolimus augments the effects of sorafenib in a syngeneic orthotopic model of hepatocellular carcinoma. Mol. Cancer Ther. 2011, 10, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Giovannini, C.; Fornari, F.; Piscaglia, F.; Gramantieri, L. Notch signaling regulation in HCC: From hepatitis virus to non-coding RNAs. Cells 2021, 10, 521. [Google Scholar] [CrossRef]
- Wang, R.; Li, Y.; Tsung, A.; Huang, H.; Du, Q.; Yang, M.; Deng, M.; Xiong, S.; Wang, X.; Zhang, L.; et al. iNOS promotes CD24(+)CD133(+) liver cancer stem cell phenotype through a TACE/ADAM17-dependent Notch signaling pathway. Proc. Natl. Acad. Sci. USA 2018, 115, E10127–E10136. [Google Scholar] [CrossRef]
- Kumar, H.; Gupta, S.P.; Anees, A. Advances in Design and development of inhibitors of nitric oxide synthases. Curr. Enzym. Inhib. 2013, 9, 117–141. [Google Scholar] [CrossRef]
- Yu, D.D.; Jing, Y.Y.; Guo, S.W.; Ye, F.; Lu, W.; Li, Q.; Dong, Y.L.; Gal, L.; Yang, Y.T.; Yang, Y.; et al. Overexpression of hepatocyte nuclear factor-1beta predicting poor prognosis is associated with biliary phenotype in patients with hepatocellular carcinoma. Sci. Rep. 2015, 5, 13319. [Google Scholar] [CrossRef]
- Zhu, J.N.; Jiang, L.; Jiang, J.H.; Yang, X.; Li, X.Y.; Zeng, J.X.; Shi, R.Y.; Shi, Y.; Pan, X.R.; Han, Z.P.; et al. Hepatocyte nuclear factor-1beta enhances the stemness of hepatocellular carcinoma cells through activation of the Notch pathway. Sci. Rep. 2017, 7, 4793. [Google Scholar] [CrossRef]
- Fang, S.; Liu, M.; Li, L.; Zhang, F.F.; Li, Y.; Yan, Q.; Cui, Y.Z.; Zhu, Y.H.; Yuan, Y.F.; Guan, X.Y. Lymphoid enhancer-binding factor-1 promotes stemness and poor differentiation of hepatocellular carcinoma by directly activating the NOTCH pathway. Oncogene 2019, 38, 4061–4074. [Google Scholar] [CrossRef] [PubMed]
- Moore, G.; Annett, S.; McClements, L.; Robson, T. Top notch targeting strategies in cancer: A detailed, overview of recent insights and current perspectives. Cells 2020, 9, 1503. [Google Scholar] [CrossRef] [PubMed]
- Mansour, F.A.; Al-Mazrou, A.; Al-Mohanna, F.; Al-Alwan, M.; Ghebeh, H. PD-L1 is overexpressed on breast cancer stem cells through notch3/mTOR axis. Oncoimmunology 2020, 9, 1729299. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Gramantieri, L.; Ferracin, M.; Fornari, F.; Veronese, A.; Sabbioni, S.; Liu, C.G.; Calin, G.A.; Giovannini, C.; Ferrazzi, E.; Grazi, G.L.; et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007, 67, 6092–6099. [Google Scholar] [CrossRef]
- Murakami, Y.; Yasuda, T.; Saigo, K.; Urashima, T.; Toyoda, H.; Okanoue, T.; Shimotohno, K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene 2006, 25, 2537–2545. [Google Scholar] [CrossRef] [PubMed]
- Pineau, P.; Volinia, S.; McJunkin, K.; Marchio, A.; Battiston, C.; Terris, B.; Mazzaferro, V.; Lowe, S.W.; Croce, C.M.; Dejean, A. miR-221 overexpression contributes to liver tumorigenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 264–269. [Google Scholar] [CrossRef]
- Fornari, F.; Gramantieri, L.; Ferracin, M.; Veronese, A.; Sabbioni, S.; Calin, G.A.; Grazi, G.L.; Giovannini, C.; Croce, C.M.; Bolondi, L.; et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 2008, 27, 5651–5661. [Google Scholar] [CrossRef]
- Gong, J.; Zhang, J.P.; Li, B.; Zeng, C.; You, K.; Chen, M.X.; Yuan, Y.; Zhuang, S.M. MicroRNA-125b promotes apoptosis by regulating the expression of Mcl-1, Bcl-w and IL-6R. Oncogene 2013, 32, 3071–3079. [Google Scholar] [CrossRef]
- Garofalo, M.; Di Leva, G.; Romano, G.; Nuovo, G.; Suh, S.S.; Ngankeu, A.; Taccioli, C.; Pichiorri, F.; Alder, H.; Secchiero, P.; et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 2009, 16, 498–509. [Google Scholar] [PubMed]
- Budhu, A.; Jia, H.L.; Forgues, M.; Liu, C.G.; Goldstein, D.; Lam, A.; Zanetti, K.A.; Ye, Q.H.; Qin, L.X.; Croce, C.M.; et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology 2008, 47, 897–907. [Google Scholar] [CrossRef]
- Ji, J.; Yamashita, T.; Budhu, A.; Forgues, M.; Jia, H.L.; Li, C.; Deng, C.; Wauthier, E.; Reid, L.M.; Ye, Q.H.; et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology 2009, 50, 472–480. [Google Scholar] [CrossRef]
- Lin, X.; Zuo, S.; Luo, R.; Li, Y.; Yu, G.; Zou, Y.; Zhou, Y.; Liu, Z.; Liu, Y.; Hu, Y.; et al. HBX-induced miR-5188 impairs FOXO1 to stimulate beta-catenin nuclear translocation and promotes tumor stemness in hepatocellular carcinoma. Theranostics 2019, 9, 7583–7598. [Google Scholar] [CrossRef] [PubMed]
- Chai, S.; Ng, K.Y.; Tong, M.; Lau, E.Y.; Lee, T.K.; Chan, K.W.; Yuan, Y.F.; Cheung, T.T.; Cheung, S.T.; Wang, X.Q.; et al. Octamer 4/microRNA-1246 signaling axis drives Wnt/beta-catenin activation in liver cancer stem cells. Hepatology 2016, 64, 2062–2076. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.X.; Wang, J.; Yang, F.; Tao, Q.F.; Zhang, J.; Wang, L.L.; Yang, Y.; Liu, H.; Wang, Z.G.; Xu, Q.G.; et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology 2016, 63, 499–511. [Google Scholar] [CrossRef]
- Mao, Y.; Chen, W.; Wu, H.; Liu, C.; Zhang, J.; Chen, S. Mechanisms and functions of MiR-200 family in hepatocellular carcinoma. OncoTargets Ther. 2020, 13, 13479–13490. [Google Scholar] [CrossRef]
- Tsai, S.C.; Lin, C.C.; Shih, T.C.; Tseng, R.J.; Yu, M.C.; Lin, Y.J.; Hsieh, S.Y. The miR-200b-ZEB1 circuit regulates diverse stemness of human hepatocellular carcinoma. Mol. Carcinog. 2017, 56, 2035–2047. [Google Scholar] [CrossRef] [PubMed]
- Brabletz, S.; Bajdak, K.; Meidhof, S.; Burk, U.; Niedermann, G.; Firat, E.; Wellner, U.; Dimmler, A.; Faller, G.; Schubert, J.; et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 2011, 30, 770–782. [Google Scholar] [CrossRef]
- Fragoso, R.; Mao, T.; Wang, S.; Schaffert, S.; Gong, X.; Yue, S.; Luong, R.; Min, H.; Yashiro-Ohtani, Y.; Davis, M.; et al. Modulating the strength and threshold of NOTCH oncogenic signals by mir-181a-1/b-1. PLoS Genet. 2012, 8, e1002855. [Google Scholar] [CrossRef]
- Zhang, J.; Jiao, J.; Cermelli, S.; Muir, K.; Jung, K.H.; Zou, R.; Rashid, A.; Gagea, M.; Zabludoff, S.; Kalluri, R.; et al. miR-21 inhibition reduces liver fibrosis and prevents tumor development by inducing apoptosis of CD24+ progenitor cells. Cancer Res. 2015, 75, 1859–1867. [Google Scholar] [CrossRef]
- Jung, K.H.; Zhang, J.; Zhou, C.; Shen, H.; Gagea, M.; Rodriguez-Aguayo, C.; Lopez-Berestein, G.; Sood, A.K.; Beretta, L. Differentiation therapy for hepatocellular carcinoma: Multifaceted effects of miR-148a on tumor growth and phenotype and liver fibrosis. Hepatology 2016, 63, 864–879. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Kwon, H.; Han, C.; Zhang, J.; Dash, S.; Lim, K.; Wu, T. Active glycolytic metabolism in CD133(+) hepatocellular cancer stem cells: Regulation by MIR-122. Oncotarget 2015, 6, 40822–40835. [Google Scholar] [CrossRef] [PubMed]
- Turato, C.; Fornari, F.; Pollutri, D.; Fassan, M.; Quarta, S.; Villano, G.; Ruvoletto, M.; Bolondi, L.; Gramantieri, L.; Pontisso, P. MiR-122 targets serpinB3 and is involved in sorafenib resistance in hepatocellular carcinoma. J. Clin. Med. 2019, 8, 171. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Tang, K.H.; Chan, Y.P.; Lee, T.K.; Kwan, P.S.; Castilho, A.; Ng, I.; Man, K.; Wong, N.; To, K.F.; et al. miR-130b promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell 2010, 7, 694–707. [Google Scholar] [CrossRef]
- Gu, Y.; Wei, X.; Sun, Y.; Gao, H.; Zheng, X.; Wong, L.L.; Jin, L.; Liu, N.; Hernandez, B.; Peplowska, K.; et al. miR-192-5p silencing by genetic aberrations is a key event in hepatocellular carcinomas with cancer stem cell features. Cancer Res. 2019, 79, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Gramantieri, L.; Pollutri, D.; Gagliardi, M.; Giovannini, C.; Quarta, S.; Ferracin, M.; Casadei-Gardini, A.; Callegari, E.; De Carolis, S.; Marinelli, S.; et al. MiR-30e-3p influences tumor phenotype through MDM2/TP53 axis and predicts sorafenib resistance in hepatocellular carcinoma. Cancer Res. 2020, 80, 1720–1734. [Google Scholar] [CrossRef] [PubMed]
- Luk, J.M.; Burchard, J.; Zhang, C.; Liu, A.M.; Wong, K.F.; Shek, F.H.; Lee, N.P.; Fan, S.T.; Poon, R.T.; Ivanovska, I.; et al. DLK1-DIO3 genomic imprinted microRNA cluster at 14q32.2 defines a stemlike subtype of hepatocellular carcinoma associated with poor survival. J. Biol. Chem. 2011, 286, 30706–30713. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.R.; Xu, M.; Toffanin, S.; Li, Y.; Llovet, J.M.; Russell, D.W. Induction of hepatocellular carcinoma by in vivo gene targeting. Proc. Natl. Acad. Sci. USA 2012, 109, 11264–11269. [Google Scholar] [CrossRef]
- Pollutri, D.; Patrizi, C.; Marinelli, S.; Giovannini, C.; Trombetta, E.; Giannone, F.A.; Baldassarre, M.; Quarta, S.; Vandewynckel, Y.P.; Vandierendonck, A.; et al. The epigenetically regulated miR-494 associates with stem-cell phenotype and induces sorafenib resistance in hepatocellular carcinoma. Cell Death Dis. 2018, 9, 4. [Google Scholar] [CrossRef]
- Lim, L.; Balakrishnan, A.; Huskey, N.; Jones, K.D.; Jodari, M.; Ng, R.; Song, G.; Riordan, J.; Anderton, B.; Cheung, S.T.; et al. MicroRNA-494 within an oncogenic microRNA megacluster regulates G1/S transition in liver tumorigenesis through suppression of mutated in colorectal cancer. Hepatology 2014, 59, 202–215. [Google Scholar] [CrossRef]
- Augello, C.; Vaira, V.; Caruso, L.; Destro, A.; Maggioni, M.; Park, Y.N.; Montorsi, M.; Santambrogio, R.; Roncalli, M.; Bosari, S.; et al. MicroRNA profiling of hepatocarcinogenesis identifies C19MC cluster as a novel prognostic biomarker in hepatocellular carcinoma. Liver Int. 2012, 32, 772–782. [Google Scholar] [CrossRef]
- Long, J.; Jiang, C.; Liu, B.; Dai, Q.; Hua, R.; Chen, C.; Zhang, B.; Li, H. Maintenance of stemness by miR-589-5p in hepatocellular carcinoma cells promotes chemoresistance via STAT3 signaling. Cancer Lett. 2018, 423, 113–126. [Google Scholar] [CrossRef]
- Wei, X.; Zhao, L.; Ren, R.; Ji, F.; Xue, S.; Zhang, J.; Liu, Z.; Ma, Z.; Wang, X.W.; Wong, L.; et al. MiR-125b Loss activated hif1alpha/pakt loop, leading to transarterial chemoembolization resistance in hepatocellular carcinoma. Hepatology 2020, 73, 1381–1398. [Google Scholar] [CrossRef]
- Zhou, J.N.; Zeng, Q.; Wang, H.Y.; Zhang, B.; Li, S.T.; Nan, X.; Cao, N.; Fu, C.J.; Yan, X.L.; Jia, Y.L.; et al. MicroRNA-125b attenuates epithelial-mesenchymal transitions and targets stem-like liver cancer cells through small mothers against decapentaplegic 2 and 4. Hepatology 2015, 62, 801–815. [Google Scholar] [CrossRef]
- Dong, K.S.; Chen, Y.; Yang, G.; Liao, Z.B.; Zhang, H.W.; Liang, H.F.; Chen, X.P.; Dong, H.H. TGF-beta1 accelerates the hepatitis B virus X-induced malignant transformation of hepatic progenitor cells by upregulating miR-199a-3p. Oncogene 2020, 39, 1807–1820. [Google Scholar] [CrossRef] [PubMed]
- Callegari, E.; D’Abundo, L.; Guerriero, P.; Simioni, C.; Elamin, B.K.; Russo, M.; Cani, A.; Bassi, C.; Zagatti, B.; Giacomelli, L.; et al. miR-199a-3p modulates MTOR and PAK4 pathways and inhibits tumor growth in a hepatocellular carcinoma transgenic mouse model. Mol. Ther. Nucleic Acids 2018, 11, 485–493. [Google Scholar] [CrossRef]
- Varshney, A.; Panda, J.J.; Singh, A.K.; Yadav, N.; Bihari, C.; Biswas, S.; Sarin, S.K.; Chauhan, V.S. Targeted delivery of microRNA-199a-3p using self-assembled dipeptide nanoparticles efficiently reduces hepatocellular carcinoma in mice. Hepatology 2018, 67, 1392–1407. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wang, N.; Gong, Z.; Liu, L.; Yang, S.; Chen, G.G.; Lai, P.B.S. Cytochrome, P450 1A2 overcomes nuclear factor kappa B-mediated sorafenib resistance in hepatocellular carcinoma. Oncogene 2021, 40, 492–507. [Google Scholar] [CrossRef]
- Willingham, S.B.; Volkmer, J.P.; Gentles, A.J.; Sahoo, D.; Dalerba, P.; Mitra, S.S.; Wang, J.; Contreras-Trujillo, H.; Martin, R.; Cohen, J.D.; et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc. Natl. Acad. Sci. USA 2012, 109, 6662–6667. [Google Scholar] [CrossRef]
- Lo, J.; Lau, E.Y.; Ching, R.H.; Cheng, B.Y.; Ma, M.K.; Ng, I.O.; Lee, T.K. Nuclear factor kappa B-mediated CD47 up-regulation promotes sorafenib resistance and its blockade synergizes the effect of sorafenib in hepatocellular carcinoma in mice. Hepatology 2015, 62, 534–545. [Google Scholar] [CrossRef]
- Marquardt, J.U.; Gomez-Quiroz, L.; Arreguin Camacho, L.O.; Pinna, F.; Lee, Y.H.; Kitade, M.; Dominguez, M.P.; Castven, D.; Breuhahn, K.; Conner, E.A.; et al. Curcumin effectively inhibits oncogenic NF-kappaB signaling and restrains stemness features in liver cancer. J. Hepathol. 2015, 63, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Fischer, P.M.; Gianella-Borradori, A. CDK inhibitors in clinical development for the treatment of cancer. Expert Opin. Investig. Drug 2003, 12, 955–970. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.X.; Wang, X.Q.; Chok, S.H.; Man, K.; Tsang, S.H.Y.; Chan, A.C.Y.; Ma, K.W.; Xia, W.; Cheung, T.T. Blocking CDK1/PDK1/beta-Catenin signaling by CDK1 inhibitor RO3306 increased the efficacy of sorafenib treatment by targeting cancer stem cells in a preclinical model of hepatocellular carcinoma. Theranostics 2018, 8, 3737–3750. [Google Scholar] [CrossRef] [PubMed]
- Bollard, J.; Miguela, V.; Ruiz de Galarreta, M.; Venkatesh, A.; Bian, C.B.; Roberto, M.P.; Tovar, V.; Sia, D.; Molina-Sanchez, P.; Nguyen, C.B.; et al. Palbociclib (PD-0332991), a selective CDK4/6 inhibitor, restricts tumour growth in preclinical models of hepatocellular carcinoma. Gut 2017, 66, 1286–1296. [Google Scholar] [CrossRef]
- Kahraman, D.C.; Kahraman, T.; Cetin-Atalay, R. Targeting PI3K/Akt/mTOR pathway identifies differential expression and functional role of IL8 in liver cancer stem cell enrichment. Mol. Cancer. Ther. 2019, 18, 2146–2157. [Google Scholar]
- Kudo, M.; Finn, R.S.; Qin, S.; Han, K.H.; Ikeda, K.; Piscaglia, F.; Baron, A.; Park, J.W.; Han, G.; Jassem, J.; et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: A randomised phase 3 non-inferiority trial. Lancet 2018, 391, 1163–1173. [Google Scholar] [CrossRef]
- Shigesawa, T.; Maehara, O.; Suda, G.; Natsuizaka, M.; Kimura, M.; Shimazaki, T.; Yamamoto, K.; Yamada, R.; Kitagataya, T.; Nakamura, A.; et al. Lenvatinib suppresses cancer stem-like cells in HCC by inhibiting FGFR1-3 signaling, but not FGFR4 signaling. Carcinogenesis 2021, 42, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.Z.; Pan, K.; Wang, Q.J.; Weng, D.S.; Zhao, J.J.; Zheng, H.X.; Zhang, X.F.; Jiang, S.S.; Lv, L.; Tang, Y.; et al. Annexin A3 as a potential target for immunotherapy of liver cancer stem-like cells. Stem Cells 2015, 33, 354–366. [Google Scholar] [CrossRef]
- Sun, Y.; Wu, L.; Zhong, Y.; Zhou, K.; Hou, Y.; Wang, Z.; Zhang, Z.; Xie, J.; Wang, C.; Chen, D.; et al. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell 2021, 184, 404–421.e16. [Google Scholar] [CrossRef]
- Cohen, S.J.; Punt, C.J.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed]
- de Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.; Pienta, K.J.; Raghavan, D.; et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef]
- Liu, M.C.; Shields, P.G.; Warren, R.D.; Cohen, P.; Wilkinson, M.; Ottaviano, Y.L.; Rao, S.B.; Eng-Wong, J.; Seillier-Moiseiwitsch, F.; Noone, A.M.; et al. Circulating tumor cells: A useful predictor of treatment efficacy in metastatic breast cancer. J. Clin. Oncol. 2009, 27, 5153–5159. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Wang, W.S.; Zhu, X.L.; Ni, C.F. High epithelial cell adhesion molecule-positive circulating tumor cell count predicts poor survival of patients with unresectable hepatocellular carcinoma treated with transcatheter arterial chemoembolization. J. Vasc. Interv. Radiol. 2018, 29, 1678–1684. [Google Scholar] [CrossRef]
- Sun, Y.F.; Xu, Y.; Yang, X.R.; Guo, W.; Zhang, X.; Qiu, S.J.; Shi, R.Y.; Hu, B.; Zhou, J.; Fan, J. Circulating stem cell-like epithelial cell adhesion molecule-positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology 2013, 57, 1458–1468. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Forgues, M.; Wang, W.; Kim, J.W.; Ye, Q.; Jia, H.; Budhu, A.; Zanetti, K.A.; Chen, Y.; Qin, L.X.; et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008, 68, 1451–1461. [Google Scholar] [CrossRef]
- Gorges, T.M.; Tinhofer, I.; Drosch, M.; Rose, L.; Zollner, T.M.; Krahn, T.; von Ahsen, O. Circulating tumour cells escape from EpCAM-based detection due to epithelial-to-mesenchymal transition. BMC Cancer 2012, 12, 178. [Google Scholar] [CrossRef]
- Miller, M.C.; Robinson, P.S.; Wagner, C.; O’Shannessy, D.J. The Parsortix Cell Separation System—A versatile liquid biopsy platform. Cytometry 2018, 93, 1234–1239. [Google Scholar] [CrossRef] [PubMed]
- Halabi, S.; Small, E.J.; Hayes, D.F.; Vogelzang, N.J.; Kantoff, P.W. Prognostic significance of reverse transcriptase polymerase chain reaction for prostate-specific antigen in metastatic prostate cancer: A nested study within CALGB 9583. J. Clin. Oncol. 2003, 21, 490–495. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Kallergi, G.; Ntoulia, M.; Perraki, M.; Apostolaki, S.; Kafousi, M.; Chlouverakis, G.; Stathopoulos, E.; Lianidou, E.; Georgoulias, V.; et al. Prognostic value of the molecular detection of circulating tumor cells using a multimarker reverse transcription-PCR assay for cytokeratin 19, mammaglobin A, and HER2 in early breast cancer. Clin. Cancer Res. 2008, 14, 2593–2600. [Google Scholar] [CrossRef]
- Uen, Y.H.; Lin, S.R.; Wu, D.C.; Su, Y.C.; Wu, J.Y.; Cheng, T.L.; Chi, C.W.; Wang, J.Y. Prognostic significance of multiple molecular markers for patients with stage II colorectal cancer undergoing curative resection. Ann. Surg. 2007, 246, 1040–1046. [Google Scholar] [CrossRef]
- Fan, S.T.; Yang, Z.F.; Ho, D.W.; Ng, M.N.; Yu, W.C.; Wong, J. Prediction of posthepatectomy recurrence of hepatocellular carcinoma by circulating cancer stem cells: A prospective study. Ann. Surg. 2011, 254, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Chan, K.W.; Hu, L.; Lee, T.K.; Wo, J.Y.; Ng, I.O.; Zheng, B.J.; Guan, X.Y. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology 2007, 132, 2542–2556. [Google Scholar] [CrossRef]
- Yin, S.; Li, J.; Hu, C.; Chen, X.; Yao, M.; Yan, M.; Jiang, G.; Ge, C.; Xie, H.; Wan, D.; et al. CD133 positive hepatocellular carcinoma cells possess high capacity for tumorigenicity. Int. J. Cancer 2007, 120, 1444–1450. [Google Scholar] [CrossRef]
- Mathew, J.; Hines, J.E.; Obafunwa, J.O.; Burr, A.W.; Toole, K.; Burt, A.D. CD44 is expressed in hepatocellular carcinomas showing vascular invasion. J. Pathol. 1996, 179, 74–79. [Google Scholar] [CrossRef]
- Washington, K.; Telen, M.J.; Gottfried, M.R. Expression of cell adhesion molecule CD44 in primary tumors of the liver: An immunohistochemical study. Liver 1997, 17, 17–23. [Google Scholar] [CrossRef]
- Hou, Y.; Zou, Q.; Ge, R.; Shen, F.; Wang, Y. The critical role of CD133(+)CD44(+/high) tumor cells in hematogenous metastasis of liver cancers. Cell Res. 2012, 22, 259–272. [Google Scholar] [CrossRef]
- Zekri, A.N.; El-Sisi, E.R.; Youssef, A.S.E.; Kamel, M.M.; Nassar, A.; Ahmed, O.S.; El Kassas, M.; Barakat, A.B.; Abd El-Motaleb, A.I.; Bahnassy, A.A.; et al. MicroRNA Signatures for circulating CD133-positive cells in hepatocellular carcinoma with HCV infection. PLoS ONE 2018, 13, e0193709. [Google Scholar] [CrossRef]
- Boltin, D.; Kamenetsky, Z.; Perets, T.T.; Snir, Y.; Sapoznikov, B.; Schmilovitz-Weiss, H.; Ablin, J.N.; Dickman, R.; Niv, Y. Circulating Bone Marrow-Derived CD45-/CD34+/CD133+/VEGF+ Endothelial Progenitor Cells in Adults with Crohn’s Disease. Dig. Dis. Sci. 2017, 62, 633–638. [Google Scholar] [CrossRef]
- Xu, W.; Cao, L.; Chen, L.; Li, J.; Zhang, X.F.; Qian, H.H.; Kang, X.Y.; Zhang, Y.; Liao, J.; Shi, L.H.; et al. Isolation of circulating tumor cells in patients with hepatocellular carcinoma using a novel cell separation strategy. Clin. Cancer Res. 2011, 17, 3783–3793. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, L.; Zhang, X.; Zhang, Y.; Liu, H.; Sun, B.; Zhao, L.; Ge, N.; Qian, H.; Yang, Y.; et al. Detection of circulating tumor cells in hepatocellular carcinoma using antibodies against asialoglycoprotein receptor, carbamoyl phosphate synthetase 1 and pan-cytokeratin. PLoS ONE 2014, 9, e96185. [Google Scholar] [CrossRef] [PubMed]
- Hamaoka, M.; Kobayashi, T.; Tanaka, Y.; Mashima, H.; Ohdan, H. Clinical significance of glypican-3-positive circulating tumor cells of hepatocellular carcinoma patients: A prospective study. PLoS ONE 2019, 14, e0217586. [Google Scholar] [CrossRef]
- Mitra, A.; Satelli, A.; Xia, X.; Cutrera, J.; Mishra, L.; Li, S. Cell-surface Vimentin: A mislocalized protein for isolating csVimentin(+) CD133(-) novel stem-like hepatocellular carcinoma cells expressing EMT markers. Int. J. Cancer 2015, 137, 491–496. [Google Scholar] [CrossRef] [PubMed]
- Court, C.M.; Hou, S.; Winograd, P.; Segel, N.H.; Li, Q.W.; Zhu, Y.; Sadeghi, S.; Finn, R.S.; Ganapathy, E.; Song, M.; et al. A novel multimarker assay for the phenotypic profiling of circulating tumor cells in hepatocellular carcinoma. Liver Transpl. 2018, 24, 946–960. [Google Scholar] [CrossRef] [PubMed]
- Brocco, D.; Lanuti, P.; Simeone, P.; Bologna, G.; Pieragostino, D.; Cufaro, M.C.; Graziano, V.; Peri, M.; Di Marino, P.; De Tursi, M.; et al. Circulating cancer stem cell-derived extracellular vesicles as a novel biomarker for clinical outcome evaluation. J. Oncol. 2019, 2019, 5879616. [Google Scholar] [CrossRef] [PubMed]
- Grosse-Gehling, P.; Fargeas, C.A.; Dittfeld, C.; Garbe, Y.; Alison, M.R.; Corbeil, D.; Kunz-Schughart, L.A. CD133 as a biomarker for putative cancer stem cells in solid tumours: Limitations, problems and challenges. J. Pathol. 2013, 229, 355–378. [Google Scholar] [CrossRef]
- Michalopoulos, G.K.; Barua, L.; Bowen, W.C. Transdifferentiation of rat hepatocytes into biliary cells after bile duct ligation and toxic biliary injury. Hepatology 2005, 41, 535–544. [Google Scholar] [CrossRef]
- Keller, L.; Pantel, K. Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat. Rev. Cancer 2019, 19, 553–567. [Google Scholar] [CrossRef]
- Tovar, V.; Cornella, H.; Moeini, A.; Vidal, S.; Hoshida, Y.; Sia, D.; Peix, J.; Cabellos, L.; Alsinet, C.; Torrecilla, S.; et al. Tumour initiating cells and IGF/FGF signalling contribute to sorafenib resistance in hepatocellular carcinoma. Gut 2017, 66, 530–540. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gramantieri, L.; Giovannini, C.; Suzzi, F.; Leoni, I.; Fornari, F. Hepatic Cancer Stem Cells: Molecular Mechanisms, Therapeutic Implications, and Circulating Biomarkers. Cancers 2021, 13, 4550. https://doi.org/10.3390/cancers13184550
Gramantieri L, Giovannini C, Suzzi F, Leoni I, Fornari F. Hepatic Cancer Stem Cells: Molecular Mechanisms, Therapeutic Implications, and Circulating Biomarkers. Cancers. 2021; 13(18):4550. https://doi.org/10.3390/cancers13184550
Chicago/Turabian StyleGramantieri, Laura, Catia Giovannini, Fabrizia Suzzi, Ilaria Leoni, and Francesca Fornari. 2021. "Hepatic Cancer Stem Cells: Molecular Mechanisms, Therapeutic Implications, and Circulating Biomarkers" Cancers 13, no. 18: 4550. https://doi.org/10.3390/cancers13184550
APA StyleGramantieri, L., Giovannini, C., Suzzi, F., Leoni, I., & Fornari, F. (2021). Hepatic Cancer Stem Cells: Molecular Mechanisms, Therapeutic Implications, and Circulating Biomarkers. Cancers, 13(18), 4550. https://doi.org/10.3390/cancers13184550





