The Role of Oncogenes and Redox Signaling in the Regulation of PD-L1 in Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Structure of Human PD-L1 and Evolution
3. Tissue Distribution and Subcellular Localization
4. Biological Functions and Related Diseases
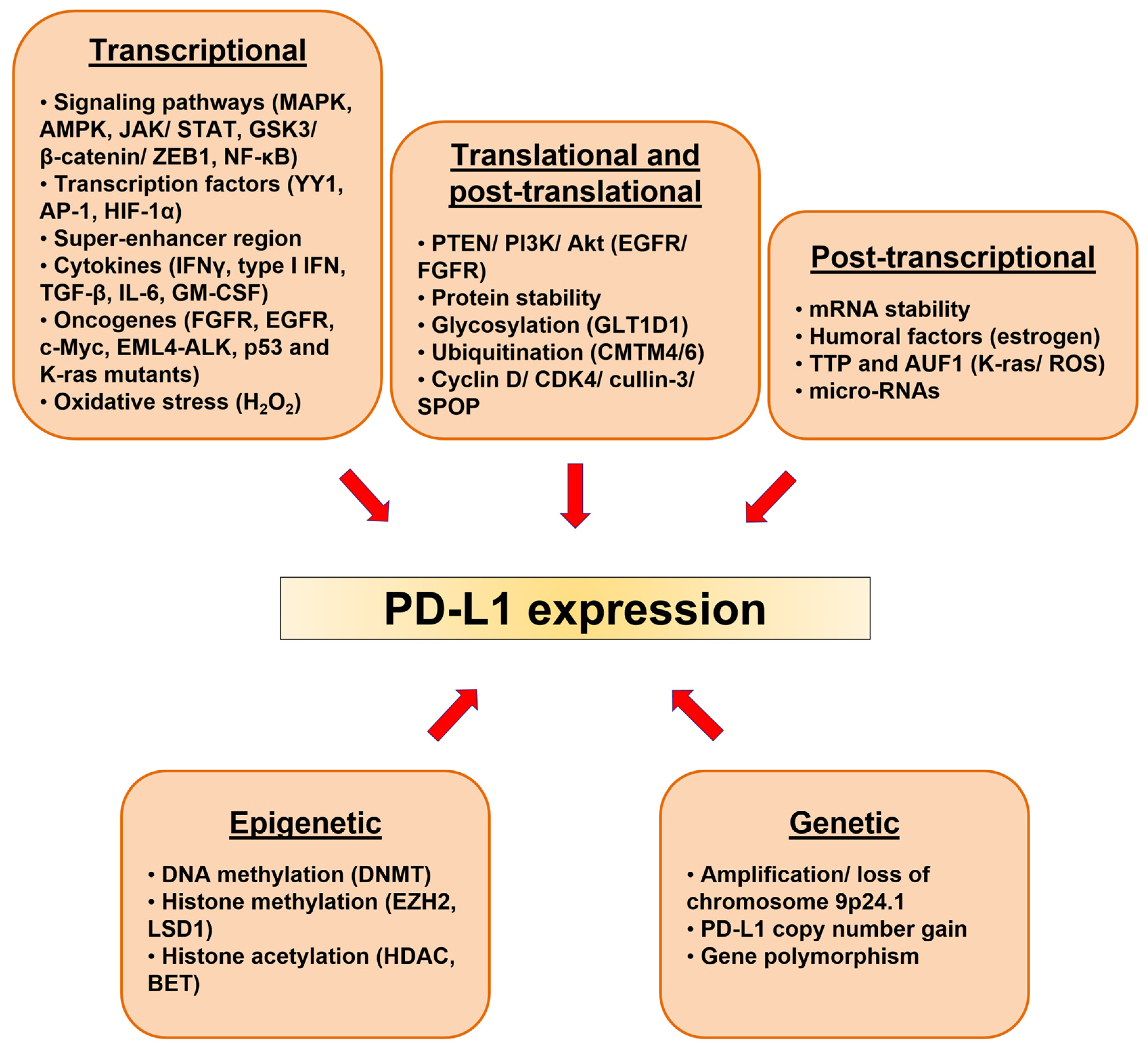
5. Regulation of PD-L1 in Cancerous Tissues
5.1. PD-L1 Expression in Cancers and Role in Tumorigenesis
5.2. Genetic Alterations
5.3. Epigenetic Alterations
5.4. Signaling Pathways and Transcriptional Regulation
5.5. Posttranscriptional and Posttranslational Regulations
5.6. Cytokines
5.7. Oncogenes
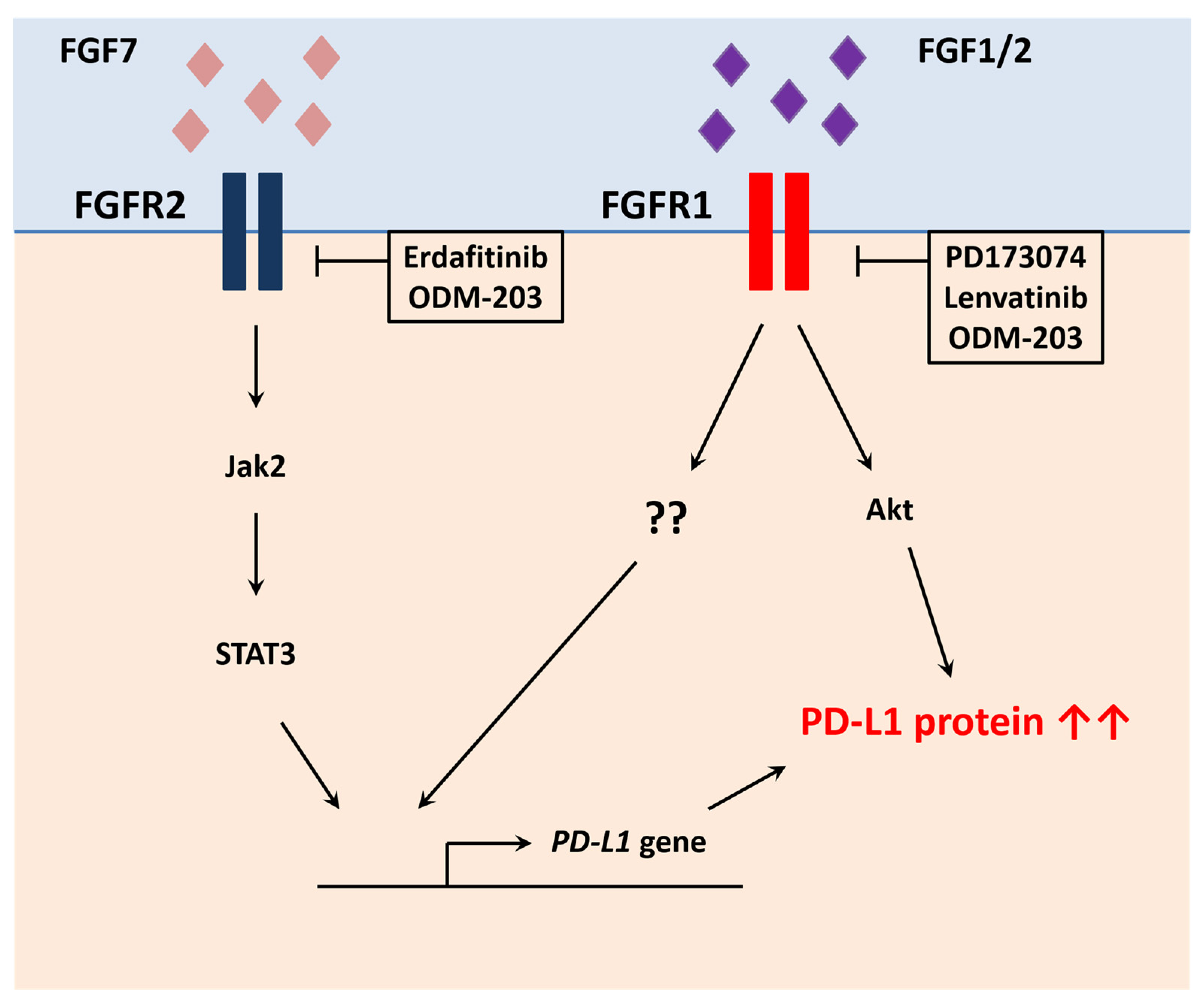
5.7.1. Fibroblast Growth Factor Receptors
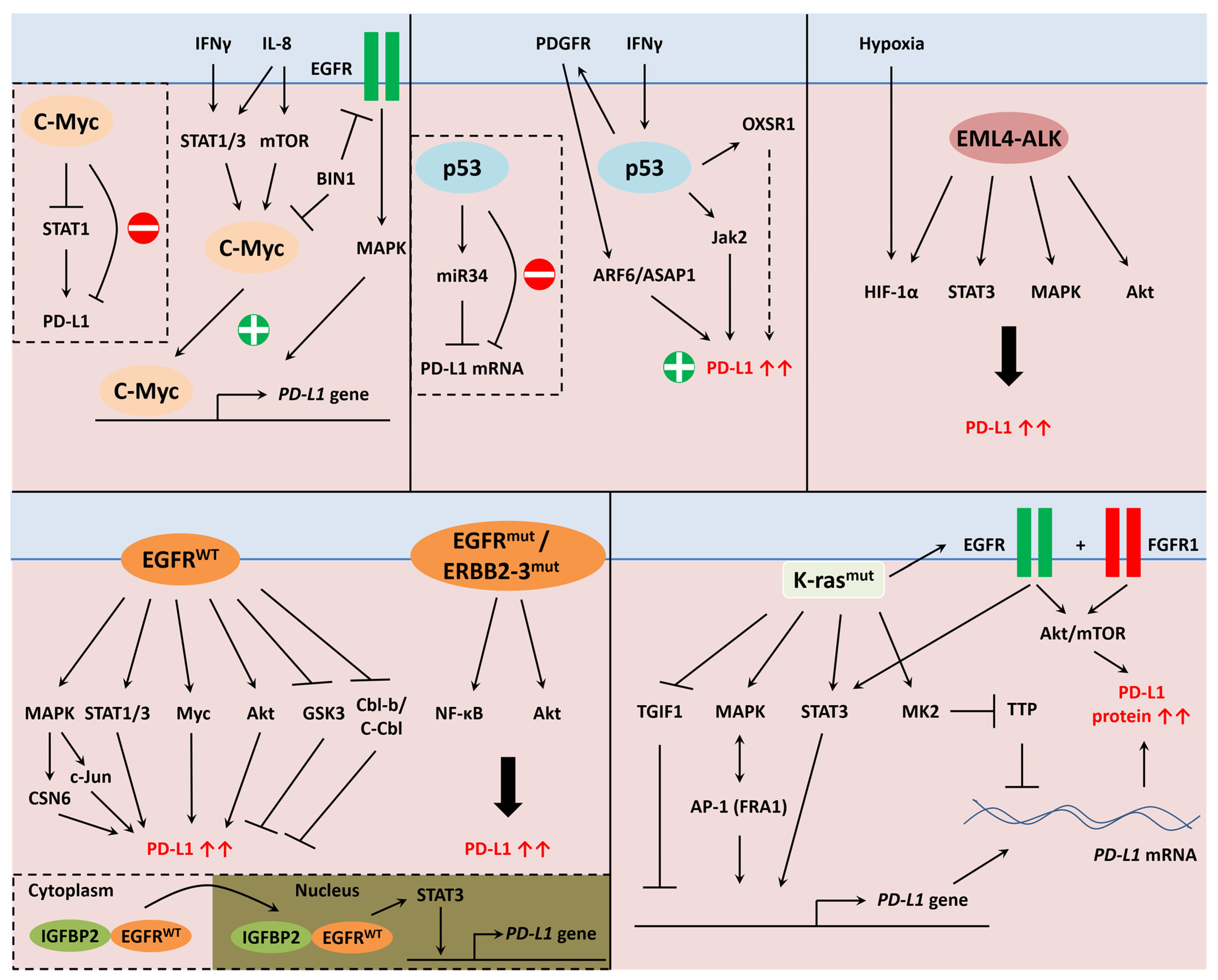
5.7.2. Epidermal Growth Factor Receptor
5.7.3. c-Myc
5.7.4. Protein p53
5.7.5. EML4-ALK
5.7.6. K-Ras
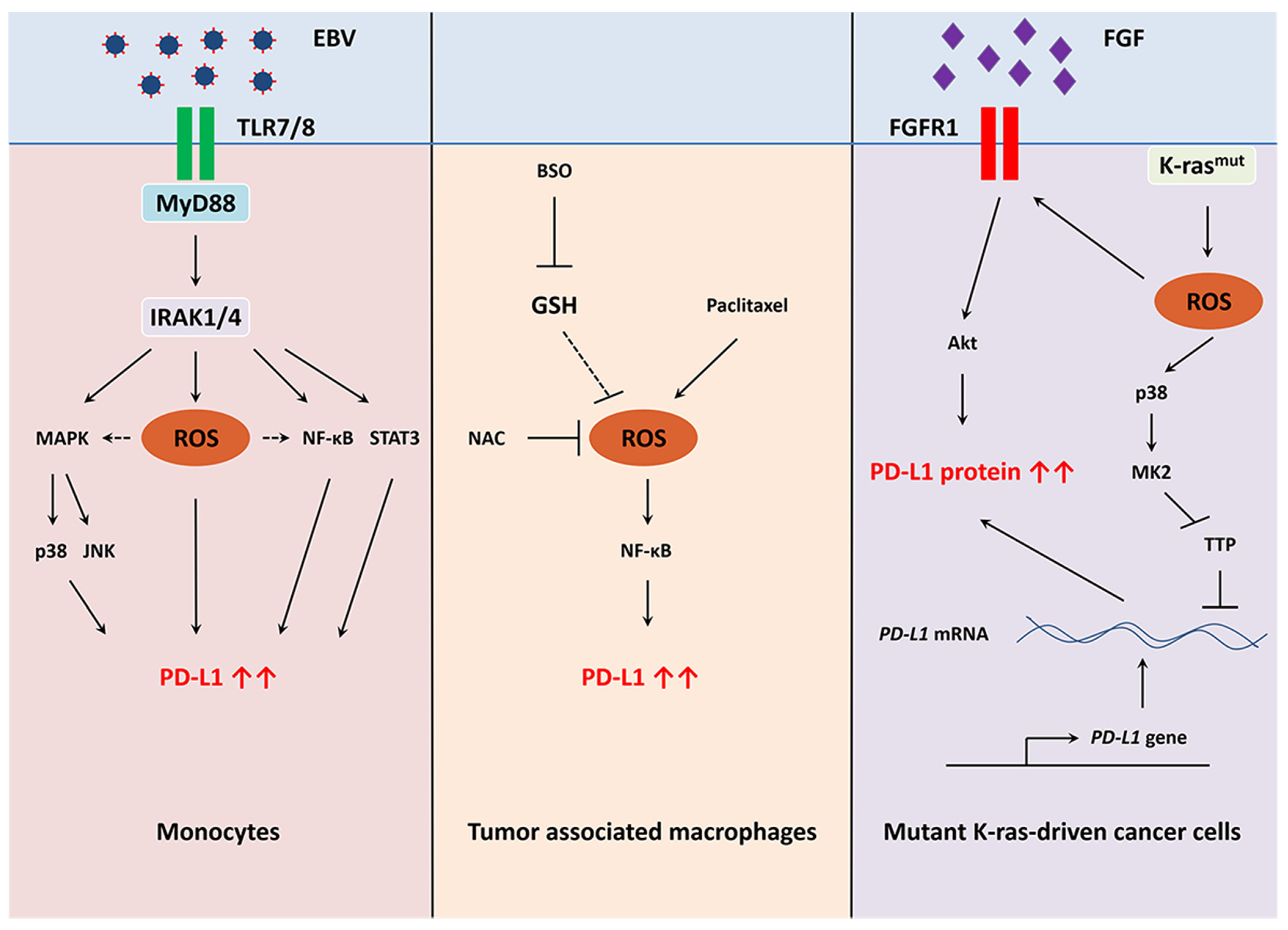
5.8. Redox Regulation of PD-L1
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Leach, D.R.; Krummel, M.F.; Allison, J.P. Enhancement of antitumor immunity by CTLA-4 blockade. Science 1996, 271, 1734–1736. [Google Scholar] [CrossRef]
- Ishida, Y.; Agata, Y.; Shibahara, K.; Honjo, T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992, 11, 3887–3895. [Google Scholar] [CrossRef]
- Dong, H.; Zhu, G.; Tamada, K.; Chen, L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat. Med. 1999, 5, 1365–1369. [Google Scholar] [CrossRef] [PubMed]
- Freeman, G.J.; Long, A.J.; Iwai, Y.; Bourque, K.; Chernova, T.; Nishimura, H.; Fitz, L.J.; Malenkovich, N.; Okazaki, T.; Byrne, M.C.; et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J. Exp. Med. 2000, 192, 1027–1034. [Google Scholar] [CrossRef]
- Latchman, Y.; Wood, C.R.; Chernova, T.; Chaudhary, D.; Borde, M.; Chernova, I.; Iwai, Y.; Long, A.J.; Brown, J.A.; Nunes, R.; et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat. Immunol. 2001, 2, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Larsen, T.V.; Hussmann, D.; Nielsen, A.L. PD-L1 and PD-L2 expression correlated genes in non-small-cell lung cancer. Cancer Commun. 2019, 39, 30. [Google Scholar] [CrossRef]
- Iwai, Y.; Ishida, M.; Tanaka, Y.; Okazaki, T.; Honjo, T.; Minato, N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. USA 2002, 99, 12293–12297. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, T.; Honjo, T. PD-1 and PD-1 ligands: From discovery to clinical application. Int. Immunol. 2007, 19, 813–824. [Google Scholar] [CrossRef]
- Lin, D.Y.; Tanaka, Y.; Iwasaki, M.; Gittis, A.G.; Su, H.P.; Mikami, B.; Okazaki, T.; Honjo, T.; Minato, N.; Garboczi, D.N. The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors. Proc. Natl. Acad. Sci. USA 2008, 105, 3011–3016. [Google Scholar] [CrossRef]
- Collins, M.; Ling, V.; Carreno, B.M. The B7 family of immune-regulatory ligands. Genome Biol. 2005, 6, 223. [Google Scholar] [CrossRef][Green Version]
- Zak, K.M.; Grudnik, P.; Magiera, K.; Domling, A.; Dubin, G.; Holak, T.A. Structural Biology of the Immune Checkpoint Receptor PD-1 and Its Ligands PD-L1/PD-L2. Structure 2017, 25, 1163–1174. [Google Scholar] [CrossRef]
- Keir, M.E.; Butte, M.J.; Freeman, G.J.; Sharpe, A.H. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 2008, 26, 677–704. [Google Scholar] [CrossRef]
- Nishimura, H.; Honjo, T. PD-1: An inhibitory immunoreceptor involved in peripheral tolerance. Trends Immunol. 2001, 22, 265–268. [Google Scholar] [CrossRef]
- Oktay, E.; Yalcin, G.D.; Ekmekci, S.; Kahraman, D.S.; Yalcin, A.; Degirmenci, M.; Dirican, A.; Altin, Z.; Ozdemir, O.; Surmeli, Z.; et al. Programmed cell death ligand-1 expression in gastroenteropancreatic neuroendocrine tumors. J. BUON 2019, 24, 779–790. [Google Scholar]
- Ishida, M.; Iwai, Y.; Tanaka, Y.; Okazaki, T.; Freeman, G.J.; Minato, N.; Honjo, T. Differential expression of PD-L1 and PD-L2, ligands for an inhibitory receptor PD-1, in the cells of lymphohematopoietic tissues. Immunol. Lett. 2002, 84, 57–62. [Google Scholar] [CrossRef]
- Liang, S.C.; Latchman, Y.E.; Buhlmann, J.E.; Tomczak, M.F.; Horwitz, B.H.; Freeman, G.J.; Sharpe, A.H. Regulation of PD-1, PD-L1, and PD-L2 expression during normal and autoimmune responses. Eur. J. Immunol. 2003, 33, 2706–2716. [Google Scholar] [CrossRef]
- Curiel, T.J.; Wei, S.; Dong, H.; Alvarez, X.; Cheng, P.; Mottram, P.; Krzysiek, R.; Knutson, K.L.; Daniel, B.; Zimmermann, M.C.; et al. Blockade of B7-H1 improves myeloid dendritic cell-mediated antitumor immunity. Nat. Med. 2003, 9, 562–567. [Google Scholar] [CrossRef] [PubMed]
- Loke, P.; Allison, J.P. PD-L1 and PD-L2 are differentially regulated by Th1 and Th2 cells. Proc. Natl. Acad. Sci. USA 2003, 100, 5336–5341. [Google Scholar] [CrossRef] [PubMed]
- Trabattoni, D.; Saresella, M.; Biasin, M.; Boasso, A.; Piacentini, L.; Ferrante, P.; Dong, H.; Maserati, R.; Shearer, G.M.; Chen, L.; et al. B7-H1 is up-regulated in HIV infection and is a novel surrogate marker of disease progression. Blood 2003, 101, 2514–2520. [Google Scholar] [CrossRef] [PubMed]
- Nazareth, M.R.; Broderick, L.; Simpson-Abelson, M.R.; Kelleher, R.J., Jr.; Yokota, S.J.; Bankert, R.B. Characterization of human lung tumor-associated fibroblasts and their ability to modulate the activation of tumor-associated T cells. J. Immunol. 2007, 178, 5552–5562. [Google Scholar] [CrossRef]
- Pinchuk, I.V.; Saada, J.I.; Beswick, E.J.; Boya, G.; Qiu, S.M.; Mifflin, R.C.; Raju, G.S.; Reyes, V.E.; Powell, D.W. PD-1 ligand expression by human colonic myofibroblasts/fibroblasts regulates CD4+ T-cell activity. Gastroenterology 2008, 135, 1228–1237. [Google Scholar] [CrossRef]
- Das, S.; Suarez, G.; Beswick, E.J.; Sierra, J.C.; Graham, D.Y.; Reyes, V.E. Expression of B7-H1 on gastric epithelial cells: Its potential role in regulating T cells during Helicobacter pylori infection. J. Immunol. 2006, 176, 3000–3009. [Google Scholar] [CrossRef]
- Ding, H.; Wu, X.; Gao, W. PD-L1 is expressed by human renal tubular epithelial cells and suppresses T cell cytokine synthesis. Clin. Immunol. 2005, 115, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Stanciu, L.A.; Bellettato, C.M.; Laza-Stanca, V.; Coyle, A.J.; Papi, A.; Johnston, S.L. Expression of programmed death-1 ligand (PD-L) 1, PD-L2, B7-H3, and inducible costimulator ligand on human respiratory tract epithelial cells and regulation by respiratory syncytial virus and type 1 and 2 cytokines. J. Infect. Dis. 2006, 193, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Reynoso, E.D.; Lee, J.W.; Turley, S.J. Peripheral tolerance induction by lymph node stroma. Adv. Exp. Med. Biol. 2009, 633, 113–127. [Google Scholar] [PubMed]
- Kitano, Y.; Yamashita, Y.I.; Nakao, Y.; Itoyama, R.; Yusa, T.; Umezaki, N.; Tsukamoto, M.; Yamao, T.; Miyata, T.; Nakagawa, S.; et al. Clinical Significance of PD-L1 Expression in Both Cancer and Stroma Cells of Cholangiocarcinoma Patients. Ann. Surg. Oncol. 2019, 27, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Teramoto, K.; Igarashi, T.; Kataoka, Y.; Ishida, M.; Hanaoka, J.; Sumimoto, H.; Daigo, Y. Clinical significance of PD-L1-positive cancer-associated fibroblasts in pN0M0 non-small cell lung cancer. Lung Cancer 2019, 137, 56–63. [Google Scholar] [CrossRef]
- Van Berckelaer, C.; Rypens, C.; van Dam, P.; Pouillon, L.; Parizel, M.; Schats, K.A.; Kockx, M.; Tjalma, W.A.A.; Vermeulen, P.; van Laere, S.; et al. Infiltrating stromal immune cells in inflammatory breast cancer are associated with an improved outcome and increased PD-L1 expression. Breast Cancer Res. 2019, 21, 28. [Google Scholar] [CrossRef]
- Wyss, J.; Dislich, B.; Koelzer, V.H.; Galvan, J.A.; Dawson, H.; Hadrich, M.; Inderbitzin, D.; Lugli, A.; Zlobec, I.; Berger, M.D. Stromal PD-1/PD-L1 Expression Predicts Outcome in Colon Cancer Patients. Clin. Colorectal Cancer 2019, 18, e20–e38. [Google Scholar] [CrossRef]
- Satelli, A.; Batth, I.S.; Brownlee, Z.; Rojas, C.; Meng, Q.H.; Kopetz, S.; Li, S. Potential role of nuclear PD-L1 expression in cell-surface vimentin positive circulating tumor cells as a prognostic marker in cancer patients. Sci. Rep. 2016, 6, 28910. [Google Scholar] [CrossRef]
- Chowdhury, S.; Veyhl, J.; Jessa, F.; Polyakova, O.; Alenzi, A.; MacMillan, C.; Ralhan, R.; Walfish, P.G. Programmed death-ligand 1 overexpression is a prognostic marker for aggressive papillary thyroid cancer and its variants. Oncotarget 2016, 7, 32318–32328. [Google Scholar] [CrossRef]
- Chen, L.; Deng, H.; Lu, M.; Xu, B.; Wang, Q.; Jiang, J.; Wu, C. B7-H1 expression associates with tumor invasion and predicts patient’s survival in human esophageal cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 6015–6023. [Google Scholar] [PubMed]
- Ghebeh, H.; Lehe, C.; Barhoush, E.; Al-Romaih, K.; Tulbah, A.; Al-Alwan, M.; Hendrayani, S.F.; Manogaran, P.; Alaiya, A.; Al-Tweigeri, T.; et al. Doxorubicin downregulates cell surface B7-H1 expression and upregulates its nuclear expression in breast cancer cells: Role of B7-H1 as an anti-apoptotic molecule. Breast Cancer Res. 2010, 12, R48. [Google Scholar] [CrossRef]
- He, X.H.; Xu, L.H.; Liu, Y. Identification of a novel splice variant of human PD-L1 mRNA encoding an isoform-lacking Igv-like domain. Acta Pharmacol. Sin. 2005, 26, 462–468. [Google Scholar] [CrossRef]
- Polioudaki, H.; Chantziou, A.; Kalyvianaki, K.; Malamos, P.; Notas, G.; Mavroudis, D.; Kampa, M.; Castanas, E.; Theodoropoulos, P.A. Nuclear localization of PD-L1: Artifact or reality? Cell. Oncol. 2019, 42, 237–242. [Google Scholar] [CrossRef]
- Latchman, Y.E.; Liang, S.C.; Wu, Y.; Chernova, T.; Sobel, R.A.; Klemm, M.; Kuchroo, V.K.; Freeman, G.J.; Sharpe, A.H. PD-L1-deficient mice show that PD-L1 on T cells, antigen-presenting cells, and host tissues negatively regulates T cells. Proc. Natl. Acad. Sci. USA 2004, 101, 10691–10696. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Dorfman, D.M.; Ma, F.R.; Sullivan, E.L.; Munoz, O.; Wood, C.R.; Greenfield, E.A.; Freeman, G.J. Blockade of programmed death-1 ligands on dendritic cells enhances T cell activation and cytokine production. J. Immunol. 2003, 170, 1257–1266. [Google Scholar] [CrossRef]
- Mazanet, M.M.; Hughes, C.C. B7-H1 is expressed by human endothelial cells and suppresses T cell cytokine synthesis. J. Immunol. 2002, 169, 3581–3588. [Google Scholar] [CrossRef] [PubMed]
- Rodig, N.; Ryan, T.; Allen, J.A.; Pang, H.; Grabie, N.; Chernova, T.; Greenfield, E.A.; Liang, S.C.; Sharpe, A.H.; Lichtman, A.H.; et al. Endothelial expression of PD-L1 and PD-L2 down-regulates CD8+ T cell activation and cytolysis. Eur. J. Immunol. 2003, 33, 3117–3126. [Google Scholar] [CrossRef] [PubMed]
- Wintterle, S.; Schreiner, B.; Mitsdoerffer, M.; Schneider, D.; Chen, L.; Meyermann, R.; Weller, M.; Wiendl, H. Expression of the B7-related molecule B7-H1 by glioma cells: A potential mechanism of immune paralysis. Cancer Res. 2003, 63, 7462–7467. [Google Scholar]
- Nishimura, H.; Nose, M.; Hiai, H.; Minato, N.; Honjo, T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity 1999, 11, 141–151. [Google Scholar] [CrossRef]
- Nishimura, H.; Okazaki, T.; Tanaka, Y.; Nakatani, K.; Hara, M.; Matsumori, A.; Sasayama, S.; Mizoguchi, A.; Hiai, H.; Minato, N.; et al. Autoimmune dilated cardiomyopathy in PD-1 receptor-deficient mice. Science 2001, 291, 319–322. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yoshida, T.; Nakaki, F.; Hiai, H.; Okazaki, T.; Honjo, T. Establishment of NOD-Pdcd1-/- mice as an efficient animal model of type I diabetes. Proc. Natl. Acad. Sci. USA 2005, 102, 11823–11828. [Google Scholar] [CrossRef]
- Lau, J.; Cheung, J.; Navarro, A.; Lianoglou, S.; Haley, B.; Totpal, K.; Sanders, L.; Koeppen, H.; Caplazi, P.; McBride, J.; et al. Tumour and host cell PD-L1 is required to mediate suppression of anti-tumour immunity in mice. Nat. Commun. 2017, 8, 14572. [Google Scholar] [CrossRef]
- Blank, C.; Gajewski, T.F.; Mackensen, A. Interaction of PD-L1 on tumor cells with PD-1 on tumor-specific T cells as a mechanism of immune evasion: Implications for tumor immunotherapy. Cancer Immunol. Immunother. 2005, 54, 307–314. [Google Scholar] [CrossRef]
- Mortezaee, K. Immune escape: A critical hallmark in solid tumors. Life Sci. 2020, 258, 118110. [Google Scholar] [CrossRef]
- Hamid, O.; Carvajal, R.D. Anti-programmed death-1 and anti-programmed death-ligand 1 antibodies in cancer therapy. Expert Opin. Biol. Ther. 2013, 13, 847–861. [Google Scholar] [CrossRef]
- Wang, X.; Teng, F.; Kong, L.; Yu, J. PD-L1 expression in human cancers and its association with clinical outcomes. Onco Targets Ther. 2016, 9, 5023–5039. [Google Scholar] [PubMed]
- Brahmer, J.; Reckamp, K.L.; Baas, P.; Crino, L.; Eberhardt, W.E.; Poddubskaya, E.; Antonia, S.; Pluzanski, A.; Vokes, E.E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Carbone, D.P.; Reck, M.; Paz-Ares, L.; Creelan, B.; Horn, L.; Steins, M.; Felip, E.; van den Heuvel, M.M.; Ciuleanu, T.E.; Badin, F.; et al. First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 376, 2415–2426. [Google Scholar] [CrossRef] [PubMed]
- Hanna, G.J.; Lizotte, P.; Cavanaugh, M.; Kuo, F.C.; Shivdasani, P.; Frieden, A.; Chau, N.G.; Schoenfeld, J.D.; Lorch, J.H.; Uppaluri, R.; et al. Frameshift events predict anti-PD-1/L1 response in head and neck cancer. JCI Insight 2018, 3, e98811. [Google Scholar] [CrossRef]
- Jerby-Arnon, L.; Shah, P.; Cuoco, M.S.; Rodman, C.; Su, M.J.; Melms, J.C.; Leeson, R.; Kanodia, A.; Mei, S.; Lin, J.R.; et al. A Cancer Cell Program Promotes T Cell Exclusion and Resistance to Checkpoint Blockade. Cell 2018, 175, 984–997. [Google Scholar] [CrossRef] [PubMed]
- Weber, J. Immunotherapy for melanoma. Curr. Opin. Oncol. 2011, 23, 163–169. [Google Scholar] [CrossRef]
- Henriksen, A.; Dyhl-Polk, A.; Chen, I.; Nielsen, D. Checkpoint inhibitors in pancreatic cancer. Cancer Treat Rev. 2019, 78, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Mehrvarz Sarshekeh, A.; Overman, M.J.; Kopetz, S. Nivolumab in the treatment of microsatellite instability high metastatic colorectal cancer. Future Oncol. 2018, 14, 1869–1874. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wu, X. Study and analysis of antitumor resistance mechanism of PD1/PD-L1 immune checkpoint blocker. Cancer Med. 2020, 9, 8086–8121. [Google Scholar] [CrossRef]
- Lacour, M.; Hiltbrunner, S.; Lee, S.Y.; Soltermann, A.; Rushing, E.J.; Soldini, D.; Weder, W.; Curioni-Fontecedro, A. Adjuvant Chemotherapy Increases Programmed Death-Ligand 1 (PD-L1) Expression in Non-small Cell Lung Cancer Recurrence. Clin. Lung Cancer 2019, 20, 391–396. [Google Scholar] [CrossRef]
- Liu, Y.T.; Sun, Z.J. Turning cold tumors into hot tumors by improving T-cell infiltration. Theranostics 2021, 11, 5365–5386. [Google Scholar] [CrossRef]
- Fabrizio, F.P.; Trombetta, D.; Rossi, A.; Sparaneo, A.; Castellana, S.; Muscarella, L.A. Gene code CD274/PD-L1: From molecular basis toward cancer immunotherapy. Ther. Adv. Med. Oncol. 2018, 10, 1758835918815598. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, B.; Sun, Z. Spectrum of EGFR aberrations and potential clinical implications: Insights from integrative pan-cancer analysis. Cancer Commun. 2020, 40, 43–59. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Monti, S.; Rodig, S.J.; Juszczynski, P.; Currie, T.; O’Donnell, E.; Chapuy, B.; Takeyama, K.; Neuberg, D.; Golub, T.R.; et al. Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood 2010, 116, 3268–3277. [Google Scholar] [CrossRef]
- Roemer, M.G.; Advani, R.H.; Ligon, A.H.; Natkunam, Y.; Redd, R.A.; Homer, H.; Connelly, C.F.; Sun, H.H.; Daadi, S.E.; Freeman, G.J.; et al. PD-L1 and PD-L2 Genetic Alterations Define Classical Hodgkin Lymphoma and Predict Outcome. J. Clin. Oncol. 2016, 34, 2690–2697. [Google Scholar] [CrossRef]
- Barrett, M.T.; Anderson, K.S.; Lenkiewicz, E.; Andreozzi, M.; Cunliffe, H.E.; Klassen, C.L.; Dueck, A.C.; McCullough, A.E.; Reddy, S.K.; Ramanathan, R.K.; et al. Genomic amplification of 9p24.1 targeting JAK2, PD-L1, and PD-L2 is enriched in high-risk triple negative breast cancer. Oncotarget 2015, 6, 26483–26493. [Google Scholar] [CrossRef]
- Chen, M.; Andreozzi, M.; Pockaj, B.; Barrett, M.T.; Ocal, I.T.; McCullough, A.E.; Linnaus, M.E.; Chang, J.M.; Yearley, J.H.; Annamalai, L.; et al. Development and validation of a novel clinical fluorescence in situ hybridization assay to detect JAK2 and PD-L1 amplification: A fluorescence in situ hybridization assay for JAK2 and PD-L1 amplification. Mod. Pathol. 2017, 30, 1516–1526. [Google Scholar] [CrossRef]
- George, J.; Saito, M.; Tsuta, K.; Iwakawa, R.; Shiraishi, K.; Scheel, A.H.; Uchida, S.; Watanabe, S.I.; Nishikawa, R.; Noguchi, M.; et al. Genomic Amplification of CD274 (PD-L1) in Small-Cell Lung Cancer. Clin. Cancer Res. 2017, 23, 1220–1226. [Google Scholar] [CrossRef]
- Lamberti, G.; Spurr, L.F.; Li, Y.; Ricciuti, B.; Recondo, G.; Umeton, R.; Nishino, M.; Sholl, L.M.; Meyerson, M.L.; Cherniack, A.D.; et al. Clinicopathological and genomic correlates of programmed cell death ligand 1 (PD-L1) expression in nonsquamous non-small-cell lung cancer. Ann. Oncol. 2020, 31, 807–814. [Google Scholar] [CrossRef]
- Kula, A.; Dawidowicz, M.; Kiczmer, P.; Prawdzic Senkowska, A.; Swietochowska, E. The role of genetic polymorphism within PD-L1 gene in cancer. Review. Exp. Mol. Pathol. 2020, 116, 104494. [Google Scholar] [CrossRef]
- Kumar, S.; Sharawat, S.K. Epigenetic regulators of programmed death-ligand 1 expression in human cancers. Transl. Res. 2018, 202, 129–145. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Yao, H.; Li, C.; Fang, J.Y.; Xu, J. Regulation of PD-L1: Emerging Routes for Targeting Tumor Immune Evasion. Front. Pharmacol. 2018, 9, 536. [Google Scholar] [CrossRef]
- Gevensleben, H.; Holmes, E.E.; Goltz, D.; Dietrich, J.; Sailer, V.; Ellinger, J.; Dietrich, D.; Kristiansen, G. PD-L1 promoter methylation is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients following radical prostatectomy. Oncotarget 2016, 7, 79943–79955. [Google Scholar] [CrossRef]
- Chatterjee, A.; Rodger, E.J.; Ahn, A.; Stockwell, P.A.; Parry, M.; Motwani, J.; Gallagher, S.J.; Shklovskaya, E.; Tiffen, J.; Eccles, M.R.; et al. Marked Global DNA Hypomethylation Is Associated with Constitutive PD-L1 Expression in Melanoma. Iscience 2018, 4, 312–325. [Google Scholar] [CrossRef]
- Micevic, G.; Thakral, D.; McGeary, M.; Bosenberg, M.W. PD-L1 methylation regulates PD-L1 expression and is associated with melanoma survival. Pigment. Cell Melanoma Res. 2019, 32, 435–440. [Google Scholar] [CrossRef]
- Elashi, A.A.; Sasidharan Nair, V.; Taha, R.Z.; Shaath, H.; Elkord, E. DNA methylation of immune checkpoints in the peripheral blood of breast and colorectal cancer patients. Oncoimmunology 2019, 8, e1542918. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan Nair, V.; Toor, S.M.; Taha, R.Z.; Shaath, H.; Elkord, E. DNA methylation and repressive histones in the promoters of PD-1, CTLA-4, TIM-3, LAG-3, TIGIT, PD-L1, and galectin-9 genes in human colorectal cancer. Clin. Epigenetics 2018, 10, 104. [Google Scholar] [CrossRef]
- Emran, A.A.; Chatterjee, A.; Rodger, E.J.; Tiffen, J.C.; Gallagher, S.J.; Eccles, M.R.; Hersey, P. Targeting DNA Methylation and EZH2 Activity to Overcome Melanoma Resistance to Immunotherapy. Trends Immunol. 2019, 40, 328–344. [Google Scholar] [CrossRef]
- Li, X.; Wang, Z.; Huang, J.; Luo, H.; Zhu, S.; Yi, H.; Zheng, L.; Hu, B.; Yu, L.; Li, L.; et al. Specific zinc finger-induced methylation of PD-L1 promoter inhibits its expression. FEBS Open Bio 2019, 9, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- De Beck, L.; Melhaoui, S.; De Veirman, K.; Menu, E.; De Bruyne, E.; Vanderkerken, K.; Breckpot, K.; Maes, K. Epigenetic treatment of multiple myeloma mediates tumor intrinsic and extrinsic immunomodulatory effects. Oncoimmunology 2018, 7, e1484981. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.; Jin, L.L.; Liu, C.Q.; Wang, Y.C.; Meng, Y.M.; Zhou, Z.G.; Chen, J.; Yu, X.J.; Zhang, Y.J.; Xu, J.; et al. EZH2 negatively regulates PD-L1 expression in hepatocellular carcinoma. J. Immunother. Cancer 2019, 7, 300. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, X.X.; Wu, W.; Long, H.; Huang, J.; Wang, Z.; Li, T.; Tang, S.; Zhu, B.; Chen, D. EZH2 regulates PD-L1 expression via HIF-1alpha in non-small cell lung cancer cells. Biochem. Biophys. Res. Commun. 2019, 517, 201–209. [Google Scholar] [CrossRef]
- Mao, W.; Ghasemzadeh, A.; Freeman, Z.T.; Obradovic, A.; Chaimowitz, M.G.; Nirschl, T.R.; McKiernan, E.; Yegnasubramanian, S.; Drake, C.G. Immunogenicity of prostate cancer is augmented by BET bromodomain inhibition. J. Immunother. Cancer 2019, 7, 277. [Google Scholar] [CrossRef]
- Li, N.; Yang, L.; Qi, X.K.; Lin, Y.X.; Xie, X.; He, G.P.; Feng, Q.S.; Liu, L.R.; Xie, X.; Zeng, Y.X.; et al. BET bromodomain inhibitor JQ1 preferentially suppresses EBV-positive nasopharyngeal carcinoma cells partially through repressing c-Myc. Cell Death Dis. 2018, 9, 761. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Tan, J. Co-inhibition of BET proteins and PD-L1 as a potential therapy for OSCC through synergistic inhibition of FOXM1 and PD-L1 expressions. J. Oral. Pathol. Med. 2019, 48, 817–825. [Google Scholar] [CrossRef] [PubMed]
- Erkes, D.A.; Field, C.O.; Capparelli, C.; Tiago, M.; Purwin, T.J.; Chervoneva, I.; Berger, A.C.; Hartsough, E.J.; Villanueva, J.; Aplin, A.E. The next-generation BET inhibitor, PLX51107, delays melanoma growth in a CD8-mediated manner. Pigment Cell Melanoma Res. 2019, 32, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Ebine, K.; Kumar, K.; Pham, T.N.; Shields, M.A.; Collier, K.A.; Shang, M.; DeCant, B.T.; Urrutia, R.; Hwang, R.F.; Grimaldo, S.; et al. Interplay between interferon regulatory factor 1 and BRD4 in the regulation of PD-L1 in pancreatic stellate cells. Sci. Rep. 2018, 8, 13225. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Hu, Q.; Zhang, H.; Yang, F.; Peng, C.; Huang, C. HDAC3 Inhibition Upregulates PD-L1 Expression in B-Cell Lymphomas and Augments the Efficacy of Anti-PD-L1 Therapy. Mol. Cancer Ther. 2019, 18, 900–908. [Google Scholar] [CrossRef]
- Hicks, K.C.; Fantini, M.; Donahue, R.N.; Schwab, A.; Knudson, K.M.; Tritsch, S.R.; Jochems, C.; Clavijo, P.E.; Allen, C.T.; Hodge, J.W.; et al. Epigenetic priming of both tumor and NK cells augments antibody-dependent cellular cytotoxicity elicited by the anti-PD-L1 antibody avelumab against multiple carcinoma cell types. Oncoimmunology 2018, 7, e1466018. [Google Scholar] [CrossRef]
- Li, T.; Zhang, C.; Hassan, S.; Liu, X.; Song, F.; Chen, K.; Zhang, W.; Yang, J. Histone deacetylase 6 in cancer. J. Hematol. Oncol. 2018, 11, 111. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, T.; Fatkhutdinov, N.; Zundell, J.A.; Tcyganov, E.N.; Nacarelli, T.; Karakashev, S.; Wu, S.; Liu, Q.; Gabrilovich, D.I.; Zhang, R. HDAC6 Inhibition Synergizes with Anti-PD-L1 Therapy in ARID1A-Inactivated Ovarian Cancer. Cancer Res. 2019, 79, 5482–5489. [Google Scholar] [CrossRef]
- Qin, Y.; Vasilatos, S.N.; Chen, L.; Wu, H.; Cao, Z.; Fu, Y.; Huang, M.; Vlad, A.M.; Lu, B.; Oesterreich, S.; et al. Inhibition of histone lysine-specific demethylase 1 elicits breast tumor immunity and enhances antitumor efficacy of immune checkpoint blockade. Oncogene 2019, 38, 390–405. [Google Scholar] [CrossRef] [PubMed]
- Parsa, A.T.; Waldron, J.S.; Panner, A.; Crane, C.A.; Parney, I.F.; Barry, J.J.; Cachola, K.E.; Murray, J.C.; Tihan, T.; Jensen, M.C.; et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat. Med. 2007, 13, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Cretella, D.; Digiacomo, G.; Giovannetti, E.; Cavazzoni, A. PTEN Alterations as a Potential Mechanism for Tumor Cell Escape from PD-1/PD-L1 Inhibition. Cancers 2019, 11, 1318. [Google Scholar] [CrossRef]
- Xu, C.; Fillmore, C.M.; Koyama, S.; Wu, H.; Zhao, Y.; Chen, Z.; Herter-Sprie, G.S.; Akbay, E.A.; Tchaicha, J.H.; Altabef, A.; et al. Loss of Lkb1 and Pten leads to lung squamous cell carcinoma with elevated PD-L1 expression. Cancer Cell 2014, 25, 590–604. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Fang, W.; Zhan, J.; Hong, S.; Tang, Y.; Kang, S.; Zhang, Y.; He, X.; Zhou, T.; Qin, T.; et al. Upregulation of PD-L1 by EGFR Activation Mediates the Immune Escape in EGFR-Driven NSCLC: Implication for Optional Immune Targeted Therapy for NSCLC Patients with EGFR Mutation. J. Thorac. Oncol. 2015, 10, 910–923. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Zhou, J.; Giobbie-Hurder, A.; Wargo, J.; Hodi, F.S. The activation of MAPK in melanoma cells resistant to BRAF inhibition promotes PD-L1 expression that is reversible by MEK and PI3K inhibition. Clin. Cancer Res. 2013, 19, 598–609. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.H.; Yang, W.H.; Xia, W.; Wei, Y.; Chan, L.C.; Lim, S.O.; Li, C.W.; Kim, T.; Chang, S.S.; Lee, H.H.; et al. Metformin Promotes Antitumor Immunity via Endoplasmic-Reticulum-Associated Degradation of PD-L1. Mol. Cell 2018, 71, 606–620. [Google Scholar] [CrossRef]
- Bellucci, R.; Martin, A.; Bommarito, D.; Wang, K.; Hansen, S.H.; Freeman, G.J.; Ritz, J. Interferon-gamma-induced activation of JAK1 and JAK2 suppresses tumor cell susceptibility to NK cells through upregulation of PD-L1 expression. Oncoimmunology 2015, 4, e1008824. [Google Scholar] [CrossRef] [PubMed]
- Wolfle, S.J.; Strebovsky, J.; Bartz, H.; Sahr, A.; Arnold, C.; Kaiser, C.; Dalpke, A.H.; Heeg, K. PD-L1 expression on tolerogenic APCs is controlled by STAT-3. Eur. J. Immunol. 2011, 41, 413–424. [Google Scholar] [CrossRef]
- Tuo, Z.; Zong, Y.; Li, J.; Xiao, G.; Zhang, F.; Li, G.; Wang, S.; Lv, Y.; Xia, J.; Liu, J. PD-L1 regulation by SDH5 via beta-catenin/ZEB1 signaling. Oncoimmunology 2019, 8, 1655361. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Wen, Q.; Zhao, Y.; Gao, Q.; Bai, Y. NF-kappaB plays a key role in inducing CD274 expression in human monocytes after lipopolysaccharide treatment. PLoS ONE 2013, 8, e61602. [Google Scholar]
- Kondo, A.; Yamashita, T.; Tamura, H.; Zhao, W.; Tsuji, T.; Shimizu, M.; Shinya, E.; Takahashi, H.; Tamada, K.; Chen, L.; et al. Interferon-gamma and tumor necrosis factor-alpha induce an immunoinhibitory molecule, B7-H1, via nuclear factor-kappaB activation in blasts in myelodysplastic syndromes. Blood 2010, 116, 1124–1131. [Google Scholar] [CrossRef]
- Jabbarzadeh Kaboli, P.; Salimian, F.; Aghapour, S.; Xiang, S.; Zhao, Q.; Li, M.; Wu, X.; Du, F.; Zhao, Y.; Shen, J.; et al. Akt-targeted therapy as a promising strategy to overcome drug resistance in breast cancer—A comprehensive review from chemotherapy to immunotherapy. Pharmacol. Res. 2020, 156, 104806. [Google Scholar] [CrossRef] [PubMed]
- Casey, S.C.; Tong, L.; Li, Y.; Do, R.; Walz, S.; Fitzgerald, K.N.; Gouw, A.M.; Baylot, V.; Gutgemann, I.; Eilers, M.; et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 2016, 352, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Zhang, Q.; Goradia, A.; Raghunath, P.N.; Liu, X.; Paessler, M.; Wang, H.Y.; Wysocka, M.; Cheng, M.; Ruggeri, B.A.; et al. Oncogenic kinase NPM/ALK induces through STAT3 expression of immunosuppressive protein CD274 (PD-L1, B7-H1). Proc. Natl. Acad. Sci. USA 2008, 105, 20852–20857. [Google Scholar] [CrossRef] [PubMed]
- Hays, E.; Bonavida, B. YY1 regulates cancer cell immune resistance by modulating PD-L1 expression. Drug Resist. Updates 2019, 43, 10–28. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Zhang, J.; Hong, S.; Zhan, J.; Chen, N.; Qin, T.; Tang, Y.; Zhang, Y.; Kang, S.; Zhou, T.; et al. EBV-driven LMP1 and IFN-gamma up-regulate PD-L1 in nasopharyngeal carcinoma: Implications for oncotargeted therapy. Oncotarget 2014, 5, 12189–12202. [Google Scholar] [CrossRef]
- Green, M.R.; Rodig, S.; Juszczynski, P.; Ouyang, J.; Sinha, P.; O’Donnell, E.; Neuberg, D.; Shipp, M.A. Constitutive AP-1 activity and EBV infection induce PD-L1 in Hodgkin lymphomas and posttransplant lymphoproliferative disorders: Implications for targeted therapy. Clin. Cancer Res. 2012, 18, 1611–1618. [Google Scholar] [CrossRef]
- Labiano, S.; Palazon, A.; Melero, I. Immune response regulation in the tumor microenvironment by hypoxia. Semin. Oncol. 2015, 42, 378–386. [Google Scholar] [CrossRef]
- Noman, M.Z.; Desantis, G.; Janji, B.; Hasmim, M.; Karray, S.; Dessen, P.; Bronte, V.; Chouaib, S. PD-L1 is a novel direct target of HIF-1alpha, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J. Exp. Med. 2014, 211, 781–790. [Google Scholar] [CrossRef]
- Xu, Y.; Wu, Y.; Zhang, S.; Ma, P.; Jin, X.; Wang, Z.; Yao, M.; Zhang, E.; Tao, B.; Qin, Y.; et al. A Tumor-Specific Super-Enhancer Drives Immune Evasion by Guiding Synchronous Expression of PD-L1 and PD-L2. Cell Rep. 2019, 29, 3435–3447. [Google Scholar] [CrossRef]
- Kim, D.J.; Jang, J.H.; Ham, S.Y.; Choi, S.H.; Park, S.S.; Jeong, S.Y.; Kim, B.C.; Jeon, D.Y.; Lee, B.J.; Ko, B.K.; et al. Doxorubicin inhibits PD-L1 expression by enhancing TTP-mediated decay of PD-L1 mRNA in cancer cells. Biochem. Biophys. Res. Commun. 2020, 522, 402–407. [Google Scholar] [CrossRef]
- Wang, T.; Chen, S.; Wang, X.; Huang, Y.; Qiu, J.; Fei, Y.; Chaulagain, A.; Chen, Y.; Wang, Y.; Lin, L.; et al. Aberrant PD-1 ligand expression contributes to the myocardial inflammatory injury caused by Coxsackievirus B infection. Antiviral Res. 2019, 166, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Omar, H.A.; El-Serafi, A.T.; Hersi, F.; Arafa, E.A.; Zaher, D.M.; Madkour, M.; Arab, H.H.; Tolba, M.F. Immunomodulatory MicroRNAs in cancer: Targeting immune checkpoints and the tumor microenvironment. FEBS J. 2019, 286, 3540–3557. [Google Scholar] [CrossRef] [PubMed]
- Li, C.W.; Lim, S.O.; Xia, W.; Lee, H.H.; Chan, L.C.; Kuo, C.W.; Khoo, K.H.; Chang, S.S.; Cha, J.H.; Kim, T.; et al. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat. Commun. 2016, 7, 12632. [Google Scholar] [CrossRef] [PubMed]
- Li, C.W.; Lim, S.O.; Chung, E.M.; Kim, Y.S.; Park, A.H.; Yao, J.; Cha, J.H.; Xia, W.; Chan, L.C.; Kim, T.; et al. Eradication of Triple-Negative Breast Cancer Cells by Targeting Glycosylated PD-L1. Cancer Cell 2018, 33, 187–201. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Y.; Han, Y.; Lu, W.; Yang, J.; Tian, J.; Sun, P.; Yu, T.; Hu, Y.; Zhang, H.; et al. Overexpression of GLT1D1 induces immunosuppression through glycosylation of PD-L1 and predicts poor prognosis in B-cell lymphoma. Mol. Oncol. 2020, 14, 1028–1044. [Google Scholar] [CrossRef]
- Cha, J.H.; Chan, L.C.; Li, C.W.; Hsu, J.L.; Hung, M.C. Mechanisms Controlling PD-L1 Expression in Cancer. Mol. Cell 2019, 76, 359–370. [Google Scholar] [CrossRef]
- Mezzadra, R.; Sun, C.; Jae, L.T.; Gomez-Eerland, R.; de Vries, E.; Wu, W.; Logtenberg, M.E.W.; Slagter, M.; Rozeman, E.A.; Hofland, I.; et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 2017, 549, 106–110. [Google Scholar] [CrossRef]
- Zhang, J.; Bu, X.; Wang, H.; Zhu, Y.; Geng, Y.; Nihira, N.T.; Tan, Y.; Ci, Y.; Wu, F.; Dai, X.; et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 2018, 553, 91–95. [Google Scholar] [CrossRef]
- Yang, L.; Huang, F.; Mei, J.; Wang, X.; Zhang, Q.; Wang, H.; Xi, M.; You, Z. Posttranscriptional Control of PD-L1 Expression by 17beta-Estradiol via PI3K/Akt Signaling Pathway in ERalpha-Positive Cancer Cell Lines. Int. J. Gynecol. Cancer 2017, 27, 196–205. [Google Scholar] [CrossRef]
- Dreher, L.S.; Hoppe, T. ERADicate Tumor Progression with Metformin. Mol. Cell 2018, 71, 481–482. [Google Scholar] [CrossRef]
- Gong, A.Y.; Zhou, R.; Hu, G.; Li, X.; Splinter, P.L.; O’Hara, S.P.; LaRusso, N.F.; Soukup, G.A.; Dong, H.; Chen, X.M. MicroRNA-513 regulates B7-H1 translation and is involved in IFN-gamma-induced B7-H1 expression in cholangiocytes. J. Immunol. 2009, 182, 1325–1333. [Google Scholar] [CrossRef]
- Liu, J.; Hamrouni, A.; Wolowiec, D.; Coiteux, V.; Kuliczkowski, K.; Hetuin, D.; Saudemont, A.; Quesnel, B. Plasma cells from multiple myeloma patients express B7-H1 (PD-L1) and increase expression after stimulation with IFN-{gamma} and TLR ligands via a MyD88-, TRAF6-, and MEK-dependent pathway. Blood 2007, 110, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Klement, J.D.; Lu, C.; Ibrahim, M.L.; Liu, K. IFNAR1 Controls Autocrine Type I IFN Regulation of PD-L1 Expression in Myeloid-Derived Suppressor Cells. J. Immunol. 2018, 201, 264–277. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Yuan, P.; Wu, H.; Chen, J.; Fu, J.; Li, P.; Lu, J.; Wei, W. Dendritic cells with an increased PD-L1 by TGF-beta induce T cell anergy for the cytotoxicity of hepatocellular carcinoma cells. Int. Immunopharmacol. 2014, 20, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Tamura, H.; Ishibashi, M.; Yamashita, T.; Tanosaki, S.; Okuyama, N.; Kondo, A.; Hyodo, H.; Shinya, E.; Takahashi, H.; Dong, H.; et al. Marrow stromal cells induce B7-H1 expression on myeloma cells, generating aggressive characteristics in multiple myeloma. Leukemia 2013, 27, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Liu, P.; Wang, K.; Glorieux, C.; Hu, Y.; Wen, S.; Jiang, W.; Huang, P. Chemotherapy induces tumor immune evasion by upregulation of programmed cell death ligand 1 expression in bone marrow stromal cells. Mol. Oncol. 2017, 11, 358–372. [Google Scholar] [CrossRef]
- Spranger, S.; Gajewski, T.F. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat. Rev. Cancer 2018, 18, 139–147. [Google Scholar] [CrossRef]
- McNiel, E.A.; Tsichlis, P.N. Analyses of publicly available genomics resources define FGF-2-expressing bladder carcinomas as EMT-prone, proliferative tumors with low mutation rates and high expression of CTLA-4, PD-1 and PD-L1. Signal Transduct. Target. Ther. 2017, 2, 16045. [Google Scholar] [CrossRef]
- Li, P.; Huang, T.; Zou, Q.; Liu, D.; Wang, Y.; Tan, X.; Wei, Y.; Qiu, H. FGFR2 Promotes Expression of PD-L1 in Colorectal Cancer via the JAK/STAT3 Signaling Pathway. J. Immunol. 2019, 202, 3065–3075. [Google Scholar] [CrossRef]
- Palakurthi, S.; Kuraguchi, M.; Zacharek, S.J.; Zudaire, E.; Huang, W.; Bonal, D.M.; Liu, J.; Dhaneshwar, A.; DePeaux, K.; Gowaski, M.R.; et al. The Combined Effect of FGFR Inhibition and PD-1 Blockade Promotes Tumor-Intrinsic Induction of Antitumor Immunity. Cancer Immunol. Res. 2019, 7, 1457–1471. [Google Scholar] [CrossRef]
- Holmstrom, T.H.; Moilanen, A.M.; Ikonen, T.; Bjorkman, M.L.; Linnanen, T.; Wohlfahrt, G.; Karlsson, S.; Oksala, R.; Korjamo, T.; Samajdar, S.; et al. ODM-203, a Selective Inhibitor of FGFR and VEGFR, Shows Strong Antitumor Activity, and Induces Antitumor Immunity. Mol. Cancer Ther. 2019, 18, 28–38. [Google Scholar] [CrossRef]
- Deng, H.; Kan, A.; Lyu, N.; Mu, L.; Han, Y.; Liu, L.; Zhang, Y.; Duan, Y.; Liao, S.; Li, S.; et al. Dual Vascular Endothelial Growth Factor Receptor and Fibroblast Growth Factor Receptor Inhibition Elicits Antitumor Immunity and Enhances Programmed Cell Death-1 Checkpoint Blockade in Hepatocellular Carcinoma. Liver Cancer 2020, 9, 338–357. [Google Scholar] [CrossRef]
- Glorieux, C.; Xia, X.; He, Y.Q.; Hu, Y.; Cremer, K.; Robert, A.; Liu, J.; Wang, F.; Ling, J.; Chiao, P.J.; et al. Regulation of PD-L1 expression in K-ras-driven cancers through ROS-mediated FGFR1 signaling. Redox Biol. 2021, 38, 101780. [Google Scholar] [CrossRef]
- Akhand, S.S.; Liu, Z.; Purdy, S.C.; Abdullah, A.; Lin, H.; Cresswell, G.M.; Ratliff, T.L.; Wendt, M. Pharmacologic Inhibition of FGFR Modulates the Metastatic Immune Microenvironment and Promotes Response to Immune Checkpoint Blockade. Cancer Immunol. Res. 2020, 8, 1542–1553. [Google Scholar] [CrossRef]
- Siefker-Radtke, A.O.; Necchi, A.; Rosenbaum, E.; Culine, S.; Burgess, E.F.; O’Donnell, P.H.; Tagawa, S.T.; Zakharia, Y.; OHagan, A.; Avadhani, A.N.; et al. Efficacy of programmed death 1 (PD-1) and programmed death 1 ligand (PD-L1) inhibitors in patients with FGFR mutations and gene fusions: Results from a data analysis of an ongoing phase 2 study of erdafitinib (JNJ-42756493) in patients (pts) with advanced urothelial cancer (UC). J. Clin. Oncol. 2018, 36, 450. [Google Scholar]
- Wang, L.; Gong, Y.; Saci, A.; Szabo, P.M.; Martini, A.; Necchi, A.; Siefker-Radtke, A.; Pal, S.; Plimack, E.R.; Sfakianos, J.P.; et al. Fibroblast Growth Factor Receptor 3 Alterations and Response to PD-1/PD-L1 Blockade in Patients with Metastatic Urothelial Cancer. Eur. Urol. 2019, 76, 599–603. [Google Scholar] [CrossRef] [PubMed]
- Kacew, A.; Sweis, R.F. FGFR3 Alterations in the Era of Immunotherapy for Urothelial Bladder Cancer. Front. Immunol 2020, 11, 575258. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M. Fibroblast growth factor receptors as treatment targets in clinical oncology. Nat. Rev. Clin. Oncol. 2019, 16, 105–122. [Google Scholar] [CrossRef]
- Taylor, M.H.; Schmidt, E.V.; Dutcus, C.; Pinheiro, E.M.; Funahashi, Y.; Lubiniecki, G.; Rasco, D. The LEAP program: Lenvatinib plus pembrolizumab for the treatment of advanced solid tumors. Future Oncol. 2021, 17, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Akbay, E.A.; Koyama, S.; Carretero, J.; Altabef, A.; Tchaicha, J.H.; Christensen, C.L.; Mikse, O.R.; Cherniack, A.D.; Beauchamp, E.M.; Pugh, T.J.; et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 2013, 3, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Abdelhamed, S.; Ogura, K.; Yokoyama, S.; Saiki, I.; Hayakawa, Y. AKT-STAT3 Pathway as a Downstream Target of EGFR Signaling to Regulate PD-L1 Expression on NSCLC cells. J. Cancer 2016, 7, 1579–1586. [Google Scholar] [CrossRef]
- Okita, R.; Maeda, A.; Shimizu, K.; Nojima, Y.; Saisho, S.; Nakata, M. PD-L1 overexpression is partially regulated by EGFR/HER2 signaling and associated with poor prognosis in patients with non-small-cell lung cancer. Cancer Immunol. Immunother. 2017, 66, 865–876. [Google Scholar] [CrossRef]
- Stutvoet, T.S.; Kol, A.; de Vries, E.G.; de Bruyn, M.; Fehrmann, R.S.; Terwisscha van Scheltinga, A.G.; de Jong, S. MAPK pathway activity plays a key role in PD-L1 expression of lung adenocarcinoma cells. J. Pathol. 2019, 249, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xu, L.; Che, X.; Li, C.; Xu, L.; Hou, K.; Fan, Y.; Wen, T.; Qu, X.; Liu, Y. E3 ubiquitin ligases Cbl-b and c-Cbl downregulate PD-L1 in EGFR wild-type non-small cell lung cancer. FEBS Lett. 2018, 592, 621–630. [Google Scholar] [CrossRef]
- Zhang, N.; Zeng, Y.; Du, W.; Zhu, J.; Shen, D.; Liu, Z.; Huang, J.A. The EGFR pathway is involved in the regulation of PD-L1 expression via the IL-6/JAK/STAT3 signaling pathway in EGFR-mutated non-small cell lung cancer. Int. J. Oncol. 2016, 49, 1360–1368. [Google Scholar] [CrossRef]
- Jiang, X.M.; Xu, Y.L.; Huang, M.Y.; Zhang, L.L.; Su, M.X.; Chen, X.; Lu, J.J. Osimertinib (AZD9291) decreases programmed death ligand-1 in EGFR-mutated non-small cell lung cancer cells. Acta Pharmacol. Sin. 2017, 38, 1512–1520. [Google Scholar] [CrossRef]
- Concha-Benavente, F.; Srivastava, R.M.; Trivedi, S.; Lei, Y.; Chandran, U.; Seethala, R.R.; Freeman, G.J.; Ferris, R.L. Identification of the Cell-Intrinsic and -Extrinsic Pathways Downstream of EGFR and IFNgamma That Induce PD-L1 Expression in Head and Neck Cancer. Cancer Res. 2016, 76, 1031–1043. [Google Scholar] [CrossRef] [PubMed]
- Ng, H.Y.; Li, J.; Tao, L.; Lam, A.K.; Chan, K.W.; Ko, J.M.Y.; Yu, V.Z.; Wong, M.; Li, B.; Lung, M.L. Chemotherapeutic Treatments Increase PD-L1 Expression in Esophageal Squamous Cell Carcinoma through EGFR/ERK Activation. Transl. Oncol. 2018, 11, 1323–1333. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Pang, Q.; Yan, C.; Wang, Q.; Yang, J.; Yu, S.; Liu, X.; Yuan, Z.; Wang, P.; Xiao, Z. Induction of PD-L1 expression by epidermal growth factor receptor-mediated signaling in esophageal squamous cell carcinoma. Onco Targets Ther. 2017, 10, 763–771. [Google Scholar] [CrossRef][Green Version]
- Suh, K.J.; Sung, J.H.; Kim, J.W.; Han, S.H.; Lee, H.S.; Min, A.; Kang, M.H.; Kim, J.E.; Kim, J.W.; Kim, S.H.; et al. EGFR or HER2 inhibition modulates the tumor microenvironment by suppression of PD-L1 and cytokines release. Oncotarget 2017, 8, 63901–63910. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, J.; Wang, Y.; Ye, W.; Zhang, X.; Ju, H.; Xu, D.; Liu, L.; Ye, D.; Zhang, L.; et al. EGFR activation induced Snail-dependent EMT and myc-dependent PD-L1 in human salivary adenoid cystic carcinoma cells. Cell Cycle 2018, 17, 1457–1470. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.C.; Lin, H.C.; Tsai, K.J.; Chiang, Y.W.; Lim, K.H.; Chen, C.G.; Su, Y.W.; Peng, C.L.; Ho, A.S.; Huang, L.; et al. Epidermal growth factor induces STAT1 expression to exacerbate the IFNr-mediated PD-L1 axis in epidermal growth factor receptor-positive cancers. Mol. Carcinog. 2018, 57, 1588–1598. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhang, C.; Zhao, G.; Zhang, X.; Hao, M.; Hassan, S.; Zhang, M.; Zheng, H.; Yang, D.; Liu, L.; et al. IGFBP2 regulates PD-L1 expression by activating the EGFR-STAT3 signaling pathway in malignant melanoma. Cancer Lett. 2020, 477, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Guo, W.; Lou, L.; Nie, S.; Zhang, Q.; Liu, Y.; Chang, Y.; Zhang, X.; Li, Y.; Shen, H. EGFR-ERK pathway regulates CSN6 to contribute to PD-L1 expression in glioblastoma. Mol. Carcinog. 2020, 59, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Liang, Y.; Chen, Y.; Wang, L.; Li, D.; Liang, Z.; Wang, X.; Tian, D.; Yang, X.; Niu, H. Glutamine Deprivation Induces PD-L1 Expression via Activation of EGFR/ERK/c-Jun Signaling in Renal Cancer. Mol. Cancer Res. 2020, 18, 324–339. [Google Scholar] [CrossRef] [PubMed]
- D’Incecco, A.; Andreozzi, M.; Ludovini, V.; Rossi, E.; Capodanno, A.; Landi, L.; Tibaldi, C.; Minuti, G.; Salvini, J.; Coppi, E.; et al. PD-1 and PD-L1 expression in molecularly selected non-small-cell lung cancer patients. Br. J. Cancer 2015, 112, 95–102. [Google Scholar] [CrossRef]
- Lin, K.; Cheng, J.; Yang, T.; Li, Y.; Zhu, B. EGFR-TKI down-regulates PD-L1 in EGFR mutant NSCLC through inhibiting NF-kappaB. Biochem. Biophys. Res. Commun. 2015, 463, 95–101. [Google Scholar] [CrossRef]
- Li, M.; Liu, F.; Zhang, F.; Zhou, W.; Jiang, X.; Yang, Y.; Qu, K.; Wang, Y.; Ma, Q.; Wang, T.; et al. Genomic ERBB2/ERBB3 mutations promote PD-L1-mediated immune escape in gallbladder cancer: A whole-exome sequencing analysis. Gut 2019, 68, 1024–1033. [Google Scholar] [CrossRef]
- To, K.K.W.; Fong, W.; Cho, W.C.S. Immunotherapy in Treating EGFR-Mutant Lung Cancer: Current Challenges and New Strategies. Front. Oncol. 2021, 11, 635007. [Google Scholar] [CrossRef]
- Gavralidis, A.; Gainor, J.F. Immunotherapy in EGFR-Mutant and ALK-Positive Lung Cancer: Implications for Oncogene-Driven Lung Cancer. Cancer J. 2020, 26, 517–524. [Google Scholar] [CrossRef]
- Liang, H.; Liu, X.; Wang, M. Immunotherapy combined with epidermal growth factor receptor-tyrosine kinase inhibitors in non-small-cell lung cancer treatment. Onco Targets Ther. 2018, 11, 6189–6196. [Google Scholar] [CrossRef] [PubMed]
- Durand-Panteix, S.; Farhat, M.; Youlyouz-Marfak, I.; Rouaud, P.; Ouk-Martin, C.; David, A.; Faumont, N.; Feuillard, J.; Jayat-Vignoles, C. B7-H1, which represses EBV-immortalized B cell killing by autologous T and NK cells, is oppositely regulated by c-Myc and EBV latency III program at both mRNA and secretory lysosome levels. J. Immunol. 2012, 189, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Zhuang, M.; Yu, X.; Li, N.; Mao, R.; Wang, Z.; Wang, J.; Wang, X.; Zhou, H.; Zhang, L.; et al. MYC inhibition increases PD-L1 expression induced by IFN-gamma in hepatocellular carcinoma cells. Mol. Immunol. 2018, 101, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Schuhmacher, B.; Rengstl, B.; Doring, C.; Bein, J.; Newrzela, S.; Brunnberg, U.; Kvasnicka, H.M.; Vornanen, M.; Kuppers, R.; Hansmann, M.L.; et al. A strong host response and lack of MYC expression are characteristic for diffuse large B cell lymphoma transformed from nodular lymphocyte predominant Hodgkin lymphoma. Oncotarget 2016, 7, 72197–72210. [Google Scholar] [CrossRef]
- Han, H.; Jain, A.D.; Truica, M.I.; Izquierdo-Ferrer, J.; Anker, J.F.; Lysy, B.; Sagar, V.; Luan, Y.; Chalmers, Z.R.; Unno, K.; et al. Small-Molecule MYC Inhibitors Suppress Tumor Growth and Enhance Immunotherapy. Cancer Cell 2019, 36, 483–497. [Google Scholar] [CrossRef]
- Fusello, A.; Horowitz, J.; Yang-Iott, K.; Brady, B.L.; Yin, B.; Rowh, M.A.; Rappaport, E.; Bassing, C.H. Histone H2AX suppresses translocations in lymphomas of Emu-c-Myc transgenic mice that contain a germline amplicon of tumor-promoting genes. Cell Cycle 2013, 12, 2867–2875. [Google Scholar] [CrossRef]
- Kharma, B.; Baba, T.; Matsumura, N.; Kang, H.S.; Hamanishi, J.; Murakami, R.; McConechy, M.M.; Leung, S.; Yamaguchi, K.; Hosoe, Y.; et al. STAT1 drives tumor progression in serous papillary endometrial cancer. Cancer Res. 2014, 74, 6519–6530. [Google Scholar] [CrossRef]
- Atsaves, V.; Tsesmetzis, N.; Chioureas, D.; Kis, L.; Leventaki, V.; Drakos, E.; Panaretakis, T.; Grander, D.; Medeiros, L.J.; Young, K.H.; et al. PD-L1 is commonly expressed and transcriptionally regulated by STAT3 and MYC in ALK-negative anaplastic large-cell lymphoma. Leukemia 2017, 31, 1633–1637. [Google Scholar] [CrossRef]
- Sun, L.; Wang, Q.; Chen, B.; Zhao, Y.; Shen, B.; Wang, H.; Xu, J.; Zhu, M.; Zhao, X.; Xu, C.; et al. Gastric cancer mesenchymal stem cells derived IL-8 induces PD-L1 expression in gastric cancer cells via STAT3/mTOR-c-Myc signal axis. Cell Death Dis. 2018, 9, 928. [Google Scholar] [CrossRef]
- Wang, J.; Jia, Y.; Zhao, S.; Zhang, X.; Wang, X.; Han, X.; Wang, Y.; Ma, M.; Shi, J.; Liu, L. BIN1 reverses PD-L1-mediated immune escape by inactivating the c-MYC and EGFR/MAPK signaling pathways in non-small cell lung cancer. Oncogene 2017, 36, 6235–6243. [Google Scholar] [CrossRef]
- Melaiu, O.; Mina, M.; Chierici, M.; Boldrini, R.; Jurman, G.; Romania, P.; D’Alicandro, V.; Benedetti, M.C.; Castellano, A.; Liu, T.; et al. PD-L1 Is a Therapeutic Target of the Bromodomain Inhibitor JQ1 and, Combined with HLA Class I, a Promising Prognostic Biomarker in Neuroblastoma. Clin. Cancer Res. 2017, 23, 4462–4472. [Google Scholar] [CrossRef]
- Kim, E.Y.; Kim, A.; Kim, S.K.; Chang, Y.S. MYC expression correlates with PD-L1 expression in non-small cell lung cancer. Lung Cancer 2017, 110, 63–67. [Google Scholar] [CrossRef]
- Pan, Y.; Fei, Q.; Xiong, P.; Yang, J.; Zhang, Z.; Lin, X.; Pan, M.; Lu, F.; Huang, H. Synergistic inhibition of pancreatic cancer with anti-PD-L1 and c-Myc inhibitor JQ1. Oncoimmunology 2019, 8, e1581529. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.Q.; Yu, F.Q.; Chen, C. C-Myc regulates PD-L1 expression in esophageal squamous cell carcinoma. Am. J. Transl. Res. 2020, 12, 379–388. [Google Scholar] [PubMed]
- Kortlever, R.M.; Sodir, N.M.; Wilson, C.H.; Burkhart, D.L.; Pellegrinet, L.; Brown Swigart, L.; Littlewood, T.D.; Evan, G.I. Myc Cooperates with Ras by Programming Inflammation and Immune Suppression. Cell 2017, 171, 1301–1315. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Poggio, M.; Jin, H.Y.; Shi, Z.; Forester, C.M.; Wang, Y.; Stumpf, C.R.; Xue, L.; Devericks, E.; So, L.; et al. Translation control of the immune checkpoint in cancer and its therapeutic targeting. Nat. Med. 2019, 25, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Wang, W.G.; Zhou, X.Y.; Li, X.Q. EBV-positive diffuse large B-cell lymphoma features PD-L1 protein but not mRNA overexpression. Pathology 2018, 50, 725–729. [Google Scholar] [CrossRef]
- Soussi, T.; Wiman, K.G. TP53: An oncogene in disguise. Cell Death Differ. 2015, 22, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Kan, G.; Dong, W. The expression of PD-L1 APE1 and P53 in hepatocellular carcinoma and its relationship to clinical pathology. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 3063–3071. [Google Scholar] [PubMed]
- Cortez, M.A.; Ivan, C.; Valdecanas, D.; Wang, X.; Peltier, H.J.; Ye, Y.; Araujo, L.; Carbone, D.P.; Shilo, K.; Giri, D.K.; et al. PDL1 Regulation by p53 via miR-34. J. Natl. Cancer Inst. 2016, 108, djv303. [Google Scholar] [CrossRef]
- Braun, M.W.; Iwakuma, T. Regulation of cytotoxic T-cell responses by p53 in cancer. Transl. Cancer Res. 2016, 5, 692–697. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, S.; Furukawa, S.; Hashimoto, A.; Tsutaho, A.; Fukao, A.; Sakamura, Y.; Parajuli, G.; Onodera, Y.; Otsuka, Y.; Handa, H.; et al. ARF6 and AMAP1 are major targets of KRAS and TP53 mutations to promote invasion, PD-L1 dynamics, and immune evasion of pancreatic cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 17450–17459. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhou, J.; Fu, H.; Ni, X.; Shan, Y. Upregulation of oxidative stress-responsive 1(OXSR1) predicts poor prognosis and promotes hepatocellular carcinoma progression. Bioengineered 2020, 11, 958–971. [Google Scholar] [CrossRef] [PubMed]
- Thiem, A.; Hesbacher, S.; Kneitz, H.; di Primio, T.; Heppt, M.V.; Hermanns, H.M.; Goebeler, M.; Meierjohann, S.; Houben, R.; Schrama, D. IFN-gamma-induced PD-L1 expression in melanoma depends on p53 expression. J. Exp. Clin. Cancer Res. 2019, 38, 397. [Google Scholar] [CrossRef]
- Assoun, S.; Theou-Anton, N.; Nguenang, M.; Cazes, A.; Danel, C.; Abbar, B.; Pluvy, J.; Gounant, V.; Khalil, A.; Namour, C.; et al. Association of TP53 mutations with response and longer survival under immune checkpoint inhibitors in advanced non-small-cell lung cancer. Lung Cancer 2019, 132, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Liu, S.Y.; Zhou, J.Y.; Xu, J.T.; Zhang, H.K.; Yan, H.H.; Huan, J.J.; Dai, P.P.; Xu, C.R.; Su, J.; et al. Specific TP53 subtype as biomarker for immune checkpoint inhibitors in lung adenocarcinoma. EBioMedicine 2020, 60, 102990. [Google Scholar] [CrossRef]
- Zhou, X.; Singh, M.; Sanz Santos, G.; Guerlavais, V.; Carvajal, L.A.; Aivado, M.; Zhan, Y.; Oliveira, M.M.S.; Westerberg, L.S.; Annis, D.A.; et al. Pharmacological activation of p53 triggers viral mimicry response thereby abolishing tumor immune evasion and promoting anti-tumor immunity. Cancer Discov. 2021. [Google Scholar] [CrossRef]
- Koh, J.; Jang, J.Y.; Keam, B.; Kim, S.; Kim, M.Y.; Go, H.; Kim, T.M.; Kim, D.W.; Kim, C.W.; Jeon, Y.K.; et al. EML4-ALK enhances programmed cell death-ligand 1 expression in pulmonary adenocarcinoma via hypoxia-inducible factor (HIF)-1alpha and STAT3. Oncoimmunology 2016, 5, e1108514. [Google Scholar] [CrossRef]
- Ota, K.; Azuma, K.; Kawahara, A.; Hattori, S.; Iwama, E.; Tanizaki, J.; Harada, T.; Matsumoto, K.; Takayama, K.; Takamori, S.; et al. Induction of PD-L1 Expression by the EML4-ALK Oncoprotein and Downstream Signaling Pathways in Non-Small Cell Lung Cancer. Clin. Cancer Res. 2015, 21, 4014–4021. [Google Scholar] [CrossRef]
- Hong, S.; Chen, N.; Fang, W.; Zhan, J.; Liu, Q.; Kang, S.; He, X.; Liu, L.; Zhou, T.; Huang, J.; et al. Upregulation of PD-L1 by EML4-ALK fusion protein mediates the immune escape in ALK positive NSCLC: Implication for optional anti-PD-1/PD-L1 immune therapy for ALK-TKIs sensitive and resistant NSCLC patients. Oncoimmunology 2016, 5, e1094598. [Google Scholar] [CrossRef]
- Guaitoli, G.; Tiseo, M.; Di Maio, M.; Friboulet, L.; Facchinetti, F. Immune checkpoint inhibitors in oncogene-addicted non-small cell lung cancer: A systematic review and meta-analysis. Transl. Lung Cancer Res. 2021, 10, 2890–2916. [Google Scholar] [CrossRef]
- Sankar, K.; Nagrath, S.; Ramnath, N. Immunotherapy for ALK-Rearranged Non-Small Cell Lung Cancer: Challenges Inform Promising Approaches. Cancers 2021, 13, 1476. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lui, V.W.Y. Emerging Roles of ALK in Immunity and Insights for Immunotherapy. Cancers 2020, 12, 426. [Google Scholar] [CrossRef] [PubMed]
- Sumimoto, H.; Takano, A.; Teramoto, K.; Daigo, Y. RAS-Mitogen-Activated Protein Kinase Signal Is Required for Enhanced PD-L1 Expression in Human Lung Cancers. PLoS ONE 2016, 11, e0166626. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Fang, W.; Lin, Z.; Peng, P.; Wang, J.; Zhan, J.; Hong, S.; Huang, J.; Liu, L.; Sheng, J.; et al. KRAS mutation-induced upregulation of PD-L1 mediates immune escape in human lung adenocarcinoma. Cancer Immunol. Immunother. 2017, 66, 1175–1187. [Google Scholar] [CrossRef]
- Lee, M.H.; Yanagawa, J.; Tran, L.; Walser, T.C.; Bisht, B.; Fung, E.; Park, S.J.; Zeng, G.; Krysan, K.; Wallace, W.D.; et al. FRA1 contributes to MEK-ERK pathway-dependent PD-L1 upregulation by KRAS mutation in premalignant human bronchial epithelial cells. Am. J. Transl. Res. 2020, 12, 409–427. [Google Scholar]
- Lastwika, K.J.; Wilson, W., 3rd; Li, Q.K.; Norris, J.; Xu, H.; Ghazarian, S.R.; Kitagawa, H.; Kawabata, S.; Taube, J.M.; Yao, S.; et al. Control of PD-L1 Expression by Oncogenic Activation of the AKT-mTOR Pathway in Non-Small Cell Lung Cancer. Cancer Res. 2016, 76, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Weng, C.C.; Hsieh, M.J.; Wu, C.C.; Lin, Y.C.; Shan, Y.S.; Hung, W.C.; Chen, L.T.; Cheng, K.H. Loss of the transcriptional repressor TGIF1 results in enhanced Kras-driven development of pancreatic cancer. Mol. Cancer 2019, 18, 96. [Google Scholar] [CrossRef]
- Coelho, M.A.; de Carne Trecesson, S.; Rana, S.; Zecchin, D.; Moore, C.; Molina-Arcas, M.; East, P.; Spencer-Dene, B.; Nye, E.; Barnouin, K.; et al. Oncogenic RAS Signaling Promotes Tumor Immunoresistance by Stabilizing PD-L1 mRNA. Immunity 2017, 47, 1083–1099. [Google Scholar] [CrossRef]
- Booth, L.; Roberts, J.L.; Sander, C.; Lalani, A.S.; Kirkwood, J.M.; Hancock, J.F.; Poklepovic, A.; Dent, P. Neratinib and entinostat combine to rapidly reduce the expression of K-RAS, N-RAS, Galphaq and Galpha11 and kill uveal melanoma cells. Cancer Biol. Ther. 2019, 20, 700–710. [Google Scholar] [CrossRef]
- Bowers, N.L.; Helton, E.S.; Huijbregts, R.P.; Goepfert, P.A.; Heath, S.L.; Hel, Z. Immune suppression by neutrophils in HIV-1 infection: Role of PD-L1/PD-1 pathway. PLoS Pathog. 2014, 10, e1003993. [Google Scholar] [CrossRef]
- Khou, S.; Popa, A.; Luci, C.; Bihl, F.; Meghraoui-Kheddar, A.; Bourdely, P.; Salavagione, E.; Cosson, E.; Rubod, A.; Cazareth, J.; et al. Tumor-Associated Neutrophils Dampen Adaptive Immunity and Promote Cutaneous Squamous Cell Carcinoma Development. Cancers 2020, 12, 1860. [Google Scholar] [CrossRef]
- Tung, J.N.; Lin, P.L.; Wang, Y.C.; Wu, D.W.; Chen, C.Y.; Lee, H. PD-L1 confers resistance to EGFR mutation-independent tyrosine kinase inhibitors in non-small cell lung cancer via upregulation of YAP1 expression. Oncotarget 2018, 9, 4637–4646. [Google Scholar] [CrossRef]
- Vokalova, L.; Balogh, A.; Toth, E.; Van Breda, S.V.; Schafer, G.; Hoesli, I.; Lapaire, O.; Hahn, S.; Than, N.G.; Rossi, S.W. Placental Protein 13 (Galectin-13) Polarizes Neutrophils Toward an Immune Regulatory Phenotype. Front Immunol. 2020, 11, 145. [Google Scholar] [CrossRef] [PubMed]
- Wangpaichitr, M.; Kandemir, H.; Li, Y.Y.; Wu, C.; Nguyen, D.; Feun, L.G.; Kuo, M.T.; Savaraj, N. Relationship of Metabolic Alterations and PD-L1 Expression in Cisplatin Resistant Lung Cancer. Cell Dev. Biol. 2017, 6, 183. [Google Scholar] [PubMed]
- Zheng, X.; Chen, Y.; Bai, M.; Liu, Y.; Xu, B.; Sun, R.; Zeng, H. The antimetastatic effect and underlying mechanisms of thioredoxin reductase inhibitor ethaselen. Free Radic. Biol. Med. 2019, 131, 7–17. [Google Scholar] [CrossRef]
- Zhu, G.; Li, X.; Li, J.; Zhou, W.; Chen, Z.; Fan, Y.; Jiang, Y.; Zhao, Y.; Sun, G.; Mao, W. Arsenic trioxide (ATO) induced degradation of Cyclin D1 sensitized PD-1/PD-L1 checkpoint inhibitor in oral and esophageal squamous cell carcinoma. J. Cancer 2020, 11, 6516–6529. [Google Scholar] [CrossRef] [PubMed]
- Bailly, C. Regulation of PD-L1 expression on cancer cells with ROS-modulating drugs. Life Sci. 2020, 246, 117403. [Google Scholar] [CrossRef]
- Gilardini Montani, M.S.; Santarelli, R.; Falcinelli, L.; Gonnella, R.; Granato, M.; Di Renzo, L.; Cuomo, L.; Vitillo, M.; Faggioni, A.; Cirone, M. EBV up-regulates PD-L1 on the surface of primary monocytes by increasing ROS and activating TLR signaling and STAT3. J. Leukoc. Biol. 2018, 104, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Romeo, M.A.; Gilardini Montani, M.S.; Benedetti, R.; Giambelli, L.; D’Aprile, R.; Gaeta, A.; Faggioni, A.; Cirone, M. The cross-talk between STAT1/STAT3 and ROS up-regulates PD-L1 and promotes the release of pro-inflammatory/immune suppressive cytokines in primary monocytes infected by HHV-6B. Virus Res. 2020, 292, 198231. [Google Scholar] [CrossRef] [PubMed]
- Roux, C.; Jafari, S.M.; Shinde, R.; Duncan, G.; Cescon, D.W.; Silvester, J.; Chu, M.F.; Hodgson, K.; Berger, T.; Wakeham, A.; et al. Reactive oxygen species modulate macrophage immunosuppressive phenotype through the up-regulation of PD-L1. Proc. Natl. Acad. Sci. USA 2019, 116, 4326–4335. [Google Scholar] [CrossRef] [PubMed]
- Glorieux, C.; Cui, L.; Zeng, P.; Xia, X.; Huang, P. Diverse effects of chemotherapeutic agents on immune cell function and implications in immunochemotherapy. Cancer Commun. 2021, 41, 432–435. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glorieux, C.; Xia, X.; Huang, P. The Role of Oncogenes and Redox Signaling in the Regulation of PD-L1 in Cancer. Cancers 2021, 13, 4426. https://doi.org/10.3390/cancers13174426
Glorieux C, Xia X, Huang P. The Role of Oncogenes and Redox Signaling in the Regulation of PD-L1 in Cancer. Cancers. 2021; 13(17):4426. https://doi.org/10.3390/cancers13174426
Chicago/Turabian StyleGlorieux, Christophe, Xiaojun Xia, and Peng Huang. 2021. "The Role of Oncogenes and Redox Signaling in the Regulation of PD-L1 in Cancer" Cancers 13, no. 17: 4426. https://doi.org/10.3390/cancers13174426
APA StyleGlorieux, C., Xia, X., & Huang, P. (2021). The Role of Oncogenes and Redox Signaling in the Regulation of PD-L1 in Cancer. Cancers, 13(17), 4426. https://doi.org/10.3390/cancers13174426






