Altered Serum Metabolic Profile Assessed by Advanced 1H-NMR in Breast Cancer Patients
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Studied Population
2.2. Blood Sample Collection and Storage
2.3. Biochemical Analysis
2.4. Lipoprotein Analysis by NMR Spectroscopy (Advanced Lipoprotein Profile)
2.5. Low Molecular Weight Metabolites Analysis
2.6. Lipid Extraction
2.7. Statistical Analysis
3. Results
3.1. Initial Characteristics of the Study Population
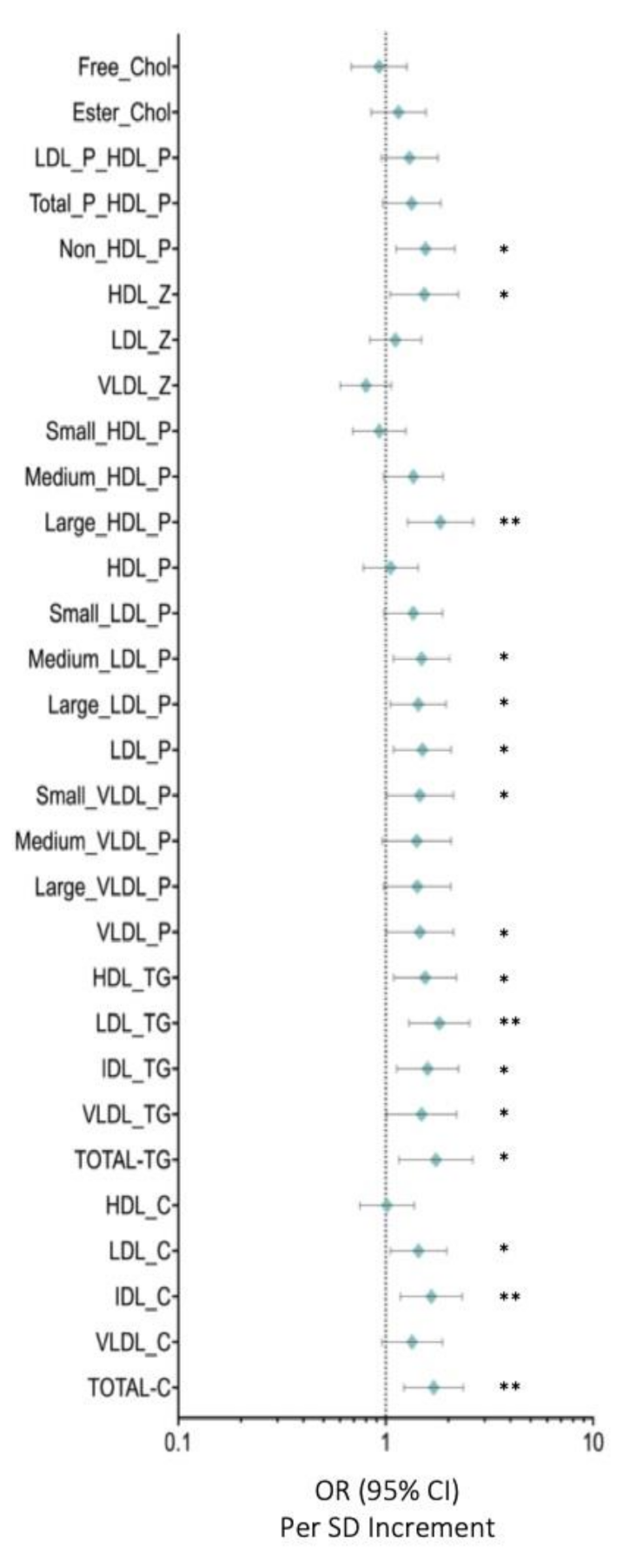
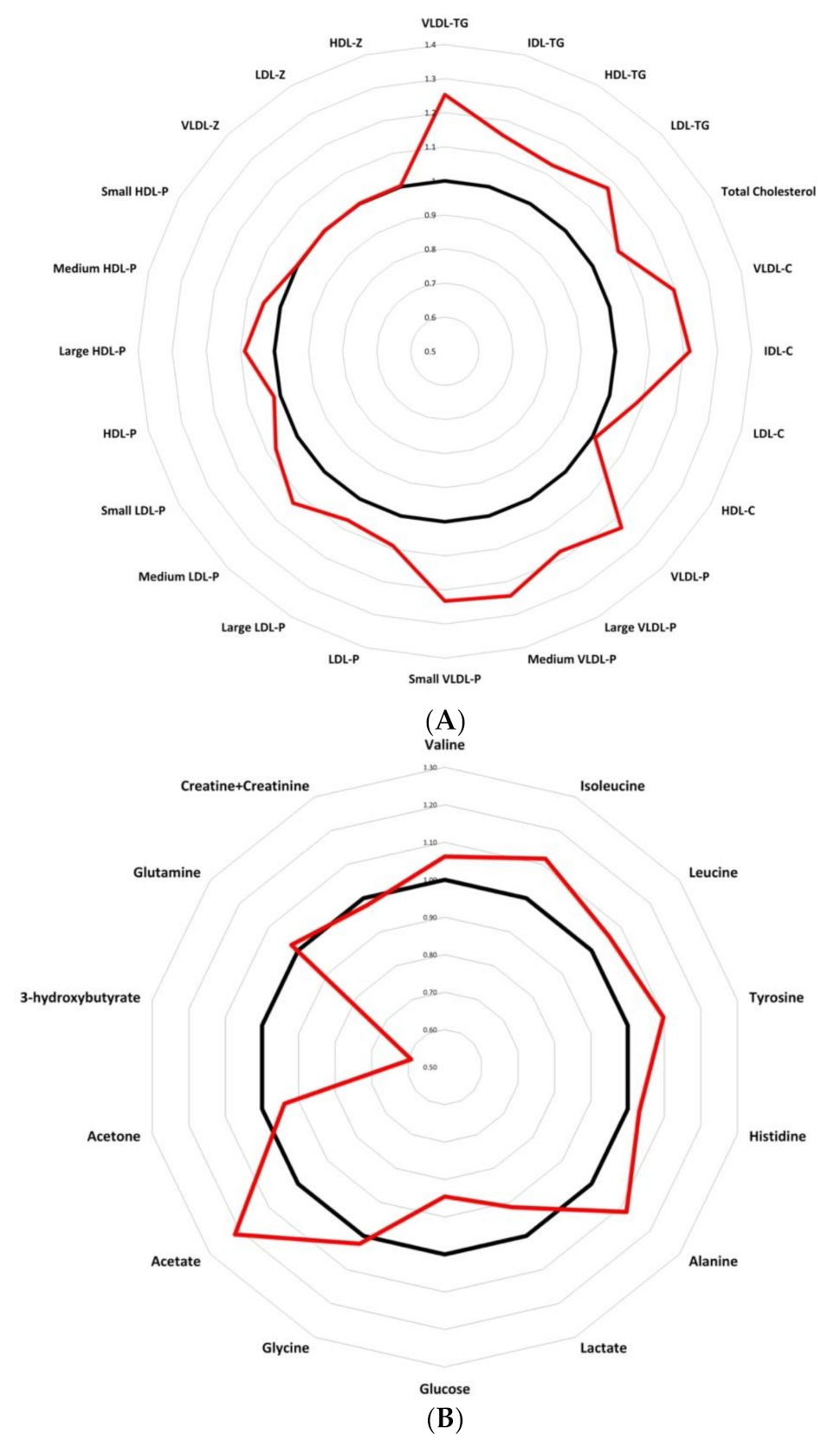
3.2. Serum Metabolome Changes
3.3. HDL Particles Are Altered and Transport a Higher Content of TG in BC Patients
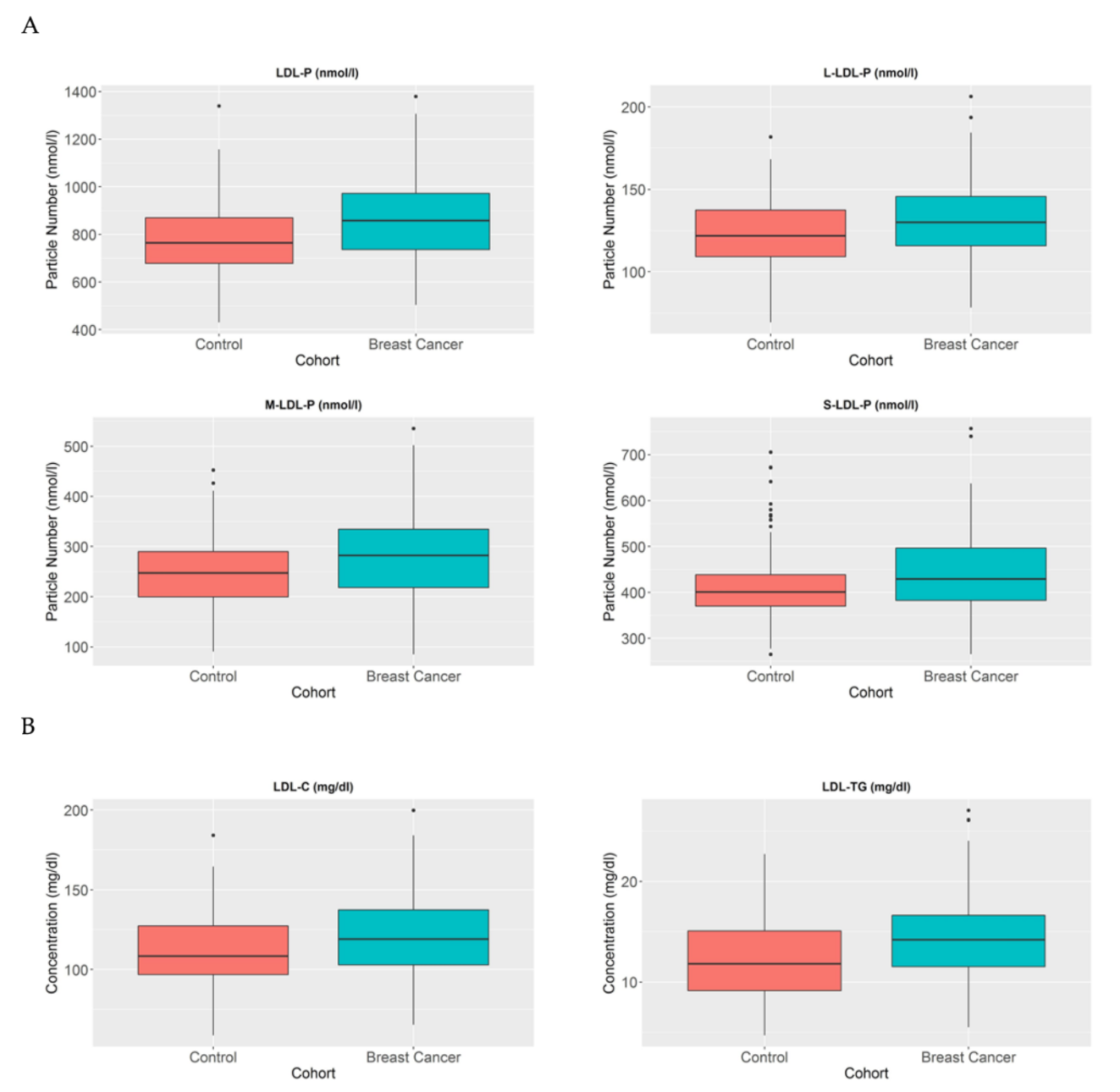
3.4. LDL-P and Their Lipid Contents Are Increased in BC Patients
3.5. Low Molecular Weight Metabolites Reveal an Imbalance in Branched Chain Amino Acids As Well As in Tyrosine and Alanine in BC Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Beloribi-Djefaflia, S.; Vasseur, S.; Guillaumond, F. Lipid metabolic reprogramming in cancer cells. Oncogenesis 2016, 5, e189. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Suo, C.; Li, S.-T.; Zhang, H.; Gao, P. Metabolic reprogramming for cancer cells and their microenvironment: Beyond the Warburg Effect. Biochim. Biophys. Acta. Rev. Cancer 2018, 1870, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Baenke, F.; Peck, B.; Miess, H.; Schulze, A. Hooked on fat: The role of lipid synthesis in cancer metabolism and tumour development. Dis. Model. Mech. 2013, 6, 1353–1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, C.; Geng, F.; Cheng, X.; Guo, D. Lipid metabolism reprogramming and its potential targets in cancer. Cancer Commun. 2018, 38, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Luo, Q.; Halim, A.; Song, G. Targeting lipid metabolism of cancer cells: A promising therapeutic strategy for cancer. Cancer Lett. 2017, 401, 39–45. [Google Scholar] [CrossRef]
- Chimento, A.; Casaburi, I.; Avena, P.; Trotta, F.; De Luca, A.; Rago, V.; Pezzi, V.; Sirianni, R. Cholesterol and Its Metabolites in Tumor Growth: Therapeutic Potential of Statins in Cancer Treatment. Front. Endocrinol. 2019, 9, 807. [Google Scholar] [CrossRef] [Green Version]
- Simons, K. How Cells Handle Cholesterol. Science 2000, 290, 1721–1726. [Google Scholar] [CrossRef] [Green Version]
- Ding, X.; Zhang, W.; Li, S.; Yang, H. The role of cholesterol metabolism in cancer. Am. J. Cancer. Res. 2019, 9, 219–227. [Google Scholar]
- Orho-Melander, M.; Hindy, G.; Borgquist, S.; Schulz, C.-A.; Manjer, J.; Melander, O.; Stocks, T. Blood lipid genetic scores, the HMGCR gene and cancer risk: A Mendelian randomization study. Int. J. Epidemiol. 2018, 47, 495–505. [Google Scholar] [CrossRef]
- Abdelsalam, K.E.A.; Hassan, I.K.; Sadig, I.A. The role of developing breast cancer in alteration of serum lipid profile. J. Res. Med. Sci. 2012, 17, 562–565. [Google Scholar] [PubMed]
- Hasija, K.; Bagga, H.K. Alterations of serum cholesterol and serum lipoprotein in breast cancer of women. Indian. J. Clin. Biochem. 2005, 20, 61–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashrafian, H.; Sounderajah, V.; Glen, R.; Ebbels, T.; Blaise, B.J.; Kalra, D.; Kultima, K.; Spjuth, O.; Tenori, L.; Salek, R.; et al. Metabolomics—the stethoscope for the 21st century. Med. Princ. Pr. 2020, 3. [Google Scholar] [CrossRef]
- Holmes, M.V.; Millwood, I.Y.; Kartsonaki, C.; Hill, M.R.; Bennett, D.A.; Boxall, R.; Guo, Y.; Xu, X.; Bian, Z.; Hu, R.; et al. Lipids, Lipoproteins, and Metabolites and Risk of Myocardial Infarction and Stroke. J. Am. Coll. Cardiol. 2018, 71, 620–632. [Google Scholar] [CrossRef]
- Guaita-Esteruelas, S.; Saavedra-García, P.; Bosquet, A.; Borràs, J.; Girona, J.; Amiliano, K.; Rodríguez-Balada, M.; Heras, M.; Masana, L.; Gumà, J. Adipose-Derived Fatty Acid-Binding Proteins Plasma Concentrations Are Increased in Breast Cancer Patients. Oncologist 2017, 22, 1309–1315. [Google Scholar] [CrossRef] [Green Version]
- Mallol, R.; Amigó, N.; Rodríguez-Gómez, M.; Ángel, H.M.; Vinaixa, M.; Plana, N.; Rock, E.; Ribalta, J.; Yanes, O.; Masana, L.; et al. Liposcale: A novel advanced lipoprotein test based on 2D diffusion-ordered 1H NMR spectroscopy. J. Lipid. Res. 2015, 56, 737–746. [Google Scholar] [CrossRef] [Green Version]
- Jeyarajah, E.J.; Cromwell, W.; Otvos, J.D. Lipoprotein Particle Analysis by Nuclear Magnetic Resonance Spectroscopy. Clin. Lab. Med. 2006, 26, 847–870. [Google Scholar] [CrossRef]
- Vinaixa, M.; Rodríguez, M.A.; Rull, A.; Beltrán, R.; Bladé, C.; Brezmes, J.; Cañellas, N.; Joven, J.; Correig, X. Metabolomic Assessment of the Effect of Dietary Cholesterol in the Progressive Development of Fatty Liver Disease. J. Proteome. Res. 2010, 9, 2527–2538. [Google Scholar] [CrossRef]
- Ouimet, M.; Barrett, T.J.; Fisher, E.A. HDL and Reverse Cholesterol Transport. Circ. Res. 2019, 124, 1505–1518. [Google Scholar] [CrossRef]
- Masana, L.; Girona, J.; Ibarretxe, D.; Rodríguez-Calvo, R.; Rosales, R.; Vallvé, J.-C.; Rodríguez-Borjabad, C.; Guardiola, M.; Rodríguez, M.; Guaita-Esteruelas, S.; et al. Clinical and pathophysiological evidence supporting the safety of extremely low LDL levels—The zero-LDL hypothesis. J. Clin. Lipidol. 2018, 12, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Cedó, L.; Reddy, S.T.; Mato, E.; Blanco-Vaca, F.; Escolà-Gil, J.C. HDL and LDL: Potential New Players in Breast Cancer Development. J. Clin. Med. 2019, 8, 853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, K.E.; Siewert, K.M.; Klarin, D.; Damrauer, S.M.; Chang, K.-M.; Tsao, P.S.; Assimes, T.L.; Maxwell, K.N.; Voight, B.F.; Program, T.V.M.V. The relationship between circulating lipids and breast cancer risk: A Mendelian randomization study. PLoS Med. 2020, 17, e1003302. [Google Scholar] [CrossRef]
- Guan, X.; Liu, Z.; Zhao, Z.; Zhang, X.; Tao, S.; Yuan, B.; Zhang, J.; Wang, D.; Liu, Q.; Ding, Y. Emerging roles of low-density lipoprotein in the development and treatment of breast cancer. Lipids. Health Dis. 2019, 18, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganjali, S.; Ricciuti, B.; Pirro, M.; Butler, A.E.; Atkin, S.L.; Banach, M.; Sahebkar, A. High-Density Lipoprotein Components and Functionality in Cancer: State-of-the-Art. Trends. Endocrinol. Metab. 2019, 30, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Ruscica, M.; Macchi, C.; Pavanello, C.; Corsini, A.; Sahebkar, A.; Sirtori, C. Appropriateness of statin prescription in the elderly. Eur. J. Intern. Med. 2018, 50, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Girona, J.; Amigó, N.; Ibarretxe, D.; Plana, N.; Rodríguez-Borjabad, C.; Heras, M.; Ferré, R.; Gil, M.; Correig, X.; Masana, L. HDL Triglycerides: A New Marker of Metabolic and Cardiovascular Risk. Int. J. Mol. Sci. 2019, 20, 3151. [Google Scholar] [CrossRef] [Green Version]
- Vettore, L.; Westbrook, R.; Tennant, D.A. New aspects of amino acid metabolism in cancer. Br. J. Cancer. 2020, 122, 150–156. [Google Scholar] [CrossRef]
- Ananieva, E.A.; Wilkinson, A.C. Branched-chain amino acid metabolism in cancer. Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 64–70. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.N.; Nguyen, H.Q.; Le, D.-H. Unveiling prognostics biomarkers of tyrosine metabolism reprogramming in liver cancer by cross-platform gene expression analyses. PLoS ONE 2020, 15, e0229276. [Google Scholar] [CrossRef]
- Choi, B.-H.; Coloff, J.L. The Diverse Functions of Non-Essential Amino Acids in Cancer. Cancers 2019, 11, 675. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirschhaeuser, F.; Sattler, U.G.; Mueller-Klieser, W. Lactate: A Metabolic Key Player in Cancer. Cancer Res. 2011, 71, 6921–6925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de la Cruz-López, K.G.; Castro-Muñoz, L.J.; Reyes-Hernández, D.O.; García-Carrancá, A.; Manzo-Merino, J. Lactate in the Regulation of Tumor Microenvironment and Therapeutic Approaches. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.-M.; Ramprasath, T.; Zou, M.-H. β-hydroxybutyrate and its metabolic effects on age-associated pathology. Exp. Mol. Med. 2020, 52, 548–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Study Group Data | Control (n = 69) | Breast Cancer (n = 171) | p Value |
|---|---|---|---|
| Clinical data | |||
| Age | 43 (38–54) | 44 (37–50) | n.s |
| Children | 2 (1–2) | 2 (1–2) | n.s |
| Age of Menarche | 12 (11–13) | 12 (12–14) | n.s |
| Menopause (Yes, %) | 30.3 | 20.2 | n.s |
| BMI (Kg/m2) | 24.02 (22.20–28.13) | 25,07 (22.72–28.36) | n.s |
| Biochemical data | |||
| Cholesterol (mg/dl) | 194.18 ± 28.31 | 208.06 ± 31.40 | <0.005 |
| Apo A1 (mg/dl) | 147.22 ± 28.66 | 147.64 ± 27.34 | n.s |
| Apo B100 (mg/dl) | 91 (78–104) | 101 (85–118) | <0.05 |
| Triglycerides (mg/dl) | 76.55 (56.83–98.74) | 86.73 (67.69–120.16) | <0.005 |
| FABP4 (ng/mL) | 13.085 (8.73–18.05) | 17.53 (13.15–23.33) | <0.001 |
| FABP5 (ng/mL) | 6.12 (5.44–7.91) | 7.00 (5.26–9.04) | n.s |
| CETP activity (pmol/µL) | 47.87 (26.95–69.54) | 51.98 (31.71–76.43) | n.s |
| CETP protein (ng/µL) | 596.51 ± 163.39 | 579.63 ± 155.76 | n.s |
| CD36 (pg/mL) | 908.67 (762.00–1037.56) | 897.00 (763.37–997.56) | n.s |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gumà, J.; Adriá-Cebrián, J.; Ruiz-Aguado, B.; Albacar, C.; Girona, J.; Rodríguez-Calvo, R.; Martínez-Micaelo, N.; Lam, E.W.F.; Masana, L.; Guaita-Esteruelas, S. Altered Serum Metabolic Profile Assessed by Advanced 1H-NMR in Breast Cancer Patients. Cancers 2021, 13, 4281. https://doi.org/10.3390/cancers13174281
Gumà J, Adriá-Cebrián J, Ruiz-Aguado B, Albacar C, Girona J, Rodríguez-Calvo R, Martínez-Micaelo N, Lam EWF, Masana L, Guaita-Esteruelas S. Altered Serum Metabolic Profile Assessed by Advanced 1H-NMR in Breast Cancer Patients. Cancers. 2021; 13(17):4281. https://doi.org/10.3390/cancers13174281
Chicago/Turabian StyleGumà, Josep, Jose Adriá-Cebrián, Belen Ruiz-Aguado, Cinta Albacar, Josefa Girona, Ricardo Rodríguez-Calvo, Neus Martínez-Micaelo, Eric W. F. Lam, Luis Masana, and Sandra Guaita-Esteruelas. 2021. "Altered Serum Metabolic Profile Assessed by Advanced 1H-NMR in Breast Cancer Patients" Cancers 13, no. 17: 4281. https://doi.org/10.3390/cancers13174281
APA StyleGumà, J., Adriá-Cebrián, J., Ruiz-Aguado, B., Albacar, C., Girona, J., Rodríguez-Calvo, R., Martínez-Micaelo, N., Lam, E. W. F., Masana, L., & Guaita-Esteruelas, S. (2021). Altered Serum Metabolic Profile Assessed by Advanced 1H-NMR in Breast Cancer Patients. Cancers, 13(17), 4281. https://doi.org/10.3390/cancers13174281






