Hot Spot Analysis of YAP-TEAD Protein-Protein Interaction Using the Fragment Molecular Orbital Method and Its Application for Inhibitor Discovery
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Structure Preparation
2.2. FMO Calculations
2.3. Virtual Screening
2.4. Molecular Docking
2.5. SPR Assay
2.6. Cell Culture and Stable Cell Lines
2.7. Luciferase Reporter Assay
2.8. mRNA Expression
2.9. Proximity Ligation Assay (PLA)
3. Results
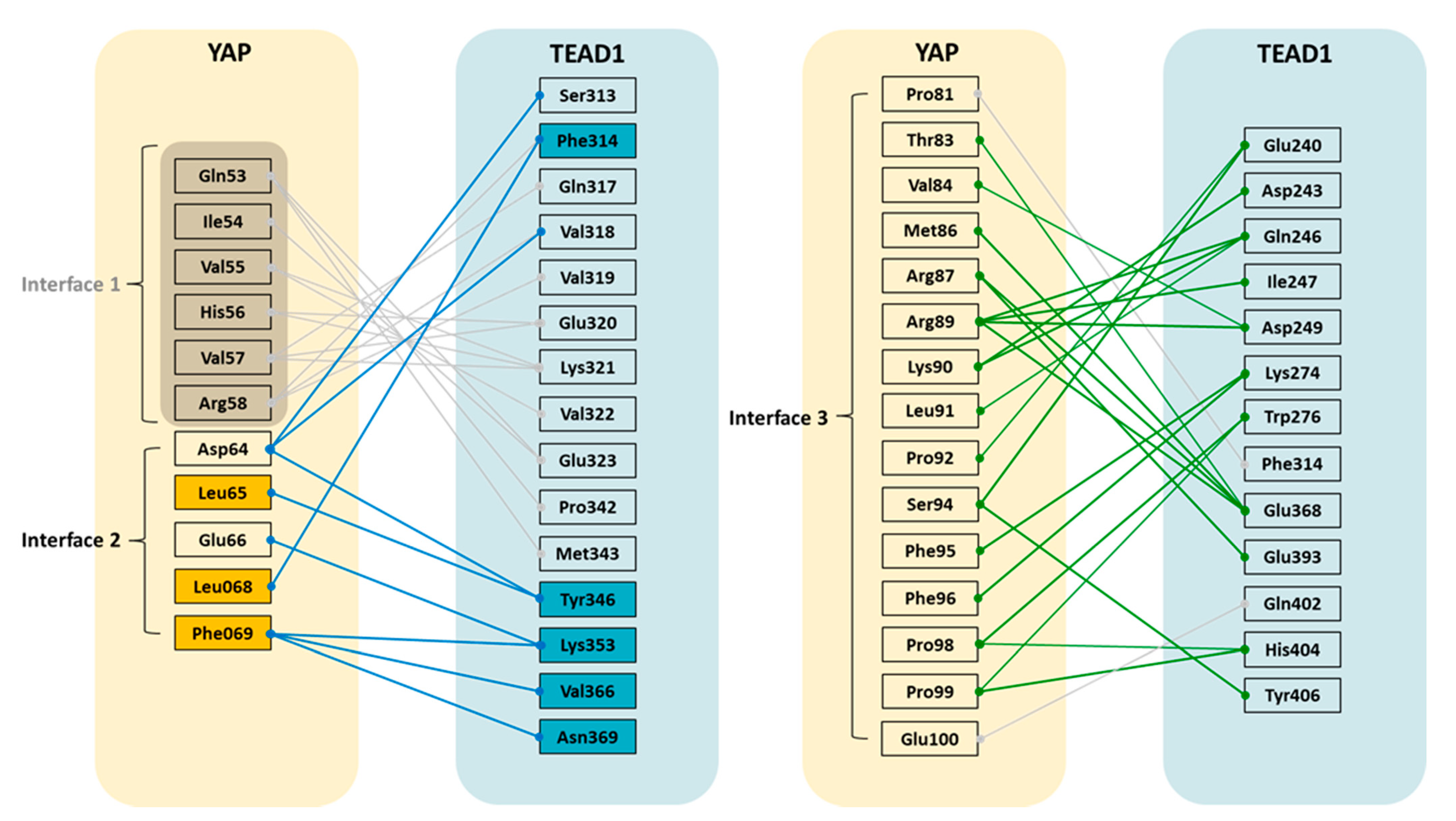
3.1. FMO Study of the YAP-TEAD1 Complex
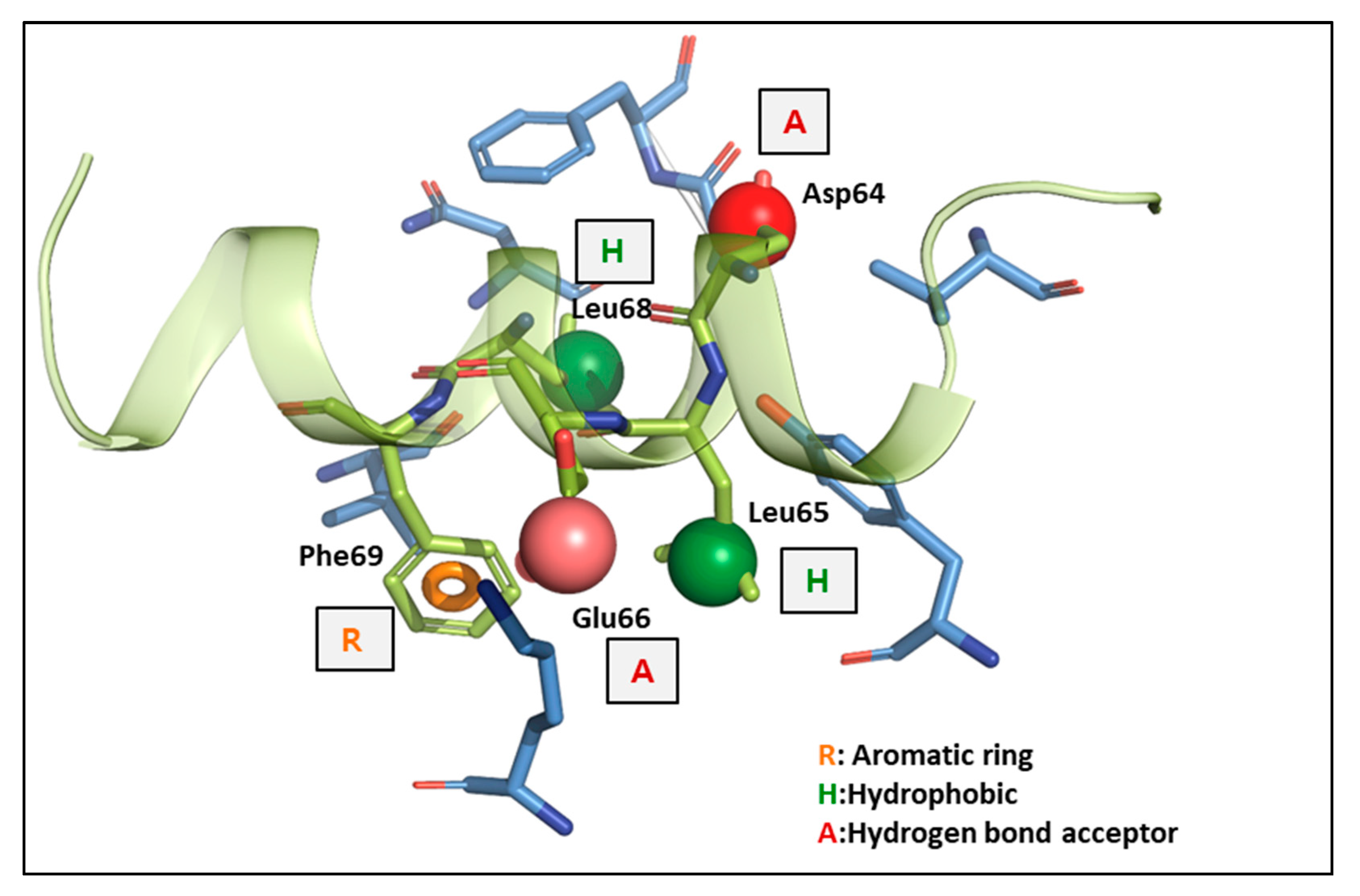
3.1.1. Analysis of the YAP-TEAD1 Interaction at FMO Study on Interface 2 with FMO Calculation
3.1.2. Analysis of the YAP-TEAD1 Interaction at Interface 3 with FMO Calculation
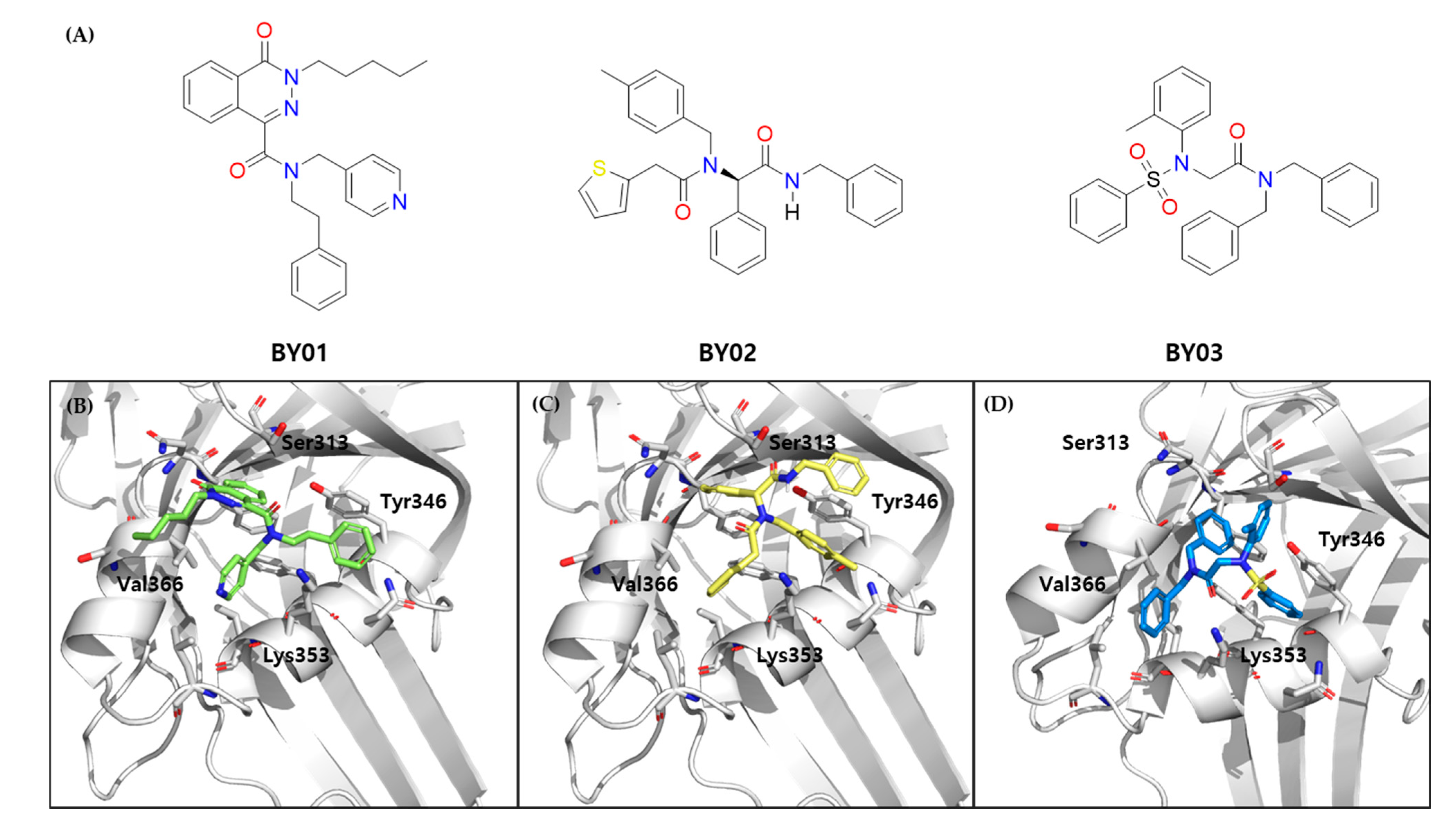
3.2. Hot Spot-Focused Virtual Screening
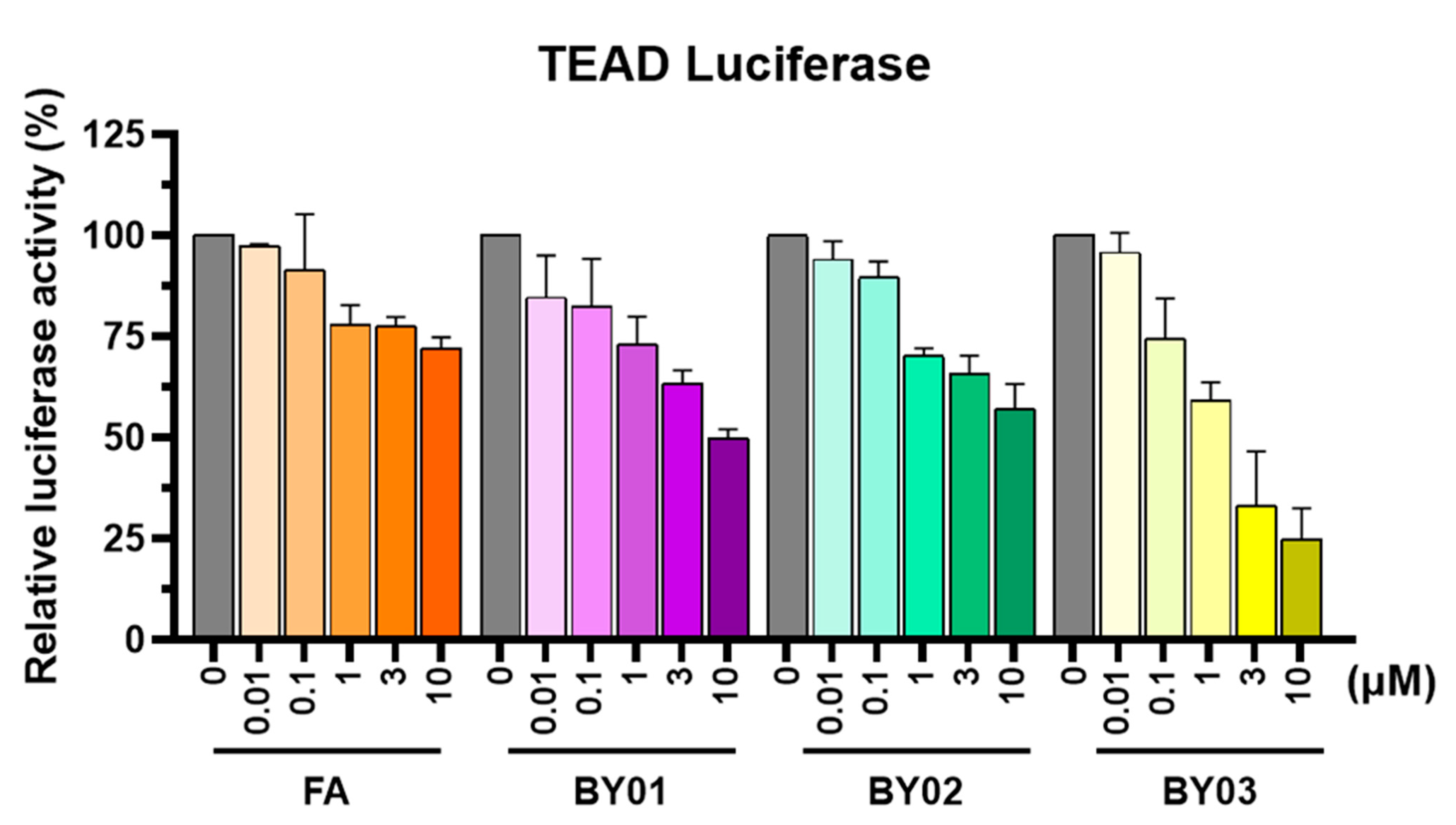
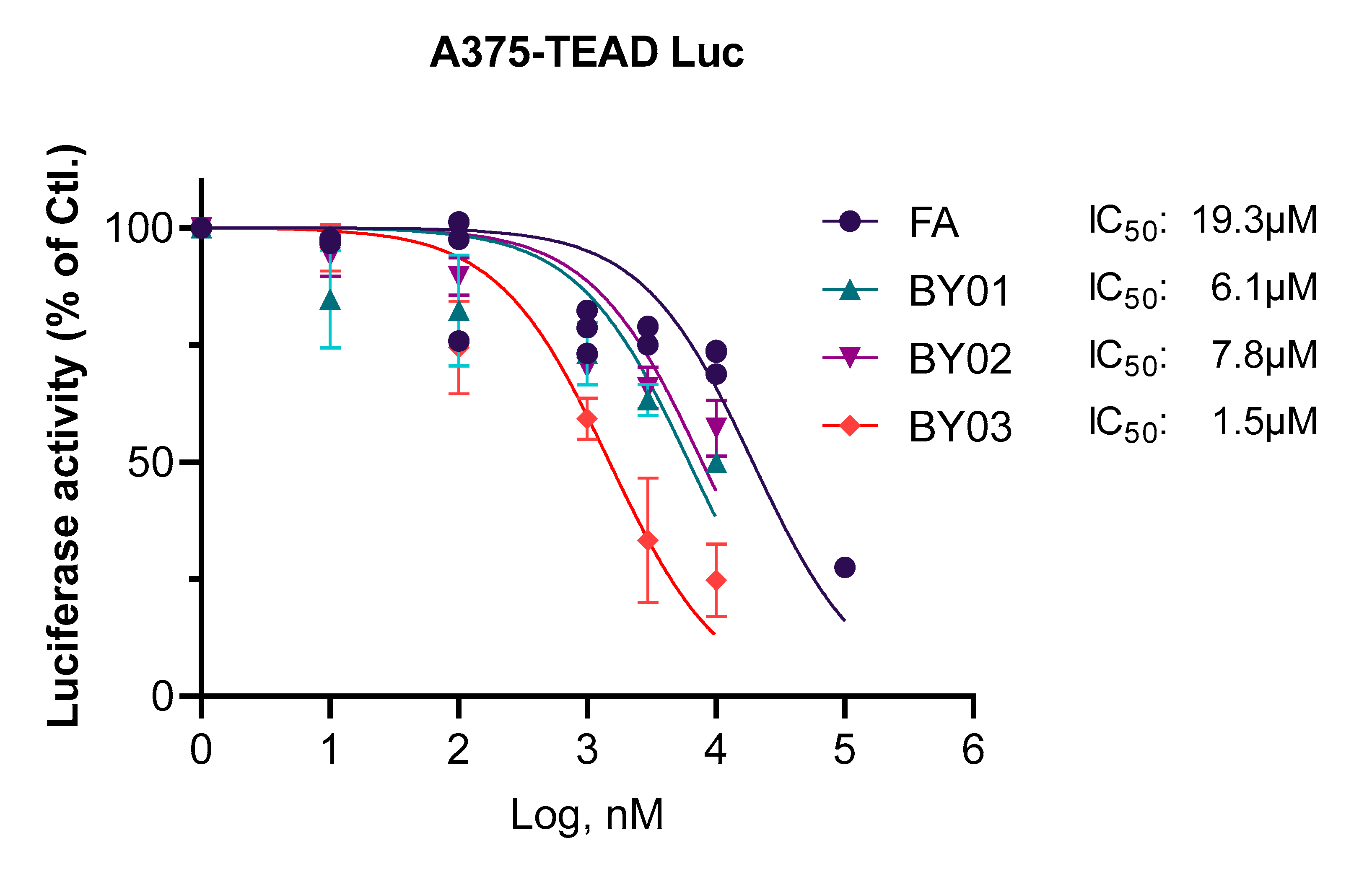
3.3. Luciferase Report Assay for Ligands
3.4. SPR Assay for Ligands
3.5. YAP-TEAD Interaction for the Proximity Ligation Assay
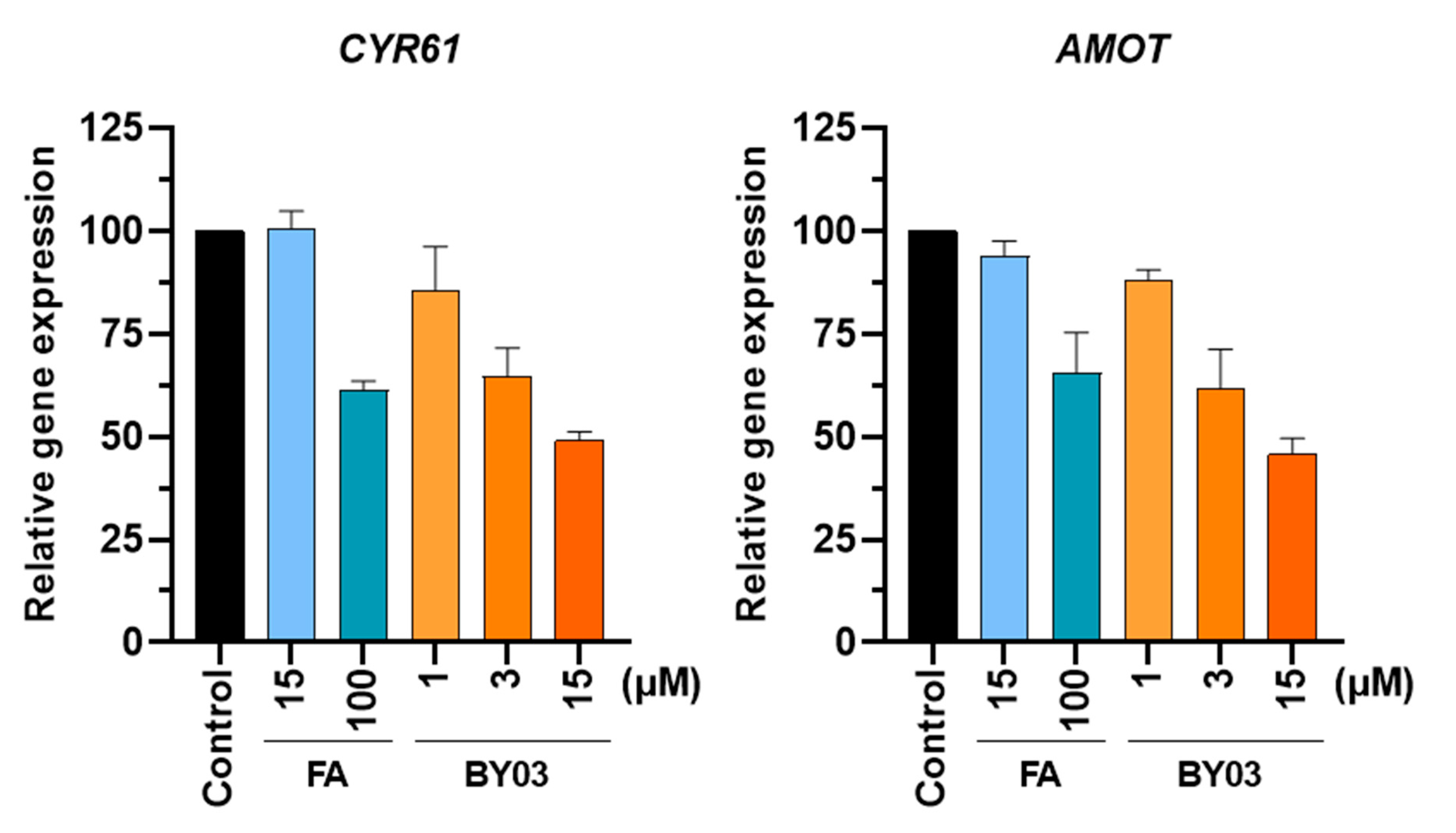
3.6. TEAD Target Gene Expression
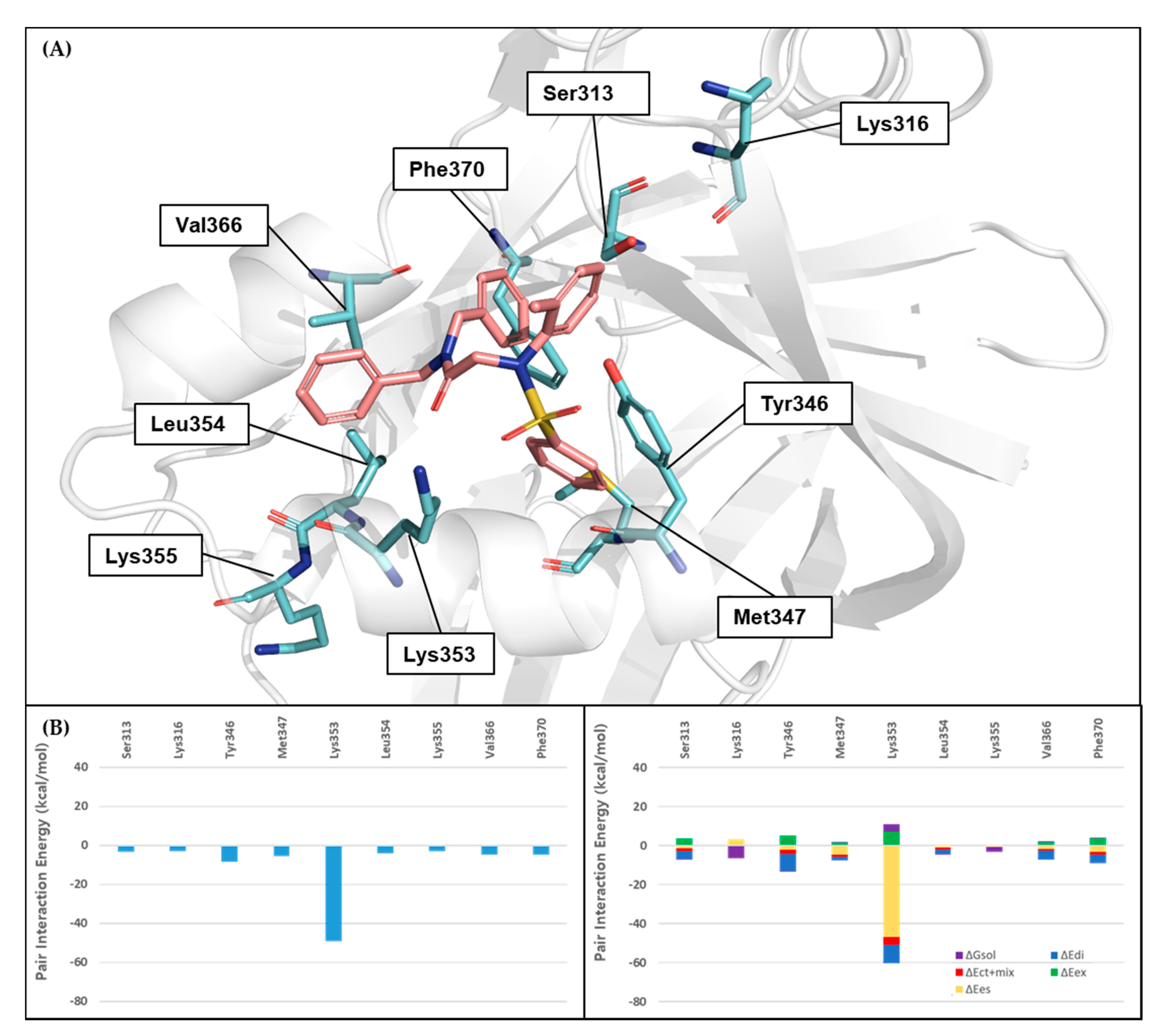
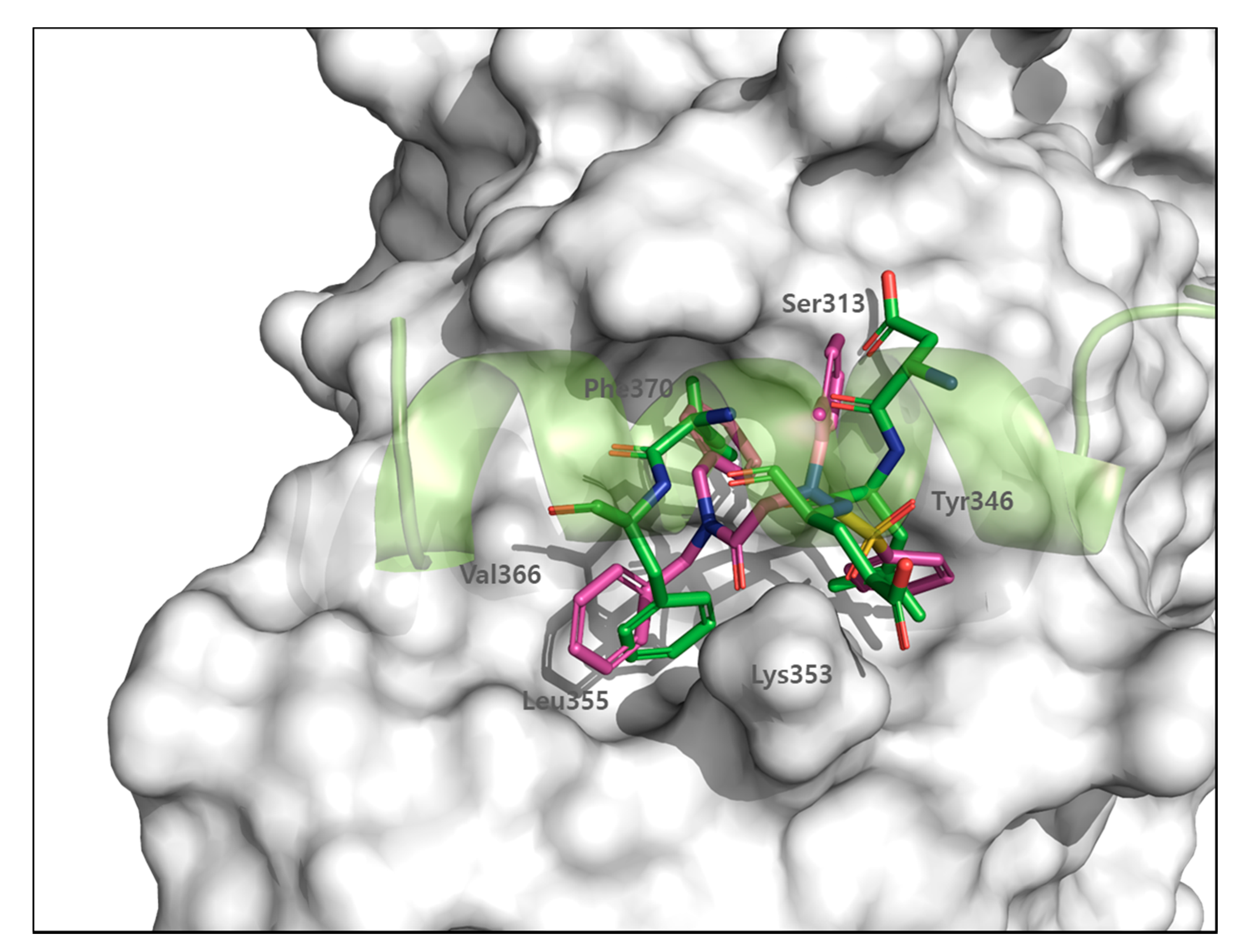
3.7. FMO Analysis of Specific Interactions between BY03 and TEAD1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Rensburg, H.J.J.; Yang, X. The roles of the Hippo pathway in cancer metastasis. Cell. Signal. 2016, 28, 1761–1772. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xu, T. Molecular mechanism of size control in development and human diseases. Cell Res. 2011, 21, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.; Varelas, X.; Guan, K.L. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat. Rev. Drug Discov. 2020, 19, 480–494. [Google Scholar] [CrossRef] [PubMed]
- Zygulska, A.L.; Krzemieniecki, K.; Pierzchalski, P. Hippo pathway—Brief overview of its relevance in cancer. J. Physiol. Pharmacol. 2017, 68, 311–335. [Google Scholar]
- Yeung, Y.T.; Guerrero-Castilla, A.; Cano, M.; Muñoz, M.F.; Ayala, A.; Argüelles, S. Dysregulation of the Hippo pathway signaling in aging and cancer. Pharmacol. Res. 2019, 143, 151–165. [Google Scholar] [CrossRef]
- Hong, W.; Guan, K.L. The YAP and TAZ transcription co-activators: Key downstream effectors of the mammalian Hippo pathway. Semin. Cell Dev. Biol. 2012, 23, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Chun, A.; Cheung, K.; Rashidi, B.; Yang, X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 2008, 283, 5496–5509. [Google Scholar] [CrossRef]
- Meng, Z.; Moroishi, T.; Guan, K.L. Mechanisms of Hippo pathway regulation. Genes Dev. 2016, 30, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Piccolo, S.; Dupont, S.; Cordenonsi, M. The biology of YAP/TAZ: Hippo signaling and beyond. Physiol. Rev. 2014, 94, 1287–1312. [Google Scholar] [CrossRef]
- Moon, S.; Yeon Park, S.; Woo Park, H. Regulation of the Hippo pathway in cancer biology. Cell. Mol. Life Sci. 2018, 75, 2303–2319. [Google Scholar] [CrossRef]
- Azad, T.; Nouri, K.; Janse van Rensburg, H.J.; Maritan, S.M.; Wu, L.; Hao, Y.; Montminy, T.; Yu, J.; Khanal, P.; Mulligan, L.M.; et al. A gain-of-functional screen identifies the Hippo pathway as a central mediator of receptor tyrosine kinases during tumorigenesis. Oncogene 2020, 39, 334–355. [Google Scholar] [CrossRef] [PubMed]
- Taha, Z.; Janse van Rensburg, H.J.; Yang, X. The hippo pathway: Immunity and cancer. Cancers 2018, 10, 94. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Hu, T.; Xu, Z.; Lin, Z.; Zhang, Z.; Feng, T.; Zhu, L.; Rong, Y.; Shen, H.; Luk, J.M.; et al. Targeting Hippo pathway by specific interruption of YAP-TEAD interaction using cyclic YAP-like peptides. FASEB J. 2015, 29, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Holden, J.K.; Cunningham, C.N. Targeting the hippo pathway and cancer through the TEAD family of transcription factors. Cancers 2018, 10, 81. [Google Scholar] [CrossRef] [PubMed]
- Gong, R.; Yu, F.-X. Targeting the Hippo Pathway for Anti-cancer Therapies. Curr. Med. Chem. 2015, 22, 4104–4117. [Google Scholar] [CrossRef]
- Basu, D.; Lettan, R.; Damodaran, K.; Strellec, S.; Reyes-Mugica, M.; Rebbaa, A. Identification, mechanism of action, and antitumor activity of a small molecule inhibitor of Hippo, TGF-β, and Wnt signaling pathways. Mol. Cancer Ther. 2014, 13, 1457–1467. [Google Scholar] [CrossRef]
- Yu, F.X.; Zhang, Y.; Park, H.W.; Jewell, J.L.; Chen, Q.; Deng, Y.; Pan, D.; Taylor, S.S.; Lai, Z.C.; Guan, K.L. Protein kinase A activates the Hippo pathway to modulate cell proliferation and differentiation. Genes Dev. 2013, 27, 1223–1232. [Google Scholar] [CrossRef]
- Yu, F.X.; Zhao, B.; Panupinthu, N.; Jewell, J.L.; Lian, I.; Wang, L.H.; Zhao, J.; Yuan, H.; Tumaneng, K.; Li, H.; et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 2012, 150, 780–791. [Google Scholar] [CrossRef]
- Gibault, F.; Sturbaut, M.; Bailly, F.; Melnyk, P.; Cotelle, P. Targeting Transcriptional Enhanced Associate Domains (TEADs). J. Med. Chem. 2018, 61, 5057–5072. [Google Scholar] [CrossRef]
- Liu-Chittenden, Y.; Huang, B.; Shim, J.S.; Chen, Q.; Lee, S.J.; Anders, R.A.; Liu, J.O.; Pan, D. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012, 26, 1300–1305. [Google Scholar] [CrossRef]
- Zhang, H.; Ramakrishnan, S.K.; Triner, D.; Centofanti, B.; Maitra, D.; Gyorffy, B.; Sebolt-Leopold, J.S.; Dame, M.K.; Varani, J.; Brenner, D.E.; et al. Tumor-selective proteotoxicity of verteporfin inhibits colon cancer progression independently of YAP1. Sci. Signal. 2015, 8, ra98. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Yang, X. Targeting the hippo pathway for breast cancer therapy. Cancers 2018, 10, 422. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Li, X. The Role of Photoactivated and Non-Photoactivated Verteporfin on Tumor. Front. Pharmacol. 2020, 11, 1626. [Google Scholar] [CrossRef]
- Jiao, S.; Wang, H.; Shi, Z.; Dong, A.; Zhang, W.; Song, X.; He, F.; Wang, Y.; Zhang, Z.; Wang, W.; et al. A Peptide Mimicking VGLL4 Function Acts as a YAP Antagonist Therapy against Gastric Cancer. Cancer Cell 2014, 25, 166–180. [Google Scholar] [CrossRef]
- Crook, Z.R.; Sevilla, G.P.; Friend, D.; Brusniak, M.Y.; Bandaranayake, A.D.; Clarke, M.; Gewe, M.; Mhyre, A.J.; Baker, D.; Strong, R.K.; et al. Mammalian display screening of diverse cystine-dense peptides for difficult to drug targets. Nat. Commun. 2017, 8, 1–15. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, B.; Wang, P.; Chen, F.; Dong, Z.; Yang, H.; Guan, K.L.; Xu, Y. Structural insights into the YAP and TEAD complex. Genes Dev. 2010, 24, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Kaan, H.Y.K.; Sim, A.Y.L.L.; Tan, S.K.J.; Verma, C.; Song, H. Targeting YAP/TAZ-TEAD protein-protein interactions using fragment-based and computational modeling approaches. PLoS ONE 2017, 12, e0178381. [Google Scholar] [CrossRef] [PubMed]
- Bokhovchuk, F.; Mesrouze, Y.; Meyerhofer, M.; Zimmermann, C.; Fontana, P.; Erdmann, D.; Jemth, P.; Chène, P. An Early Association between the α-Helix of the TEAD Binding Domain of YAP and TEAD Drives the Formation of the YAP:TEAD Complex. Biochemistry 2020, 59, 1804–1812. [Google Scholar] [CrossRef]
- Chen, L.; Chan, S.W.; Zhang, X.Q.; Walsh, M.; Lim, C.J.; Hong, W.; Song, H. Structural basis of YAP recognition by TEAD4 in the Hippo pathway. Genes Dev. 2010, 24, 290–300. [Google Scholar] [CrossRef]
- Tian, W.; Yu, J.; Tomchick, D.R.; Pan, D.; Luo, X. Structural and functional analysis of the YAP-binding domain of human TEAD2. Proc. Natl. Acad. Sci. USA 2010, 107, 7293–7298. [Google Scholar] [CrossRef]
- Sheng, C.; Dong, G.; Miao, Z.; Zhang, W.; Wang, W. State-of-the-art strategies for targeting protein-protein interactions by small-molecule inhibitors. Chem. Soc. Rev. 2015, 44, 8238–8259. [Google Scholar] [CrossRef]
- Clackson, T.; Wells, J.A. A hot spot of binding energy in a hormone-receptor interface. Science 1995, 267, 383–386. [Google Scholar] [CrossRef]
- Smith, M.C.; Gestwicki, J.E. Features of protein-protein interactions that translate into potent inhibitors: Topology, surface area and affinity. Expert Rev. Mol. Med. 2012, 14, e16. [Google Scholar] [CrossRef]
- Kitaura, K.; Ikeo, E.; Asada, T.; Nakano, T.; Uebayasi, M. Fragment molecular orbital method: An approximate computational method for large molecules. Chem. Phys. Lett. 1999, 313, 701–706. [Google Scholar] [CrossRef]
- Lim, H.; Jin, X.; Kim, J.; Hwang, S.; Shin, K.B.; Choi, J.; Nam, K.-Y.; No, K.T. Investigation of Hot Spot Region in XIAP Inhibitor Binding Site by Fragment Molecular Orbital Method. Comput. Struct. Biotechnol. J. 2019, 17, 1217–1225. Available online: https://pubmed.ncbi.nlm.nih.gov/31673305/ (accessed on 21 May 2020). [CrossRef]
- Lim, H.; Chun, J.; Jin, X.; Kim, J.; Yoon, J.H.; No, K.T. Investigation of protein-protein interactions and hot spot region between PD-1 and PD-L1 by fragment molecular orbital method. Sci. Rep. 2019, 9, 16727. [Google Scholar] [CrossRef]
- Heifetz, A. (Ed.) Quantum Mechanics in Drug Discovery; Methods in Molecular Biology; Springer: New York, NY, USA, 2020; Volume 2114, ISBN 978-1-0716-0281-2. [Google Scholar]
- Lim, H.; Baek, A.; Kim, J.; Kim, M.S.; Liu, J.; Nam, K.-Y.; Yoon, J.H.; No, K.T. Hot spot profiles of SARS-CoV-2 and human ACE2 receptor protein protein interaction obtained by density functional tight binding fragment molecular orbital method. Sci. Rep. 2020, 10, 16862. [Google Scholar] [CrossRef]
- Heifetz, A.; Sladek, V.; Townsend-Nicholson, A.; Fedorov, D.G. Characterizing Protein-Protein Interactions with the Fragment Molecular Orbital Method. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2020; Volume 2114, pp. 187–205. [Google Scholar]
- Tanaka, S.; Watanabe, C.; Honma, T.; Fukuzawa, K.; Ohishi, K.; Maruyama, T. Identification of correlated inter-residue interactions in protein complex based on the fragment molecular orbital method. J. Mol. Graph. Model. 2020, 100, 107650. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, T. A novel method for analysis of the electrostatic complementarity of protein-protein interaction based on fragment molecular orbital method. Chem. Phys. Lett. 2020, 761, 138103. [Google Scholar] [CrossRef]
- Neumann, T.; Junker, H.-D.; Schmidt, K.; Sekul, R. SPR-based Fragment Screening: Advantages and Applications. Curr. Top. Med. Chem. 2007, 7, 1630–1642. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, M.P.; Friesner, R.A.; Xiang, Z.; Honig, B. On the role of the crystal environment in determining protein side-chain conformations. J. Mol. Biol. 2002, 320, 597–608. [Google Scholar] [CrossRef]
- Olsson, M.H.M.; SØndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent treatment of internal and surface residues in empirical p K a predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef]
- Alexeev, Y.P.; Mazanetz, M.; Ichihara, O.G.; Fedorov, D. GAMESS As a Free Quantum-Mechanical Platform for Drug Research. Curr. Top. Med. Chem. 2013, 12, 2013–2033. [Google Scholar] [CrossRef]
- Nakano, T.; Kaminuma, T.; Sato, T.; Akiyama, Y.; Uebayasi, M.; Kitaura, K. Fragment molecular orbital method: Application to polypeptides. Chem. Phys. Lett. 2000, 318, 614–618. [Google Scholar] [CrossRef]
- Fedorov, D.G.; Nagata, T.; Kitaura, K. Exploring chemistry with the fragment molecular orbital method. Phys. Chem. Chem. Phys. 2012, 14, 7562–7577. [Google Scholar] [CrossRef]
- Tanaka, S.; Mochizuki, Y.; Komeiji, Y.; Okiyama, Y.; Fukuzawa, K. Electron-correlated fragment-molecular-orbital calculations for biomolecular and nano systems. Phys. Chem. Chem. Phys. 2014, 16, 10310–10344. [Google Scholar] [CrossRef]
- Fedorov, D.G.; Kitaura, K. Second order Møller-Plesset perturbation theory based upon the fragment molecular orbital method. J. Chem. Phys. 2004, 121, 2483–2490. [Google Scholar] [CrossRef]
- Fedorov, D.G.; Kitaura, K.; Li, H.; Jensen, J.H.; Gordon, M.S. The polarizable continuum model (PCM) interfaced with the fragment molecular orbital method (FMO). J. Comput. Chem. 2006, 27, 976–985. [Google Scholar] [CrossRef] [PubMed]
- Heifetz, A.; Chudyk, E.I.; Gleave, L.; Aldeghi, M.; Cherezov, V.; Fedorov, D.G.; Biggin, P.C.; Bodkin, M.J. The Fragment Molecular Orbital Method Reveals New Insight into the Chemical Nature of GPCR-Ligand Interactions. J. Chem. Inf. Model. 2016, 56, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Nishimoto, Y.; Fedorov, D.G.; Irle, S. Third-order density-functional tight-binding combined with the fragment molecular orbital method. Chem. Phys. Lett. 2015, 636, 90–96. [Google Scholar] [CrossRef][Green Version]
- Gaus, M.; Lu, X.; Elstner, M.; Cui, Q. Parameterization of DFTB3/3OB for sulfur and phosphorus for chemical and biological applications. J. Chem. Theory Comput. 2014, 10, 1518–1537. [Google Scholar] [CrossRef]
- Rappé, A.K.; Casewit, C.J.; Colwell, K.S.; Goddard, W.A.; Skiff, W.M. UFF, a Full Periodic Table Force Field for Molecular Mechanics and Molecular Dynamics Simulations. J. Am. Chem. Soc. 1992, 114, 10024–10035. [Google Scholar] [CrossRef]
- Nishimoto, Y.; Fedorov, D.G. The fragment molecular orbital method combined with density-functional tight-binding and the polarizable continuum model. Phys. Chem. Chem. Phys. 2016, 18, 22047–22061. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A New Approach for Rapid, Accurate Docking and Scoring. 1. Method and Assessment of Docking Accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Bierbaumer, L.; Katschnig, A.M.; Radic-Sarikas, B.; Kauer, M.O.; Petro, J.A.; Högler, S.; Gurnhofer, E.; Pedot, G.; Schäfer, B.W.; Schwentner, R.; et al. YAP/TAZ inhibition reduces metastatic potential of Ewing sarcoma cells. Oncogenesis 2021, 10. [Google Scholar] [CrossRef] [PubMed]
- Mesrouze, Y.; Bokhovchuk, F.; Meyerhofer, M.; Fontana, P.; Zimmermann, C.; Martin, T.; Delaunay, C.; Erdmann, D.; Schmelzle, T.; Chène, P. Dissection of the interaction between the intrinsically disordered YAP protein and the transcription factor TEAD. Elife 2017, 6, e25068. [Google Scholar] [CrossRef]
- Zhang, Z.Z.; Lin, Z.; Zhou, Z.; Shen, H.C.; Yan, S.F.; Mayweg, A.V.; Xu, Z.; Qin, N.; Wong, J.C.; Zhang, Z.Z.; et al. Structure-based design and synthesis of potent cyclic peptides inhibiting the YAP-TEAD protein-protein interaction. ACS Med. Chem. Lett. 2014, 5, 993–998. [Google Scholar] [CrossRef]
- Furet, P.; Salem, B.; Mesrouze, Y.; Schmelzle, T.; Lewis, I.; Kallen, J.; Chène, P. Structure-based design of potent linear peptide inhibitors of the YAP-TEAD protein-protein interaction derived from the YAP omega-loop sequence. Bioorganic Med. Chem. Lett. 2019, 29, 2316–2319. [Google Scholar] [CrossRef] [PubMed]
- Gibault, F.; Coevoet, M.; Sturbaut, M.; Farce, A.; Renault, N.; Allemand, F.; Guichou, J.F.; Drucbert, A.S.; Foulon, C.; Magnez, R.; et al. Toward the discovery of a novel class of YAP-TEAD interaction inhibitors by virtual screening approach targeting YAP-TEAD protein-protein interface. Cancers 2018, 10, 140. [Google Scholar] [CrossRef]
- Mesrouze, Y.; Bokhovchuk, F.; Izaac, A.; Meyerhofer, M.; Zimmermann, C.; Fontana, P.; Schmelzle, T.; Erdmann, D.; Furet, P.; Kallen, J.; et al. Adaptation of the bound intrinsically disordered protein YAP to mutations at the YAP:TEAD interface. Protein Sci. 2018, 27, 1810–1820. [Google Scholar] [CrossRef]
- Pobbati, A.V.; Rubin, B.P. Protein-Protein Interaction Disruptors of the YAP/TAZ-TEAD Transcriptional Complex. Molecules 2020, 25, 6001. [Google Scholar] [CrossRef]
- Shin, W.H.; Christoffer, C.W.; Kihara, D. In silico structure-based approaches to discover protein-protein interaction-targeting drugs. Methods 2017, 131, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Tokutomi, S.; Shimamura, K.; Fukuzawa, K.; Tanaka, S. Machine learning prediction of inter-fragment interaction energies between ligand and amino-acid residues on the fragment molecular orbital calculations for Janus kinase—Inhibitor complex. Chem. Phys. Lett. 2020, 757, 137883. [Google Scholar] [CrossRef]
- Paciotti, R.; Agamennone, M.; Coletti, C.; Storchi, L. Characterization of PD-L1 binding sites by a combined FMO/GRID-DRY approach. J. Comput. Aided. Mol. Des. 2020, 34, 897–914. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Lim, H.; Moon, S.; Cho, S.Y.; Kim, M.; Park, J.H.; Park, H.W.; No, K.T. Hot Spot Analysis of YAP-TEAD Protein-Protein Interaction Using the Fragment Molecular Orbital Method and Its Application for Inhibitor Discovery. Cancers 2021, 13, 4246. https://doi.org/10.3390/cancers13164246
Kim J, Lim H, Moon S, Cho SY, Kim M, Park JH, Park HW, No KT. Hot Spot Analysis of YAP-TEAD Protein-Protein Interaction Using the Fragment Molecular Orbital Method and Its Application for Inhibitor Discovery. Cancers. 2021; 13(16):4246. https://doi.org/10.3390/cancers13164246
Chicago/Turabian StyleKim, Jongwan, Hocheol Lim, Sungho Moon, Seon Yeon Cho, Minhye Kim, Jae Hyung Park, Hyun Woo Park, and Kyoung Tai No. 2021. "Hot Spot Analysis of YAP-TEAD Protein-Protein Interaction Using the Fragment Molecular Orbital Method and Its Application for Inhibitor Discovery" Cancers 13, no. 16: 4246. https://doi.org/10.3390/cancers13164246
APA StyleKim, J., Lim, H., Moon, S., Cho, S. Y., Kim, M., Park, J. H., Park, H. W., & No, K. T. (2021). Hot Spot Analysis of YAP-TEAD Protein-Protein Interaction Using the Fragment Molecular Orbital Method and Its Application for Inhibitor Discovery. Cancers, 13(16), 4246. https://doi.org/10.3390/cancers13164246





