Epigenetic Changes Affecting the Development of Hepatocellular Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. HCC Etiology and Prevalence
3. DNA Methylation Pattern in HCC
4. Function of DNA Methyltransferases in HCC
5. Histone Modifications
6. Non-Coding RNAs
7. N6-Methyladenosine mRNA Modification
8. HCC Risk Factors and Epigenetics
9. Liver Cancer Stem Cells
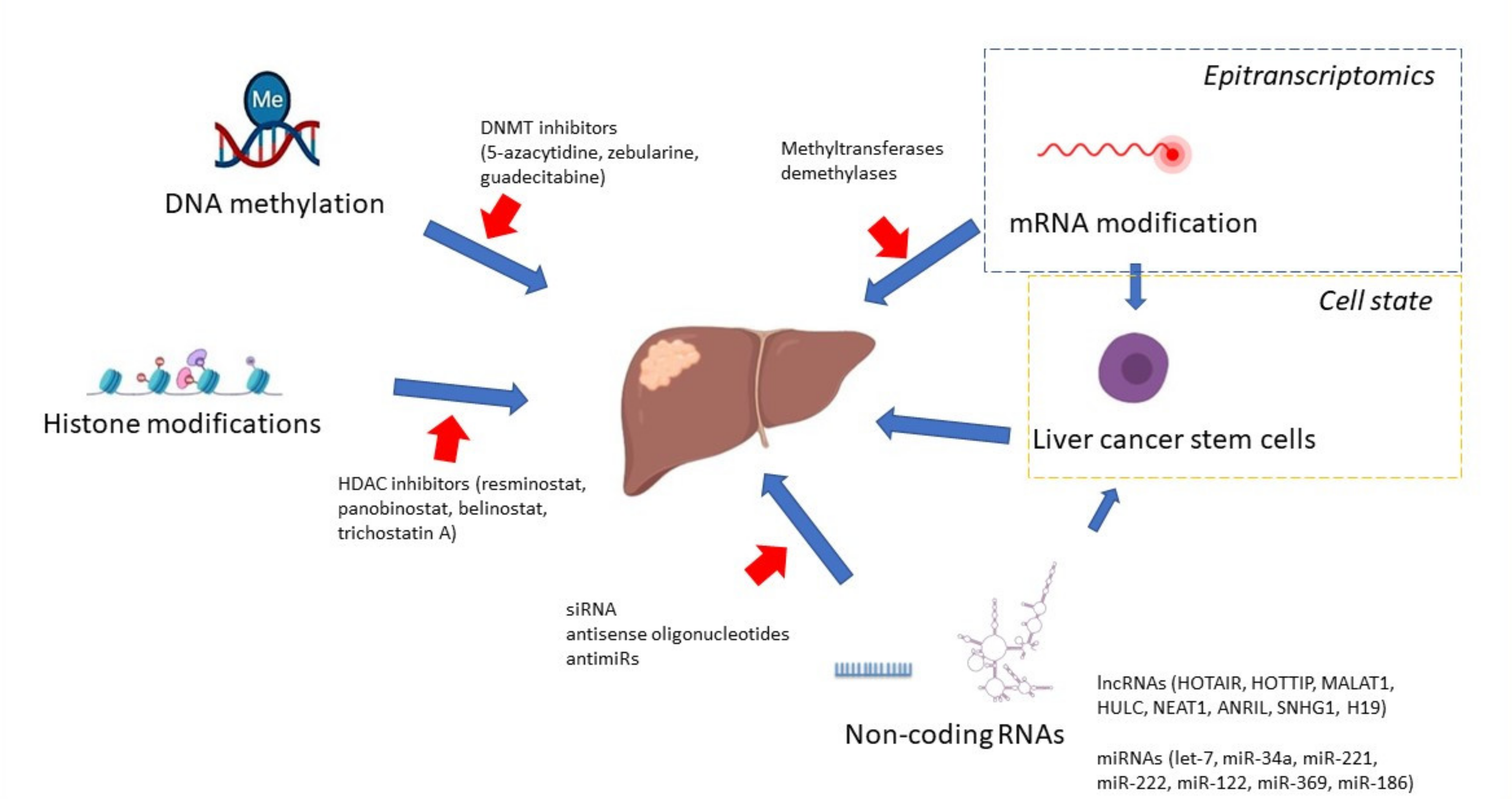
10. Targeted Therapies
11. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Bruix, J.; Sherman, M. Management of hepatocellular carcinoma. Hepatology 2005, 42, 1208–1236. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, M.; Roayaie, S.; Konstadoulakis, M. Strategies for the management of hepatocellular carcinoma. Nat. Clin. Pract. Oncol. 2007, 4, 424–432. [Google Scholar] [CrossRef] [PubMed]
- El–Serag, H.B.; Rudolph, K.L. Hepatocellular Carcinoma: Epidemiology and Molecular Carcinogenesis. Gastroenterology 2007, 132, 2557–2576. [Google Scholar] [CrossRef] [PubMed]
- El-Serag, H.B. Epidemiology of Viral Hepatitis and Hepatocellular Carcinoma. Gastroenterology 2012, 142, 1264–1273. [Google Scholar] [CrossRef]
- Larsson, S.C.; Wolk, A. Overweight, obesity and risk of liver cancer: A meta-analysis of cohort studies. Br. J. Cancer 2007, 97, 1005–1008. [Google Scholar] [CrossRef]
- Kalra, M.; Mayes, J.; Assefa, S.; Kaul, A.K.; Kaul, R. Role of sex steroid receptors in pathobiology of hepatocellular carcinoma. World J. Gastroenterol. 2008, 14, 5945–5961. [Google Scholar] [CrossRef]
- Esteller, M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007, 8, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, A.P.; Vogelstein, B. Hypomethylation distinguishes genes of some human cancers from their normal counter-parts. Nature 1983, 301, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Baylin, S.B. Mechanisms underlying epigenetically mediated gene silencing in cancer. Semin. Cancer Biol. 2002, 12, 331–337. [Google Scholar] [CrossRef]
- Hama, N.; Totoki, Y.; Miura, F.; Tatsuno, K.; Saito-Adachi, M.; Nakamura, H.; Arai, Y.; Hosoda, F.; Urushidate, T.; Ohashi, S.; et al. Epigenetic landscape influences the liver cancer genome architecture. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Calvisi, D.F.; Ladu, S.; Gorden, A.; Farina, M.; Lee, J.-S.; Conner, E.A.; Schroeder, I.; Factor, V.M.; Thorgeirsson, S.S. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J. Clin. Investig. 2007, 117, 2713–2722. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, J.P. Epigenetic mechanisms involved in the pathogenesis of hepatobiliary malignancies. Epigenomics 2010, 2, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Liew, C.T.; Li, H.-M.; Lo, K.W.; Leow, C.K.; Chan, J.Y.; Hin, L.-Y.; Lau, W.Y.; Lai, P.B.-S.; Lim, B.K.; Huang, J.; et al. High frequency of p16INK4A gene alterations in hepatocellular carcinoma. Oncogene 1999, 18, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Newell, P.; Toffanin, S.; Villanueva, A.; Chiang, D.; Minguez, B.; Cabellos, L.; Savic, R.; Hoshida, Y.; Lim, K.H.; Melgar-Lesmes, P.; et al. Ras pathway activation in hepatocellular carcinoma and anti-tumoral effect of combined sorafenib and rapamycin in vivo. J. Hepatol. 2009, 51, 725–733. [Google Scholar] [CrossRef]
- Park, J.H.; Cho, S.B.; Lee, W.S.; Park, C.H.; Joo, Y.E.; Kim, H.S.; Choi, S.K.; Rew, J.S.; Lee, J.H.; Kim, S.J. Methylation pattern of DNA repair genes and microsatellite instability in hepatocelluar carcinoma. Korean J. Gastroenterol. 2006, 48, 327–336. [Google Scholar] [PubMed]
- Liang, Y.; Ma, B.; Jiang, P.; Yang, H.-M. Identification of methylation-regulated differentially expressed genes and related pathways in hepatocellular carcinoma: A study based on tcga database and bioinformatics analysis. Front. Oncol. 2021, 11, 2040. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Tu, Y.; Chen, C.; Sun, H.; Wan, C.; Cai, X. DNA methylation biomarkers for hepatocellular carcinoma. Cancer Cell Int. 2018, 18, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-H.; Seo, D.; Choi, K.-J.; Andersen, J.B.; Won, M.-A.; Kitade, M.; Gómez-Quiroz, L.E.; Judge, A.D.; Marquardt, J.; Raggi, C.; et al. Antitumor Effects in Hepatocarcinoma of Isoform-Selective Inhibition of HDAC2. Cancer Res. 2014, 74, 4752–4761. [Google Scholar] [CrossRef]
- Richon, V.M.; Sandhoff, T.W.; Rifkind, R.A.; Marks, P.A. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acet-ylation. Proc. Natl. Acad. Sci. USA 2000, 97, 10014–10019. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.F.; Cao, X.Y.; Zhu, Y.J.; Wu, Z.R.; Zhuang, X.; Shao, M.Y.; Xu, Q.; Zhou, Y.-J.; Ji, H.-J.; Lu, Q.-R.; et al. Histone deacetylase 3 promotes liver regeneration and liver cancer cells proliferation through signal transduc-er and activator of transcription 3 signaling pathway. Cell Death Dis. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Wu, H.; Yang, T.Y.; Li, Y.; Ye, W.L.; Liu, F.; He, X.S.; Wang, J.-R.; Gan, W.-J.; Li, X.-M.; Zhang, S.; et al. Tumor necrosis factor receptor-associated factor 6 promotes hepatocarcinogenesis by interacting with histone deacetylase 3 to enhance c-Myc gene expression and protein stability. Hepatology 2020, 71, 148–163. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Zhou, Y.; Zhuang, X.; Wu, Z.; Lu, Y.; Li, S.; Zeng, Y.; Lu, Q.R.; Huo, Y.; Shi, Y.; et al. HDAC3 deficiency promotes liver cancer through a defect in H3K9ac/H3K9me3 transition. Cancer Res. 2019, 79, 3676–3688. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Nie, Q.; Dai, M.; Chen, F.; Wu, H. Histone Deacetylases Inhibit the Snail2-Mediated EMT During Metastasis of Hepatocellular Carcinoma Cells. Front. Cell Dev. Biol. 2020, 8, 752. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-T.; Chiou, S.-S.; Chai, C.-Y.; Hsi, E.; Wang, S.-N.; Huang, S.-K.; Hsu, S.-H. Aryl hydrocarbon receptor regulates histone deacetylase 8 expression to repress tumor suppressive activity in hepatocellular carcinoma. Oncotarget 2017, 8, 7489–7501. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Lou, B.; Chen, W.; Zhang, J.; Lin, S.; Lv, F.-F.; Chen, Y. Down-regulation of HDAC5 inhibits growth of human hepatocellular carcinoma by induction of apoptosis and cell cycle arrest. Tumor Biol. 2014, 35, 11523–11532. [Google Scholar] [CrossRef]
- Kanki, K.; Watanabe, R.; Thai, L.N.; Zhao, C.-H.; Naito, K. HDAC9 Is Preferentially Expressed in Dedifferentiated Hepatocellular Carcinoma Cells and Is Involved in an Anchorage-Independent Growth. Cancers 2020, 12, 2734. [Google Scholar] [CrossRef] [PubMed]
- Pineau, P.; Volinia, S.; McJunkin, K.; Marchio, A.; Battiston, C.; Terris, B.; Mazzaferro, V.M.; Lowe, S.W.; Croce, C.M.; Dejean, A. miR-221 overexpression contributes to liver tumorigenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Li, F.; Wang, J.; Hu, J.; Li, Z.; Gu, Y.; Feng, Y. miR-369 inhibits Liver Cancer progression by targeting ZEB1 pathway and predicts the prognosis of HCC patients. J. Cancer 2021, 12, 3067–3076. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yang, X.; Zhou, X.; Wu, B.; Zhu, D.; Jia, W.; Chu, J.; Wang, J.; Wu, J.; Kong, L. MiR-3174 promotes proliferation and inhibits apoptosis by targeting FOXO1 in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2020, 526, 889–897. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lu, L.; Luo, Z.; Li, W.; Lu, Y.; Tang, Q.; Pu, J. miR-383 inhibits cell growth and promotes cell apoptosis in hepatocellular carcinoma by targeting IL-17 via STAT3 signaling pathway. Biomed. Pharmacother. 2019, 120, 109551. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.-J.; Chen, G.-Y.; Xie, Z.-T. MicroRNA-361-5p Inhibits Cancer Cell Growth by Targeting CXCR6 in Hepatocellular Carcinoma. Cell. Physiol. Biochem. 2016, 38, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Yang, Z.; Lou, Y.; Huang, J.; Yang, P.; Jiang, W.; Chen, S. miR-186 Inhibits Liver Cancer Stem Cells Expansion via Targeting PTPN11. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.; Cheng, Z.; Liu, H.; Wang, X.; Han, T.; Sun, W.; Li, X.; Yang, W.; Chen, C.; Xia, M.; et al. Shp2 promotes liver cancer stem cell expansion by augmenting beta-catenin signaling and predicts chemo-therapeutic response of patients. Hepatology 2017, 65, 1566–1580. [Google Scholar] [CrossRef]
- Thakral, S.; Ghoshal, K. miR-122 is a unique molecule with great potential in diagnosis, prognosis of liver disease, and therapy both as miRNA mimic and antimir. Curr. Gene Ther. 2015, 15, 142–150. [Google Scholar] [CrossRef]
- Lanzafame, M.; Bianco, G.; Terracciano, L.M.; Ng, C.K.Y.; Piscuoglio, S. The Role of Long Non-Coding RNAs in Hepatocarcinogenesis. Int. J. Mol. Sci. 2018, 19, 682. [Google Scholar] [CrossRef]
- Jin, N.; Yang, L.Y.; Xu, Z.P. Long non-coding RNA HOTTIP is able to predict poor prognosis in various neoplasms: A me-ta-analysis. Mol. Clin. Oncol. 2017, 7, 263–266. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhou, L.; Li, H.; Sun, T.; Wen, X.; Li, X.; Meng, Y.; Li, Y.; Liu, M.; Liu, S.; et al. Nuclear-Encoded lncRNA MALAT1 Epigenetically Controls Metabolic Reprogramming in HCC Cells through the Mitophagy Pathway. Mol. Ther.-Nucleic Acids 2021, 23, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Lin, Z.; Li, X.; Xin, X.; An, J.; Zheng, Q.; Yang, Y.; Lu, D. HULC cooperates with MALAT1 to aggravate liver cancer stem cells growth through telomere repeat-binding factor 2. Sci. Rep. 2016, 6, 36045. [Google Scholar] [CrossRef]
- Li, P.; Li, Y.; Ma, L. Long noncoding RNA highly upregulated in liver cancer promotes the progression of hepatocellular carcinoma and attenuates the chemosensitivity of oxaliplatin by regulating miR-383-5p/vesicle-associated membrane pro-tein-2 axis. Pharmacol. Res. Perspect. 2021, 9, e00815. [Google Scholar] [CrossRef]
- Yan, C.; Wei, S.; Han, D.; Wu, L.; Tan, L.; Wang, H.; Dong, Y.; Hua, J.; Yang, W. LncRNA HULC shRNA disinhibits miR-377-5p to suppress the growth and invasion of hepatocellular carci-noma in vitro and hepatocarcinogenesis in vivo. Ann. Transl. Med. 2020, 8, 1294. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Yang, T.; He, W.; Jiang, S.; Zhong, D.; Xu, Z.; Wei, Q.; Zhang, Y.; Shi, C. Liver X receptor inhibits the growth of hepatocellular carcinoma cells via regulating HULC/miR-134-5p/FOXM1 axis. Cell. Signal. 2020, 74, 109720. [Google Scholar] [CrossRef] [PubMed]
- Matsukura, S.; Soejima, H.; Nakagawachi, T.; Yakushiji, H.; Ogawa, A.; Fukuhara, M.; Miyazaki, K.; Nakabeppu, Y.; Sekiguchi, M.; Mukai, T. CpG methylation of MGMT and hMLH1 promoter in hepatocellular carcinoma associated with hepatitis viral infection. Br. J. Cancer 2003, 88, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Tischoff, I.; Tannapfe, A. DNA methylation in hepatocellular carcinoma. World J. Gastroenterol. 2008, 14, 1741–1748. [Google Scholar] [CrossRef]
- Qiu, J.; Peng, B.; Tang, Y.; Qian, Y.; Guo, P.; Li, M.; Luo, J.; Chen, B.; Tang, H.; Lu, C.; et al. CpG methylation signature predicts recurrence in early-stage hepatocellular carcinoma: Results from a multicenter study. J. Clin. Oncol. 2017, 35, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Tong, W.; Xie, F.; Zhu, L.; Wu, H.; Shi, R.; Wang, L.; Yang, L.; Liu, Z.; Miao, F.; et al. DNA methylation biomarkers for diagnosis of primary liver cancer and distinguishing hepatocellular carcinoma from intrahepatic cholangiocarcinoma. Aging 2021, 13, 17592–17606. [Google Scholar] [CrossRef]
- Zopf, S.; Ocker, M.; Neureiter, D.; Alinger, B.; Gahr, S.; Neurath, M.F.; Di Fazio, P. Inhibition of DNA methyltransferase activity and expression by treatment with the pan-deacetylase inhibitor panobinostat in hepatocellular carcinoma cell lines. BMC Cancer 2012, 12, 386. [Google Scholar] [CrossRef]
- Saito, Y.; Kanai, Y.; Nakagawa, T.; Sakamoto, M.; Saito, H.; Ishii, H.; Hirohashi, S. Increased protein expression of DNA methyltransferase (DNMT) 1 is significantly correlated with the malig-nant potential and poor prognosis of human hepatocellular carcinomas. Int. J. Cancer 2003, 105, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Oh, B.-K.; Kim, H.; Park, H.-J.; Shim, Y.-H.; Choi, J.; Park, C.; Park, Y.N. DNA methyltransferase expression and DNA methylation in human hepatocellular carcinoma and their clinicopathological correlation. Int. J. Mol. Med. 2007, 20, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Venturelli, S.; Berger, A.; Weiland, T.; Zimmermann, M.; Häcker, S.; Peter, C.; Wesselborg, S.; Königsrainer, A.; Weiss, T.; Gregor, M.; et al. Dual antitumour effect of 5-azacytidine by inducing a breakdown of resistance-mediating factors and epigenetic modulation. Gut 2011, 60, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Calvisi, D.F.; Ladu, S.; Gorden, A.; Farina, M.; Conner, E.A.; Lee, J.; Factor, V.M.; Thorgeirsson, S.S. Ubiquitous Activation of Ras and Jak/Stat Pathways in Human HCC. Gastroenterology 2006, 130, 1117–1128. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.K.; Li, Y.; Pandit, H.; Li, S.; Pulliam, Z.; Zheng, Q.; Yu, Y.; Martin, R.C. Epigenetic modulation enhances immunotherapy for hepatocellular carcinoma. Cell. Immunol. 2019, 336, 66–74. [Google Scholar] [CrossRef]
- Bannister, A.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Ler, S.Y.; LEuNG, C.H.W.; Khin, L.W.; Lu, G.D.; Salto-Tellez, M.; Hartman, M.; Iau, P.T.C.; Yap, C.T.; Hooi, S.C. HDAC1 and HDAC2 independently predict mortality in hepatocellular carcinoma by a competing risk re-gression model in a Southeast Asian population. Oncol. Rep. 2015, 34, 2238–2250. [Google Scholar] [CrossRef]
- Quint, K.; Agaimy, A.; Di Fazio, P.; Montalbano, R.; Steindorf, C.; Jung, R.; Hellerbrand, C.; Hartmann, A.; Sitter, H.; Neureiter, D.; et al. Clinical significance of histone deacetylases 1, 2, 3, and 7: HDAC2 is an independent predictor of survival in HCC. Virchows Arch. 2011, 459, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Sun, L.; Tao, S.; Dai, M.; Wang, Y.; Li, Y.; Wu, J. Clinical significance of HDAC9 in hepatocellular carcinoma. Cell. Mol. Biol. 2019, 65, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Han, H.D.; Lopez-Berestein, G.; Sood, A.K. MicroRNA therapeutics: Principles, expectations, and challenges. Chin. J. Cancer 2011, 30, 368–370. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Li, Y.; Xu, K.; Chen, G.; Lu, X.; Duan, Q.; Kang, Z. RETRACTED: miR-361-5p inhibits hepatocellular carcinoma cell proliferation and invasion by targeting VEGFA. Biochem. Biophys. Res. Commun. 2016, 479, 901–906. [Google Scholar] [CrossRef]
- Ren, Q.; Xiao, X.; Leng, X.; Zhang, Q.; Zhou, X.; Ren, Z.; Xiao, H. MicroRNA-361-5p induces hepatocellular carcinoma cell apoptosis and enhances drug sensitivity by targeting MAP3K9. Exp. Ther. Med. 2021, 21, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Park, J.K.; Kogure, T.; Nuovo, G.J.; Jiang, J.; He, L.; Kim, J.H.; Phelps, M.A.; Papenfuss, T.L.; Croce, C.M.; Patel, T.; et al. miR-221 Silencing Blocks Hepatocellular Carcinoma and Promotes Survival. Cancer Res. 2011, 71, 7608–7616. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.; Hu, Q.; Wu, W.; Wang, M.; Huang, J.; Zhao, X.; Tang, G.; Liang, T. Tumor-triggered personalized microRNA cocktail therapy for hepatocellular carcinoma. Biomater. Sci. 2020, 8, 6579–6591. [Google Scholar] [CrossRef]
- Stiuso, P.; Potenza, N.; Lombardi, A.; Ferrandino, I.; Monaco, A.; Zappavigna, S.; Vanacore, D.; Mosca, N.; Castiello, F.; Porto, S.; et al. MicroRNA-423-5p Promotes Autophagy in Cancer Cells and Is Increased in Serum From Hepatocarcinoma Patients Treated With Sorafenib. Mol. Ther.-Nucleic Acids 2015, 4, e233. [Google Scholar] [CrossRef]
- Hsu, S.H.; Wang, B.; Kutay, H.; Bid, H.; Shreve, J.; Zhang, X.; Costinean, S.; Bratasz, A.; Houghton, P.; Ghoshal, K. Hepatic loss of miR-122 predisposes mice to hepatobiliary cyst and hepatocellular carcinoma upon diethyl-nitrosamine exposure. Am. J. Pathol. 2013, 183, 1719–1730. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.-C.; Hung, T.; Argani, P.; Rinn, J.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Yan, X.; Hu, Z.; Feng, Y.; Hu, X.; Yuan, J.; Zhao, S.D.; Zhang, Y.; Yang, L.; Shan, W.; He, Q.; et al. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell 2015, 28, 529–540. [Google Scholar] [CrossRef]
- Tsang, F.H.; Au, S.L.; Wei, L.; Fan, D.N.; Lee, J.M.; Wong, C.C.L.; Ng, I.O.; Wong, C.-M. Long non-coding RNA HOTTIP is frequently up-regulated in hepatocellular carcinoma and is targeted by tumour suppressive miR-125b. Liver Int. 2015, 35, 1597–1606. [Google Scholar] [CrossRef]
- Luo, F.; Sun, B.; Li, H.; Xu, Y.; Liu, Y.; Liu, X.; Lu, L.; Li, J.; Wang, Q.; Wei, S.; et al. A MALAT1/HIF-2α feedback loop contributes to arsenite carcinogenesis. Oncotarget 2016, 7, 5769–5787. [Google Scholar] [CrossRef]
- Lai, M.C.; Yang, Z.; Zhou, L.; Zhu, Q.Q.; Xie, H.Y.; Zhang, F.; Wu, L.-M.; Chen, L.-M.; Zheng, S.-S. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med. Oncol. 2012, 29, 1810–1816. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Gholipour, M.; Hussen, B.M.; Taheri, M. The Impact of Long Non-Coding RNAs in the Pathogenesis of Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 1150. [Google Scholar] [CrossRef] [PubMed]
- Pai, S.I.; Lin, Y.-Y.; Macaes, B.; Meneshian, A.; Hung, C.-F.; Wu, T.-C. Prospects of RNA interference therapy for cancer. Gene Ther. 2006, 13, 464–477. [Google Scholar] [CrossRef]
- Shen, S.; Yan, J.; Zhang, Y.; Dong, Z.; Xing, J.; He, Y. N6-methyladenosine (m6A)-mediated messenger RNA signatures and the tumor immune microenvironment can predict the prognosis of hepatocellular carcinoma. Ann. Transl. Med. 2021, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Lan, Y.; Zhao, Y.; Shi, Y.; Jin, J.; Xie, W. The emerging roles of N6-methyladenosine RNA methylation in human cancers. Biomark. Res. 2020, 8, 1–16. [Google Scholar] [CrossRef]
- Park, I.Y.; Sohn, B.H.; Yu, E.; Suh, D.J.; Chung, Y.; Lee, J.; Surzycki, S.J.; Lee, Y.I. Aberrant Epigenetic Modifications in Hepatocarcinogenesis Induced by Hepatitis B Virus X Protein. Gastroenterology 2007, 132, 1476–1494. [Google Scholar] [CrossRef]
- Zhao, P.; Malik, S.; Xing, S. Epigenetic Mechanisms Involved in HCV-Induced Hepatocellular Carcinoma (HCC). Front. Oncol. 2021, 11, 2752. [Google Scholar] [CrossRef]
- Domovitz, T.; Gal-Tanamy, M. Tracking Down the Epigenetic Footprint of HCV-Induced Hepatocarcinogenesis. J. Clin. Med. 2021, 10, 551. [Google Scholar] [CrossRef]
- Quan, H.; Zhou, F.; Nie, D.; Chen, Q.; Cai, X.; Shan, X.; Zhou, Z.; Chen, K.; Huang, A.; Li, S.; et al. Hepatitis C virus core protein epigenetically silences SFRP1 and enhances HCC aggressiveness by inducing epithelial-mesenchymal transition. Oncogene 2014, 33, 2826–2835. [Google Scholar] [CrossRef]
- Siddiqui, N.N.; ul Haq, A.; Siddiqui, O.A.; Khan, R. DNA methyltransferase 1, 3a, and 3b expression in hepatitis C associated human hepatocellular carci-noma and their clinicopathological association. Tumour Biol. 2016, 37, 10487–10497. [Google Scholar] [CrossRef] [PubMed]
- Arora, P.; Kim, E.O.; Jung, J.K.; Jang, K.L. Hepatitis C virus core protein downregulates E-cadherin expression via activation of DNA methyltransfer-ase 1 and 3b. Cancer Lett. 2008, 261, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.L.; Heo, S.; Jang, K.L. Hepatitis C virus core protein overcomes H2O2-induced apoptosis by downregulating p14 expression via DNA methylation. J. Gen. Virol. 2015, 96, 822–832. [Google Scholar] [CrossRef]
- Park, S.H.; Lim, J.S.; Lim, S.Y.; Tiwari, I.; Jang, K.L. Hepatitis C virus core protein stimulates cell growth by down-regulating p16 expression via DNA methyl-ation. Cancer Lett. 2011, 310, 61–68. [Google Scholar]
- Higgs, M.R.; Lerat, H.; Pawlotsky, J.M. Downregulation of Gadd45beta expression by hepatitis C virus leads to defective cell cycle arrest. Cancer Res. 2010, 70, 4901–4911. [Google Scholar] [CrossRef]
- Perez, S.; Kaspi, A.; Domovitz, T.; Davidovich, A.; Lavi-Itzkovitz, A.; Meirson, T.; Holmes, J.A.; Dai, C.-Y.; Huang, C.-F.; Chung, R.T.; et al. Hepatitis C virus leaves an epigenetic signature post cure of infection by direct-acting antivirals. PLoS Genet. 2019, 15, e1008181. [Google Scholar] [CrossRef] [PubMed]
- Eslam, M.; Valenti, L.; Romeo, S. Genetics and epigenetics of NAFLD and NASH: Clinical impact. J. Hepatol. 2018, 68, 268–279. [Google Scholar] [CrossRef] [PubMed]
- Hsu, S.-H.; Wang, B.; Kota, J.; Yu, J.; Costinean, S.; Kutay, H.; Yu, L.; Bai, S.; La Perle, K.; Chivukula, R.R.; et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J. Clin. Investig. 2012, 122, 2871–2883. [Google Scholar] [CrossRef] [PubMed]
- Meroni, M.; Longo, M.; Rametta, R.; Dongiovanni, P. Genetic and Epigenetic Modifiers of Alcoholic Liver Disease. Int. J. Mol. Sci. 2018, 19, 3857. [Google Scholar] [CrossRef] [PubMed]
- Ambade, A.; Satishchandran, A.; Szabo, G. Alcoholic hepatitis accelerates early hepatobiliary cancer by increasing stem-ness and miR-122-mediated HIF-1alpha activation. Sci. Rep. 2016, 6, 21340. [Google Scholar] [CrossRef]
- Sookoian, S.; Rosselli, M.S.; Gemma, C.; Burgueño, A.L.; Fernández Gianotti, T.; Castaño, G.O.; Pirola, C.J. Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: Impact of liver methylation of the peroxisome proliferator-activated receptor gamma coactivator 1alpha promoter. Hepatology 2010, 52, 1992–2000. [Google Scholar] [CrossRef]
- Johnson, N.D.; Wu, X.; Still, C.D.; Chu, X.; Petrick, A.T.; Gerhard, G.S.; Conneely, K.N.; DiStefano, J.K. Differential DNA methylation and changing cell-type proportions as fibrotic stage progresses in NAFLD. Clin. Epigenetics 2021, 13, 1–14. [Google Scholar] [CrossRef]
- Gerhard, G.S.; Malenica, I.; Llaci, L.; Chu, X.; Petrick, A.T.; Still, C.D.; Distefano, J.K. Differentially methylated loci in NAFLD cirrhosis are associated with key signaling pathways. Clin. Epigenetics 2018, 10, 1–9. [Google Scholar] [CrossRef]
- Hyun, J.; Jung, Y. DNA Methylation in Nonalcoholic Fatty Liver Disease. Int. J. Mol. Sci. 2020, 21, 8138. [Google Scholar] [CrossRef]
- Haraguchi, N.; Utsunomiya, T.; Inoue, H.; Tanaka, F.; Mimori, K.; Barnard, G.F.; Mori, M. Characterization of a Side Population of Cancer Cells from Human Gastrointestinal System. Stem Cells 2006, 24, 506–513. [Google Scholar] [CrossRef] [PubMed]
- Jordan, C.T.; Guzman, M.L.; Noble, M. Cancer stem cells. N. Engl. J. Med. 2006, 355, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Suetsugu, A.; Nagaki, M.; Aoki, H.; Motohashi, T.; Kunisada, T.; Moriwaki, H. Characterization of CD133+ hepatocellular carcinoma cells as cancer stem/progenitor cells. Biochem. Biophys. Res. Commun. 2006, 351, 820–824. [Google Scholar] [CrossRef]
- Ma, S.; Tang, K.H.; Chan, Y.P.; Lee, K.W.; Kwan, P.S.; Castilho, A.; Ng, I.O.-L.; Man, K.; Wong, N.; To, K.-F.; et al. miR-130b Promotes CD133+ Liver Tumor-Initiating Cell Growth and Self-Renewal via Tumor Protein 53-Induced Nuclear Protein 1. Cell Stem Cell 2010, 7, 694–707. [Google Scholar] [CrossRef]
- Piao, L.S.; Hur, W.; Kim, T.-K.; Hong, S.W.; Kim, S.W.; Choi, J.E.; Sung, P.S.; Song, M.J.; Lee, B.-C.; Hwang, D.; et al. CD133+ liver cancer stem cells modulate radioresistance in human hepatocellular carcinoma. Cancer Lett. 2012, 315, 129–137. [Google Scholar] [CrossRef]
- Lan, X.; Wu, Y.Z.; Wang, Y.; Wu, F.R.; Zang, C.B.; Tang, C.; Cao, S.; Li, S.-L. CD133 silencing inhibits stemness properties and enhances chemoradiosensitivity in CD133-positive liver can-cer stem cells. Int. J. Mol. Med. 2013, 31, 315–324. [Google Scholar] [CrossRef]
- Castelli, G.; Pelosi, E.; Testa, U. Liver Cancer: Molecular Characterization, Clonal Evolution and Cancer Stem Cells. Cancers 2017, 9, 127. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, J.; Tsui, Y.M.; Shi, C.; Wang, Y.; Zhang, X.; Yan, Q.; Chen, M.; Jiang, C.; Yuan, Y.-F.; et al. RALYL increases hepatocellular carcinoma stemness by sustaining the mRNA stability of TGF-beta2. Nat. Commun. 2021, 12, 1–14. [Google Scholar]
- De Mattia, E.; Cecchin, E.; Guardascione, M.; Foltran, L.; Di Raimo, T.; Angelini, F.; D’Andrea, M.; Toffoli, G. Pharmacogenetics of the systemic treatment in advanced hepatocellular carcinoma. World J. Gastroenterol. 2019, 25, 3870–3896. [Google Scholar] [CrossRef]
- Thillai, K.; Srikandarajah, K.; Ross, P. Regorafenib as treatment for patients with advanced hepatocellular cancer. Futur. Oncol. 2017, 13, 2223–2232. [Google Scholar] [CrossRef]
- Bakouny, Z.; Assi, T.; El Rassy, E.; Nasr, F. Second-line treatments of advanced hepatocellular carcinoma: Systematic review and network meta-analysis of randomized controlled trials. J. Clin. Gastroenterol. 2019, 53, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Kudo, M.; Finn, R.S.; Qin, S.; Han, K.-H.; Ikeda, K.; Piscaglia, F.; Baron, A.; Park, J.-W.; Han, G.; Jassem, J.; et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: A randomised phase 3 non-inferiority trial. Lancet 2018, 391, 1163–1173. [Google Scholar] [CrossRef]
- Gordan, J.D.; Kennedy, E.B.; Abou-Alfa, G.K.; Beg, M.S.; Brower, S.T.; Gade, T.P.; Goff, L.; Gupta, S.; Guy, J.; Harris, W.P.; et al. Systemic Therapy for Advanced Hepatocellular Carcinoma: ASCO Guideline. J. Clin. Oncol. 2020, 38, 4317–4345. [Google Scholar] [CrossRef] [PubMed]
- Marks, P.A.; Breslow, R. Dimethyl sulfoxide to vorinostat: Development of this histone deacetylase inhibitor as an anti-cancer drug. Nat. Biotechnol. 2007, 25, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Barbarotta, L.; Hurley, K. Romidepsin for the Treatment of Peripheral T-Cell Lymphoma. J. Adv. Pract. Oncol. 2015, 6, 22–36. [Google Scholar] [PubMed]
- Bitzer, M.; Horger, M.; Giannini, E.; Ganten, T.M.; Wörns, M.A.; Siveke, J.T.; Dollinger, M.M.; Gerken, G.; Scheulen, M.E.; Wege, H.; et al. Resminostat plus sorafenib as second-line therapy of advanced hepatocellular carcinoma—The SHELTER study. J. Hepatol. 2016, 65, 280–288. [Google Scholar] [CrossRef]
- Soukupova, J.; Bertran, E.; Peñuelas-Haro, I.; Urdiroz-Urricelqui, U.; Borgman, M.; Kohlhof, H.; Fabregat, I. Resminostat induces changes in epithelial plasticity of hepatocellular carcinoma cells and sensitizes them to sorafenib-induced apoptosis. Oncotarget 2017, 8, 110367–110379. [Google Scholar] [CrossRef]
- Streubel, G.; Schrepfer, S.; Kallus, H.; Parnitzke, U.; Wulff, T.; Hermann, F.; Borgmann, M.; Hamm, S. Histone deacetylase inhibitor resminostat in combination with sorafenib counteracts platelet-mediated pro-tumoral effects in hepatocellular carcinoma. Sci. Rep. 2021, 11, 1–15. [Google Scholar] [CrossRef]
- Tak, W.Y.; Ryoo, B.-Y.; Lim, H.Y.; Kim, D.-Y.; Okusaka, T.; Ikeda, M.; Hidaka, H.; Yeon, J.-E.; Mizukoshi, E.; Morimoto, M.; et al. Phase I/II study of first-line combination therapy with sorafenib plus resminostat, an oral HDAC inhibitor, versus sorafenib monotherapy for advanced hepatocellular carcinoma in east Asian patients. Investig. New Drugs 2018, 36, 1072–1084. [Google Scholar] [CrossRef]
- Lachenmayer, A.; Toffanin, S.; Cabellos, L.; Alsinet, C.; Hoshida, Y.; Villanueva, A.; Minguez, B.; Tsai, H.-W.; Ward, S.C.; Thung, S.; et al. Combination therapy for hepatocellular carcinoma: Additive preclinical efficacy of the HDAC inhib-itor panobinostat with sorafenib. J. Hepatol. 2012, 56, 1343–1350. [Google Scholar] [CrossRef]
- Yeo, W.; Chung, H.; Chan, S.; Wang, L.Z.; Lim, R.; Picus, J.; Boyer, M.; Mo, F.K.; Koh, J.; Rha, S.Y.; et al. Epigenetic Therapy Using Belinostat for Patients With Unresectable Hepatocellular Carcinoma: A Multicenter Phase I/II Study With Biomarker and Pharmacokinetic Analysis of Tumors From Patients in the Mayo Phase II Consortium and the Cancer Therapeutics Research Group. J. Clin. Oncol. 2012, 30, 3361–3367. [Google Scholar] [CrossRef] [PubMed]
- Dunn, J.; Rao, S. Epigenetics and immunotherapy: The current state of play. Mol. Immunol. 2017, 87, 227–239. [Google Scholar] [CrossRef]
- Llopiz, D.; Ruiz, M.; Villanueva, L.; Iglesias, T.; Silva, L.; Egea, J.; Lasarte, J.J.; Pivette, P.; Trochon-Joseph, V.; Vasseur, B.; et al. Enhanced anti-tumor efficacy of checkpoint inhibitors in combination with the histone deacetylase inhibitor Belinostat in a murine hepatocellular carcinoma model. Cancer Immunol. Immunother. 2019, 68, 379–393. [Google Scholar] [CrossRef]
- Shin, S.; Kim, M.; Lee, S.-J.; Park, K.-S. Trichostatin A Sensitizes Hepatocellular Carcinoma Cells to Enhanced NK Cell-mediated Killing by Regulating Immune-related Genes. Cancer Genom. Proteom. 2017, 14, 349–362. [Google Scholar] [CrossRef]
- Fenaux, P.; Mufti, G.J.; Hellström-Lindberg, E.; Santini, V.; Gattermann, N.; Germing, U.; Sanz, G.; List, A.; Gore, S.; Seymour, J.F.; et al. Azacitidine Prolongs Overall Survival Compared With Conventional Care Regimens in Elderly Patients With Low Bone Marrow Blast Count Acute Myeloid Leukemia. J. Clin. Oncol. 2010, 28, 562–569. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, L.; Li, H.; Hinoue, T.; Zhou, W.; Ohtani, H.; El-Khoueiry, A.; Daniels, J.; O’Connell, C.; Dorff, T.B.; et al. Integrative epigenetic analysis reveals therapeutic targets to the dna methyltransferase inhibitor guadecitabine (SGI-110) in hepatocellular carcinoma. Hepatology 2018, 68, 1412–1428. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Y.; El-Khoueiry, A.; Taverna, P.; Ljungman, M.; Neamati, N. Guadecitabine (SGI-110) priming sensitizes hepatocellular carcinoma cells to oxaliplatin. Mol. Oncol. 2015, 9, 1799–1814. [Google Scholar] [CrossRef] [PubMed]
| Epigenetic Changes | Mechanism Affected | Reference |
|---|---|---|
| DNA methylation | ||
| Hypomethylation | Protooncogene c-Jun and c-myc activation | [12] |
| Mitotic recombination/genomic instability | [12] | |
| Hypermethylation | WNT/β-catenin signaling activation | [13] |
| APC inactivation | [13] | |
| p16INK4A activation | [14] | |
| RASSF1A and NORE1A activation | [15] | |
| Mismatch repair system genes (hMLH1, hMSH2, and hMSH3) inactivation | [16] | |
| Cardiotrophin-1 (CTF1), FZD8, pyruvate dehydrogenase kinase 4 (PDK4), and ZNF334 activity | [17] | |
| MAD2L1, CDC20, CCNB1, CCND1, AR, and ESR1 | [18] | |
| p53 and MAPK signaling regulation | [18] | |
| Histone modification | ||
| Upregulated HDAC2 | Dysregulation of cell cycle, apoptosis, and differentiation via p27, p53, BCL-2, or PPAR γ | [19] |
| Downregulated HDAC3 | An increase in p21WAF1/cip1 expression; G1-phase arrest | [20] |
| Downregulated HDAC3 | STAT3-dependent cell proliferation | [21] |
| Downregulated HDAC3 | c-Myc protein synthesis and stability | [22] |
| Downregulated HDAC3 | Defective double-strand breaks repair | [23] |
| HDAC3 and HDAC1 | Cell migration, epithelial-mesenchymal transition (EMT), and tumor metastasis regulation | [24] |
| Upregulated HDAC8 | Downregulation of RB1 | [25] |
| Upregulated HDAC5 | Increased cell proliferation | [26] |
| Downregulation of HDAC5 | Cell apoptosis via antiapoptotic proteins (p53, bax, bcl-2, cyto C, and caspase 3), G1-phase cell cycle arrest via cell cycle regulators (cyclin D1 and CDK2/4/6) | [26] |
| Upregulated HDAC9 | Epithelial–mesenchymal transition process activation; cellular stemness properties regulation | [27] |
| Non-coding RNAs | ||
| miR-221/222 | Enhanced cell growth via p27 regulation mTOR kinase regulation | [28] |
| miR-369 | Zinc finger E-box binding homeobox 1 regulation | [29] |
| miR-3174 | FOXO1 regulation | [30] |
| miR-383 | IL-17 via STAT3 signaling pathway regulation | [31] |
| miR-361-5p | CXCR6, VEGFA, or MAP3K9 regulation | [32] |
| miR-186 | CSCs self-renewal | [33] |
| miR-186 | Protein tyrosine phosphatase non-receptor type 11 regulation | [34] |
| miR-122 | Tumor growth regulation | [35] |
| HOTAIR | Proliferation, regulation of pluripotency, metastasis, and sensitivity to chemotherapeutics | [36] |
| HOTTIP | Survival, tumor grade, and prognosis | [37] |
| MALAT1 | Regulation of mitochondrial metabolism | [38] |
| HULC | Growth of liver cancer stem cells | [39] |
| Chemosensitivity of anti-cancer drug oxaliplatin inhibition | [40] | |
| Regulation of miR-383-5p/vesicle-associated membrane protein-2 pathway; miR-377-5p/HIF-1α pathway and miR-134-5p/FOXM1 pathway | [41,42] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wolinska, E.; Skrzypczak, M. Epigenetic Changes Affecting the Development of Hepatocellular Carcinoma. Cancers 2021, 13, 4237. https://doi.org/10.3390/cancers13164237
Wolinska E, Skrzypczak M. Epigenetic Changes Affecting the Development of Hepatocellular Carcinoma. Cancers. 2021; 13(16):4237. https://doi.org/10.3390/cancers13164237
Chicago/Turabian StyleWolinska, Ewa, and Maciej Skrzypczak. 2021. "Epigenetic Changes Affecting the Development of Hepatocellular Carcinoma" Cancers 13, no. 16: 4237. https://doi.org/10.3390/cancers13164237
APA StyleWolinska, E., & Skrzypczak, M. (2021). Epigenetic Changes Affecting the Development of Hepatocellular Carcinoma. Cancers, 13(16), 4237. https://doi.org/10.3390/cancers13164237





