Raman Spectroscopy and Machine Learning for IDH Genotyping of Unprocessed Glioma Biopsies
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Experimental Design
2.2. Samples Collection and Analysis
2.3. Raman Analysis
2.4. Data Processing and Classification Models
2.4.1. Data Processing
2.4.2. Classification Models
- -
- XGB parameters were set to default except for step size shrinkage (eta) (0.01, 0.05, 0.1, 0.2, 0.3); tree method was set to ‘hist’, the learning objective was set to: binary-logistic; the evaluation metric to: negative log-likelihood (logloss); the maximum depth of a tree: (5, 8, 10, 12, 15), the minimum loss reduction required to make a further partition on a leaf node of the tree (gamma): (0.1, 0.2, 0.3, 0.4).
- -
- Grid search parameters for SVM were set default except for the kernel set to Radial Basis Function (RBF), and the regularization parameter (C):(0.01, 0.1, 1, 10, 100, 1000).
2.5. Statistical Analysis
3. Results
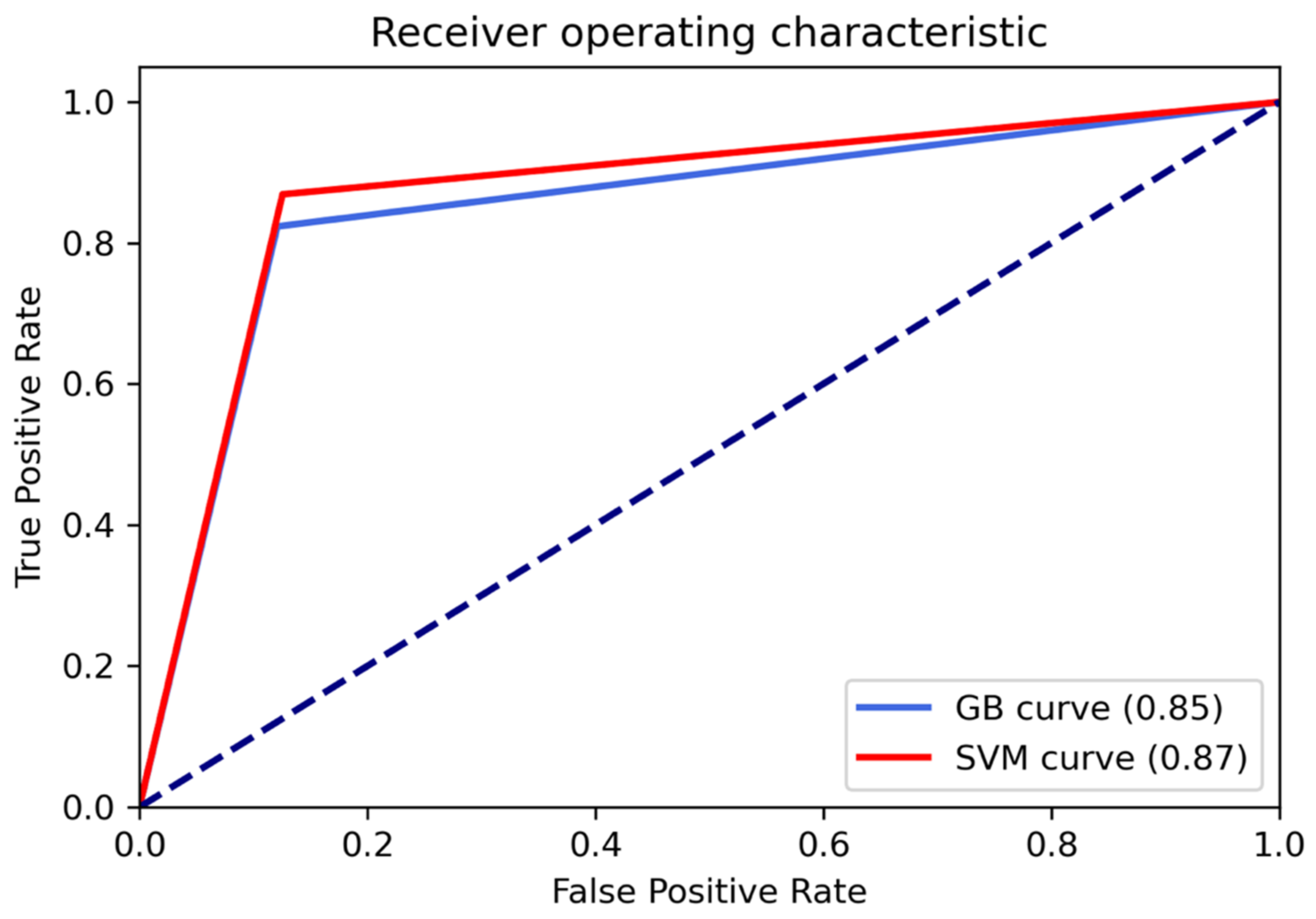
3.1. Classification Performances
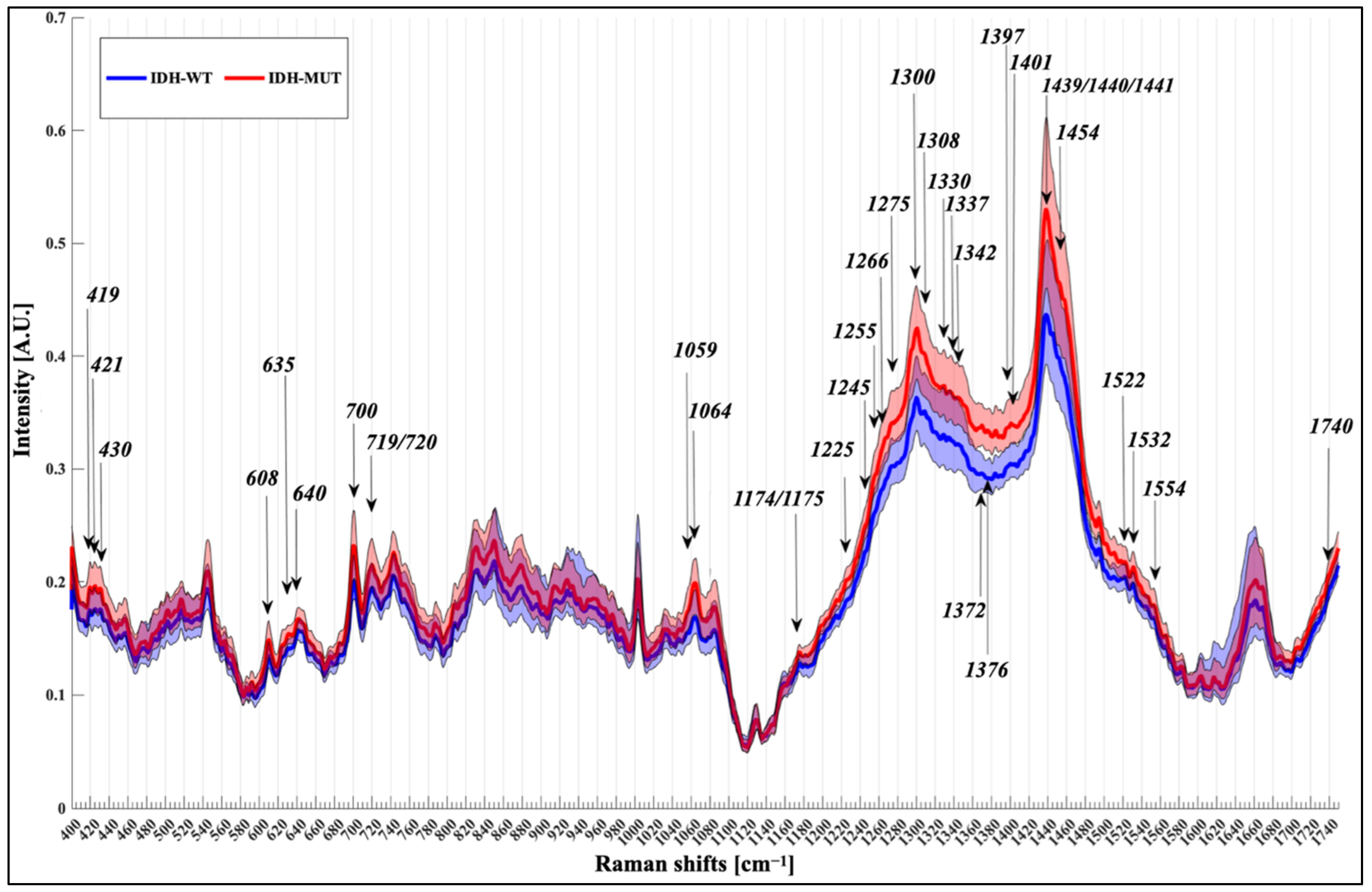
3.2. Spectral Analysis
4. Discussion
4.1. Raman Spectroscopy of Fresh Biopsies Next to the Operative Room
4.2. Raman Spectroscopy and IDH Mutation
4.3. Raman Shifts
4.4. Study Limitation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary; WHO: Geneva, Switzerland, 2016; Volume 131, pp. 803–820. [Google Scholar]
- Rossi, M.; Ambrogi, F.; Gay, L.; Gallucci, M.; Nibali, M.C.; Leonetti, A.; Puglisi, G.; Sciortino, T.; Howells, H.; Riva, M.; et al. Is Supratotal Resection Achievable in Low-Grade Gliomas? Feasibility, Putative Factors, Safety, and Functional Outcome. J. Neurosurg. 2020, 132, 1692–1705. [Google Scholar] [CrossRef] [Green Version]
- Rossi, M.; Gay, L.; Ambrogi, F.; Nibali, M.C.; Sciortino, T.; Puglisi, G.; Leonetti, A.; Mocellini, C.; Caroli, M.; Cordera, S.; et al. Association of Supratotal Resection with Progression-Free Survival, Malignant Transformation, and Overall Survival in Lower-Grade Gliomas. J. Neuro-Oncol. 2020, 23, 812–826. [Google Scholar] [CrossRef] [PubMed]
- Jakola, A.S.; Skjulsvik, A.J.; Myrmel, K.S.; Sjåvik, K.; Unsgård, G.; Torp, S.H.; Aaberg, K.; Berg, T.; Dai, H.Y.; Johnsen, K.; et al. Surgical Resection versus Watchful Waiting in Low-Grade Gliomas. Ann. Oncol. 2017, 28, 1942–1948. [Google Scholar] [CrossRef] [PubMed]
- Jermyn, M.; Mok, K.; Mercier, J.; Desroches, J.; Pichette, J.; Saint-Arnaud, K.; Bernstein, L.; Guiot, M.-C.C.; Petrecca, K.; Leblond, F. Intraoperative Brain Cancer Detection with Raman Spectroscopy in Humans. Sci. Transl. Med. 2015, 7, 274ra19. [Google Scholar] [CrossRef] [PubMed]
- Riva, M.; Sciortino, T.; Secoli, R.; D’Amico, E.; Moccia, S.; Fernandes, B.; Conti Nibali, M.; Gay, L.; Rossi, M.; De Momi, E.; et al. Glioma Biopsies Classification Using Raman Spectroscopy and Machine Learning Models on Fresh Tissue Samples. Cancers 2021, 13, 1073. [Google Scholar] [CrossRef]
- Cameron, J.M.; Conn, J.J.A.; Rinaldi, C.; Sala, A.; Brennan, P.M.; Jenkinson, M.D.; Caldwell, H.; Cinque, G.; Syed, K.; Butler, H.J.; et al. Interrogation of IDH1 Status in Gliomas by Fourier Transform Infrared Spectroscopy. Cancers 2020, 12, 3682. [Google Scholar] [CrossRef]
- Gajjar, K.; Heppenstall, L.D.; Pang, W.; Ashton, K.M.; Trevisan, J.; Patel, I.I.; Llabjani, V.; Stringfellow, H.F.; Martin-Hirsch, P.L.; Dawson, T.; et al. Diagnostic Segregation of Human Brain Tumours Using Fourier-Transform Infrared and/or Raman Spectroscopy Coupled with Discriminant Analysis. Anal. Methods 2013, 5, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Uckermann, O.; Yao, W.; Juratli, T.A.; Galli, R.; Leipnitz, E.; Meinhardt, M.; Koch, E.; Schackert, G.; Steiner, G.; Kirsch, M. IDH1 Mutation in Human Glioma Induces Chemical Alterations That Are Amenable to Optical Raman Spectroscopy. J. Neuro-Oncol. 2018, 139, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Livermore, L.J.; Isabelle, M.; Bell, I.M.; Scott, C.; Walsby-Tickle, J.; Gannon, J.; Plaha, P.; Vallance, C.; Ansorge, O. Rapid Intraoperative Molecular Genetic Classification of Gliomas Using Raman Spectroscopy. J. Neuro-Oncol. Adv. 2019, 1, vdz008. [Google Scholar] [CrossRef]
- Kalkanis, S.N.; Kast, R.E.; Rosenblum, M.L.; Mikkelsen, T.; Yurgelevic, S.M.; Nelson, K.M.; Raghunathan, A.; Poisson, L.M.; Auner, G.W. Raman Spectroscopy to Distinguish Grey Matter, Necrosis, and Glioblastoma Multiforme in Frozen Tissue Sections. J. Neuro-Oncol. 2014, 116, 477–485. [Google Scholar] [CrossRef]
- Riva, M.; Hennersperger, C.; Milletari, F.; Katouzian, A.; Pessina, F.; Gutierrez-Becker, B.; Castellano, A.; Navab, N.; Bello, L. 3D Intra-Operative Ultrasound and MR Image Guidance: Pursuing an Ultrasound-Based Management of Brainshift to Enhance Neuronavigation. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 1711–1725. [Google Scholar] [CrossRef]
- Riva, M.; Hiepe, P.; Frommert, M.; Divenuto, I.; Gay, L.G.; Sciortino, T.; Nibali, M.C.; Rossi, M.; Pessina, F.; Bello, L. Intraoperative Computed Tomography and Finite Element Modelling for Multimodal Image Fusion in Brain Surgery. Oper. Neurosurg. 2020, 18, 531–541. [Google Scholar] [CrossRef]
- Eiseman, E.; Bloom, G.; Brower, J.; Clancy, N.; Olmsted, S.S. Case Studies of Existing Human Tissue Repositories “Best Practices” for a Biospecimen Resource for the Genomic and Proteomic Era; RAND: Santa Monica, CA, USA, 2013; ISBN 2325-7776/2325-7792. [Google Scholar]
- Barman, I.; Kong, C.-R.; Singh, G.P.; Dasari, R.R. Effect of Photobleaching on Calibration Model Development in Biological Raman Spectroscopy. J. Biomed. Opt. 2011, 16, 011004. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Henson, M.J. A Practical Algorithm to Remove Cosmic Spikes in Raman Imaging Data for Pharmaceutical Applications. Appl. Spectrosc. 2007, 61, 1015–1020. [Google Scholar] [CrossRef]
- Zhao, J.; Lui, H.; McLean, D.I.; Zeng, H. Automated Autofluorescence Background Subtraction Algorithm for Biomedical Raman Spectroscopy. Appl. Spectrosc. 2007, 61, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Olson, R.S.; Cava, W.L.; Mustahsan, Z.; Varik, A.; Moore, J.H. Data-Driven Advice for Applying Machine Learning to Bioinformatics Problems. Pac. Symp. Biocomput. 2018, 23, 192–203. [Google Scholar]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Brusatori, M.; Auner, G.; Noh, T.; Scarpace, L.; Broadbent, B.; Kalkanis, S.N. Intraoperative Raman Spectroscopy. Neurosurg. Clin. N. Am. 2017, 28, 633–652. [Google Scholar] [CrossRef]
- Bergner, N.; Krafft, C.; Geiger, K.D.; Kirsch, M.; Schackert, G.; Popp, J. Unsupervised Unmixing of Raman Microspectroscopic Images for Morphochemical Analysis of Non-Dried Brain Tumor Specimens. Anal. Bioanal. Chem. 2012, 403, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Köhler, M.; MacHill, S.; Salzer, R.; Krafft, C. Characterization of Lipid Extracts from Brain Tissue and Tumors Using Raman Spectroscopy and Mass Spectrometry. Anal. Bioanal. Chem. 2009, 393, 1513–1520. [Google Scholar] [CrossRef]
- Krafft, C.; Shapoval, L.; Sobottka, S.B.; Geiger, K.D.; Schackert, G.; Salzer, R. Identification of Primary Tumors of Brain Metastases by SIMCA Classification of IR Spectroscopic Images. Biochim. Et Biophys. Acta-Biomembr. 2006, 1758, 883–891. [Google Scholar] [CrossRef] [Green Version]
- Koljenović, S.; Schut, T.B.; Vincent, A.; Kros, J.M.; Puppels, G.J. Detection of Meningioma in Dura Mater by Raman Spectroscopy. Anal. Chem. 2005, 77, 7958–7965. [Google Scholar] [CrossRef]
- Kast, R.E.; Auner, G.W.; Rosenblum, M.L.; Mikkelsen, T.; Yurgelevic, S.M.; Raghunathan, A.; Poisson, L.M.; Kalkanis, S.N. Raman Molecular Imaging of Brain Frozen Tissue Sections. J. Neuro-Oncol. 2014, 120, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, H.N.; Zhang, L. Deciphering the Finger Prints of Brain Cancer Astrocytoma in Comparison to Astrocytes by Using near Infrared Raman Spectroscopy. Mol. Cell. Biochem. 2007, 295, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Krafft, C.; Sobottka, S.B.; Schackert, G.; Salzer, R. Raman and Infrared Spectroscopic Mapping of Human Primary Intracranial Tumors: A Comparative Study. J. Raman Spectrosc. 2006, 37, 367–375. [Google Scholar] [CrossRef]
- Livermore, L.J.; Isabelle, M.; Bell, I.M.; Edgar, O.; Voets, N.L.; Stacey, R.; Ansorge, O.; Vallance, C.; Plaha, P. Raman Spectroscopy to Differentiate between Fresh Tissue Samples of Glioma and Normal Brain: A Comparison with 5-ALA–Induced Fluorescence-Guided Surgery. J. Neurosurg. 2020, 1–11. [Google Scholar] [CrossRef]
- Ali, S.M.; Bonnier, F.; Tfayli, A.; Lambkin, H.; Flynn, K.; McDonagh, V.; Healy, C.; Clive Lee, T.; Lyng, F.M.; Byrne, H.J. Raman Spectroscopic Analysis of Human Skin Tissue Sections Ex-Vivo: Evaluation of the Effects of Tissue Processing and Dewaxing. J. Biomed. Opt. 2013, 18, 061202. [Google Scholar] [CrossRef] [Green Version]
- Koivunen, P.; Lee, S.; Duncan, C.G.; Lopez, G.; Lu, G.; Ramkissoon, S.; Losman, J.A.; Joensuu, P.; Bergmann, U.; Gross, S.; et al. Transformation by the (R)-Enantiomer of 2-Hydroxyglutarate Linked to EGLN Activation. Nature 2012, 483, 484–488. [Google Scholar] [CrossRef]
- Shankar, G.M.; Kirtane, A.R.; Miller, J.J.; Mazdiyasni, H.; Rogner, J.; Tai, T.; Williams, E.A.; Higuchi, F.; Juratli, T.A.; Tateishi, K.; et al. Genotype-Targeted Local Therapy of Glioma. Proc. Natl. Acad. Sci. USA 2018, 115, E8388–E8394. [Google Scholar] [CrossRef] [Green Version]
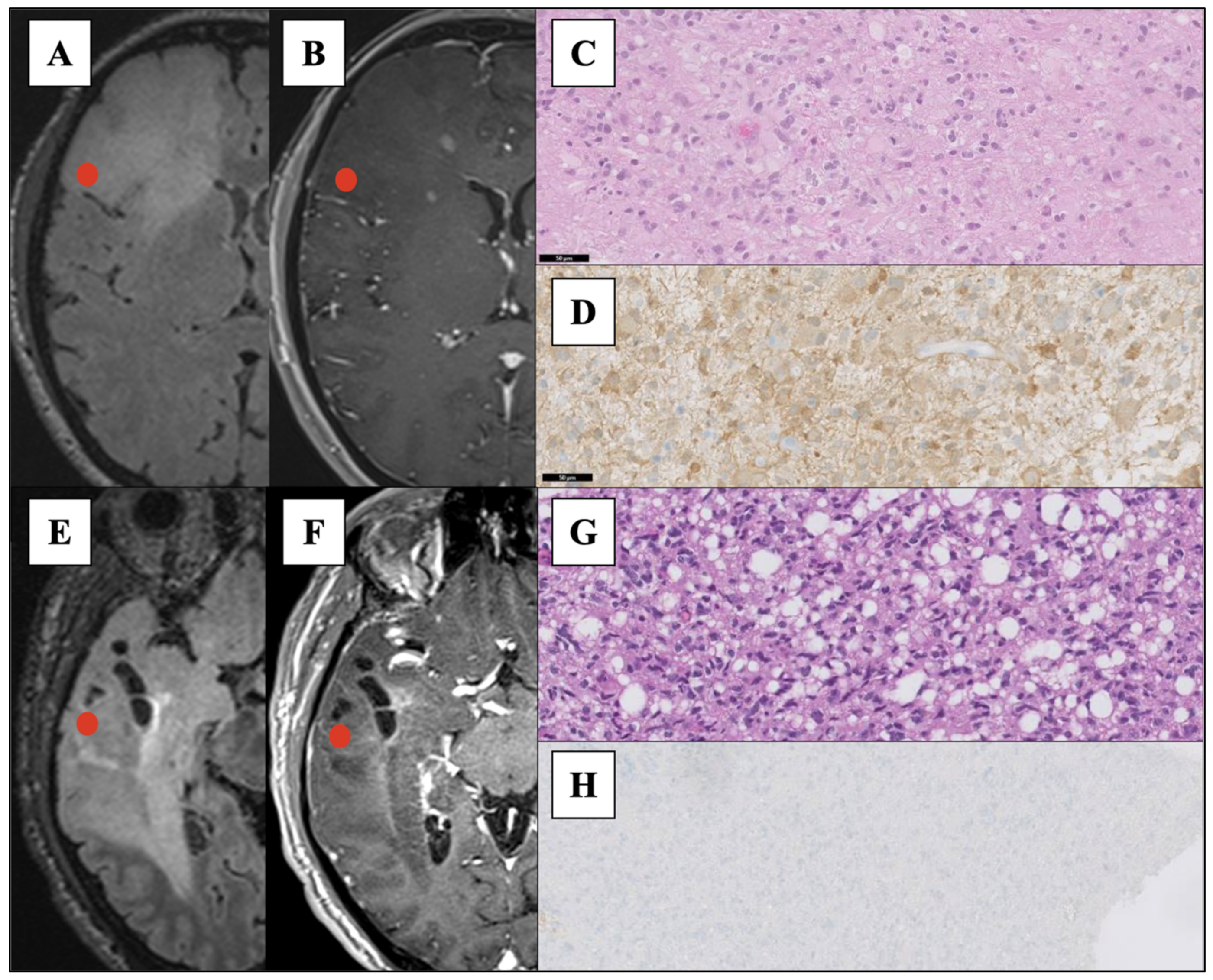
| Characteristics | IDH-MUT | IDH-WT |
|---|---|---|
| Number of samples | 21 | 17 |
| Number of total spectra acquired (mean) | 1133 (53.9) | 940 (55.3) |
| Histological classification (n° of spectra): | ||
| -Astrocytoma, WHO grade II | 1 (55) | 1 (62) |
| -Oligodendroglioma, WHO grade II | 4 (220) | - |
| -Astrocytoma, WHO grade III | 3 (141) | 3 (167) |
| -Oligodendroglioma, WHO grade III | 10(538) | - |
| -Glioblastoma, WHO grade IV | 3 (179) | 13 (711) |
| Mean Age (SD) | 42.5 (13.04) | 63.23 (9.72) |
| Sex (%) | ||
| Male | 13 (65) | 10 (58.8) |
| Female | 7 (35) | 7 (41.2) |
| Median KPS before surgery (range) | 100 (70–100) | 100 (70–100) |
| Tumor location (primary lobe involved) | ||
| Frontal | 14 | 7 |
| Temporal | 3 | 6 |
| Parietal | 3 | 3 |
| Occipital | 1 | 1 |
| Raman Shifts (cm−1) | Proposed Assignments |
|---|---|
| 419 | Cholesterol |
| 421 | Cholesterol |
| 424 | Undefined |
| 430 | Cholesterol/cholesterol ester |
| 608 | Cholesterol |
| 633 | Undefined |
| 635 | Tyrosine |
| 640 | Cysteine, tyrosine |
| 700 | Cholesterol |
| 719 | Choline in the head group of sphingomyelin and phosphatidylcholine, phosphatidylethanolamine |
| 720 | DNA |
| 743 | Adenin, DNA, Heme |
| 808 | Undefined |
| 1059 | Triglycerides/fatty acids |
| 1064 | Lipids [C-O stretch and C-O-C symmetric stretch, C-C stretch of phospholipids (side chains specifically) and cholesterol] |
| 1174 | Proteins |
| 1175 | Proteins |
| 1215 | Undefined |
| 1225 | Amide III band |
| 1245 | Amide III band |
| 1250 | Amide III, proteins |
| 1255 | Lipids |
| 1265 | Amide III band |
| 1266 | Lipids |
| 1275 | Amide III |
| 1300 | Phospholipids, fatty acid, cholesterol |
| 1305 | Triglycerides/fatty acids |
| 1308 | C-N asymmetric stretching in asymmetric aromatic amines |
| 1330 | C-H deformation or CH2 bend (proteins) |
| 1337 | Lipids and proteins |
| 1342 | Nucleic acids |
| 1354 | Undefined |
| 1366 | Undefined |
| 1372 | Nucleic acids |
| 1376 | Nucleic acids, DNA |
| 1385 | Undefined |
| 1390 | Undefined |
| 1397 | CH2/CH3 deformation of lipids and proteins |
| 1401 | Protein |
| 1412 | amino acids: aspartic & glutamic acid |
| 1439 | CH2/CH3 deformation of lipids side chains, proteins, amino acids, cholesterol/cholesterol ester |
| 1440 | CH2/CH3 deformation of lipids side chains, proteins, amino acids, cholesterol/cholesterol ester |
| 1441 | Lipids and Proteins |
| 1445 | Lipid |
| 1454 | Heme groups |
| 1495 | Undefined |
| 1502 | Undefined |
| 1522 | Proteins |
| 1532 | Carotenoid |
| 1554 | Tryptophan |
| 1705 | Undefined |
| 1740 | Lipids |
| Performance Metrics | RBF-SVM | XGB |
|---|---|---|
| Accuracy | 0.87 | 0.85 |
| Precision | 0.87 | 0.85 |
| Recall | 0.87 | 0.85 |
| F1-score | 0.87 | 0.85 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sciortino, T.; Secoli, R.; d’Amico, E.; Moccia, S.; Conti Nibali, M.; Gay, L.; Rossi, M.; Pecco, N.; Castellano, A.; De Momi, E.; et al. Raman Spectroscopy and Machine Learning for IDH Genotyping of Unprocessed Glioma Biopsies. Cancers 2021, 13, 4196. https://doi.org/10.3390/cancers13164196
Sciortino T, Secoli R, d’Amico E, Moccia S, Conti Nibali M, Gay L, Rossi M, Pecco N, Castellano A, De Momi E, et al. Raman Spectroscopy and Machine Learning for IDH Genotyping of Unprocessed Glioma Biopsies. Cancers. 2021; 13(16):4196. https://doi.org/10.3390/cancers13164196
Chicago/Turabian StyleSciortino, Tommaso, Riccardo Secoli, Ester d’Amico, Sara Moccia, Marco Conti Nibali, Lorenzo Gay, Marco Rossi, Nicolò Pecco, Antonella Castellano, Elena De Momi, and et al. 2021. "Raman Spectroscopy and Machine Learning for IDH Genotyping of Unprocessed Glioma Biopsies" Cancers 13, no. 16: 4196. https://doi.org/10.3390/cancers13164196
APA StyleSciortino, T., Secoli, R., d’Amico, E., Moccia, S., Conti Nibali, M., Gay, L., Rossi, M., Pecco, N., Castellano, A., De Momi, E., Fernandes, B., Riva, M., & Bello, L. (2021). Raman Spectroscopy and Machine Learning for IDH Genotyping of Unprocessed Glioma Biopsies. Cancers, 13(16), 4196. https://doi.org/10.3390/cancers13164196








