Silibinin Suppresses Tumor Cell-Intrinsic Resistance to Nintedanib and Enhances Its Clinical Activity in Lung Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
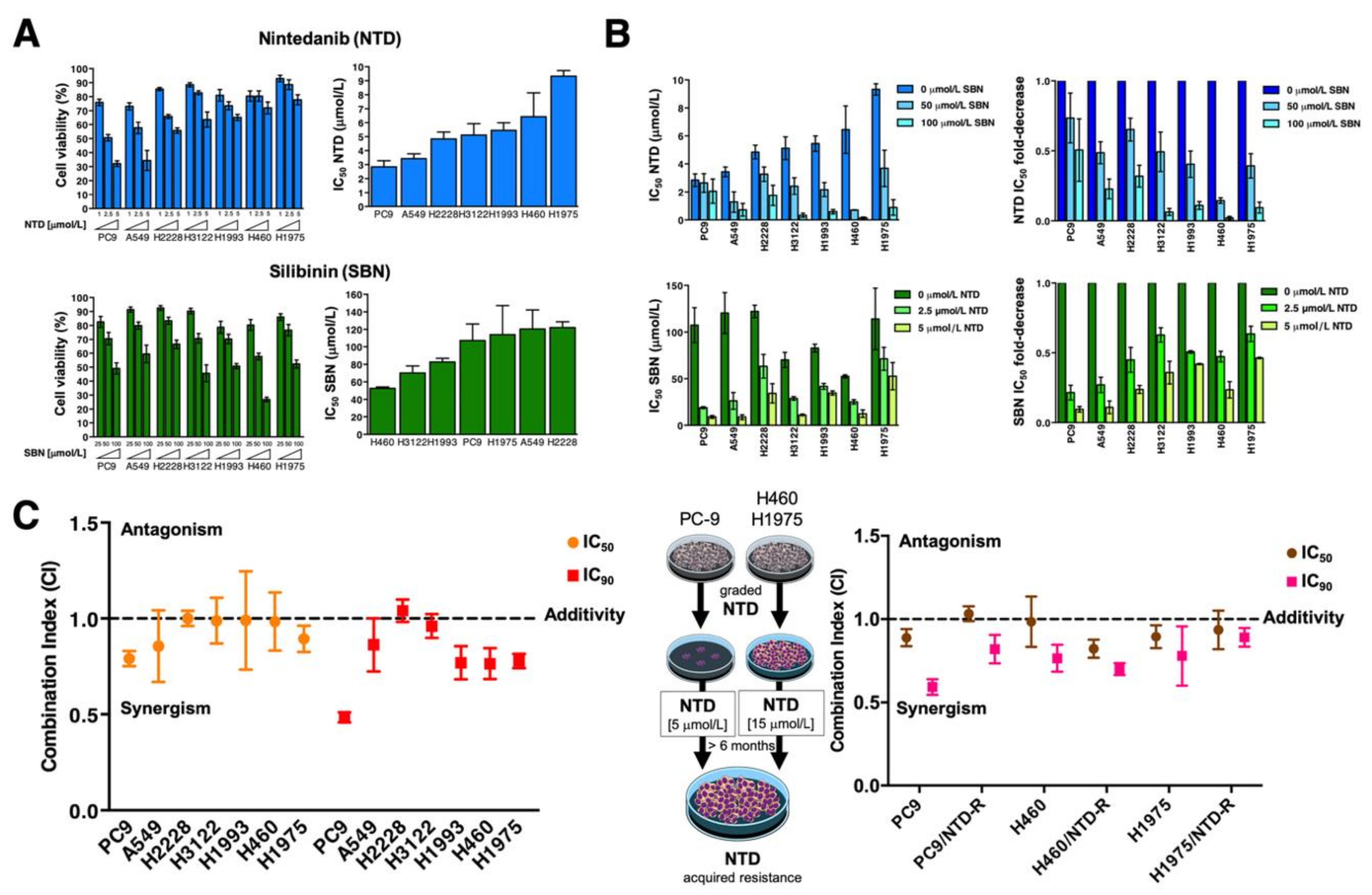
2.1. Inverse Correlation between Nintedanib and Silibinin Sensitivity in Human NSCLC Cell Lines
2.2. Synergistic Interactions between Nintedanib and Silibinin in Human NSCLC Cells
2.3. Synergistic Interactions between Silibinin and Nintedanib in NSCLC Cells with Acquired Resistance to EGFR- and ALK-Tyrosine Kinase Inhibitors
2.4. Synergistic Interactions between Nintedanib and Docetaxel in NSCLC Cells
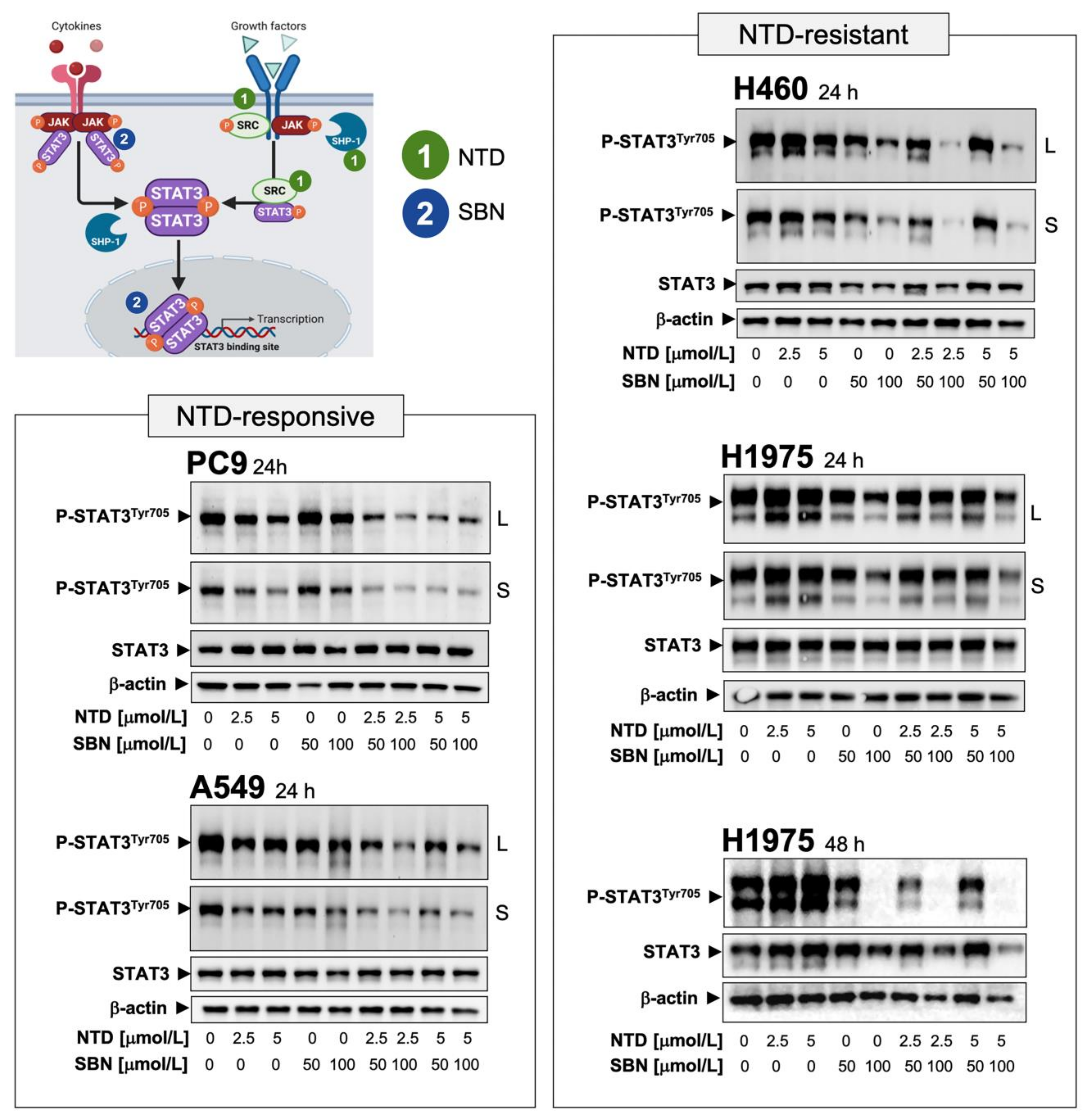
2.5. Silibinin Suppresses Nintedanib-Unresponsive STAT3 Hyperactivation in NSCLC Cells
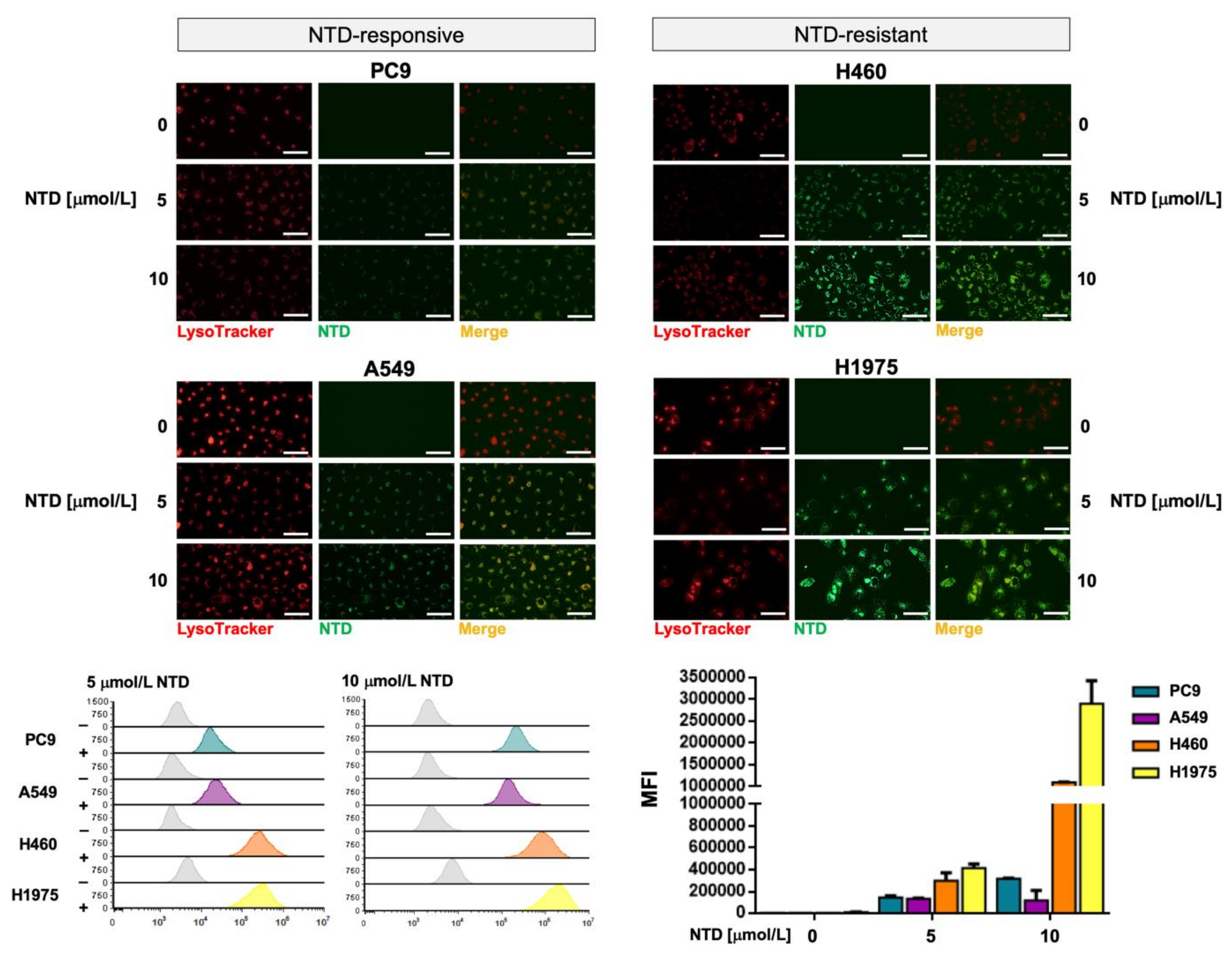
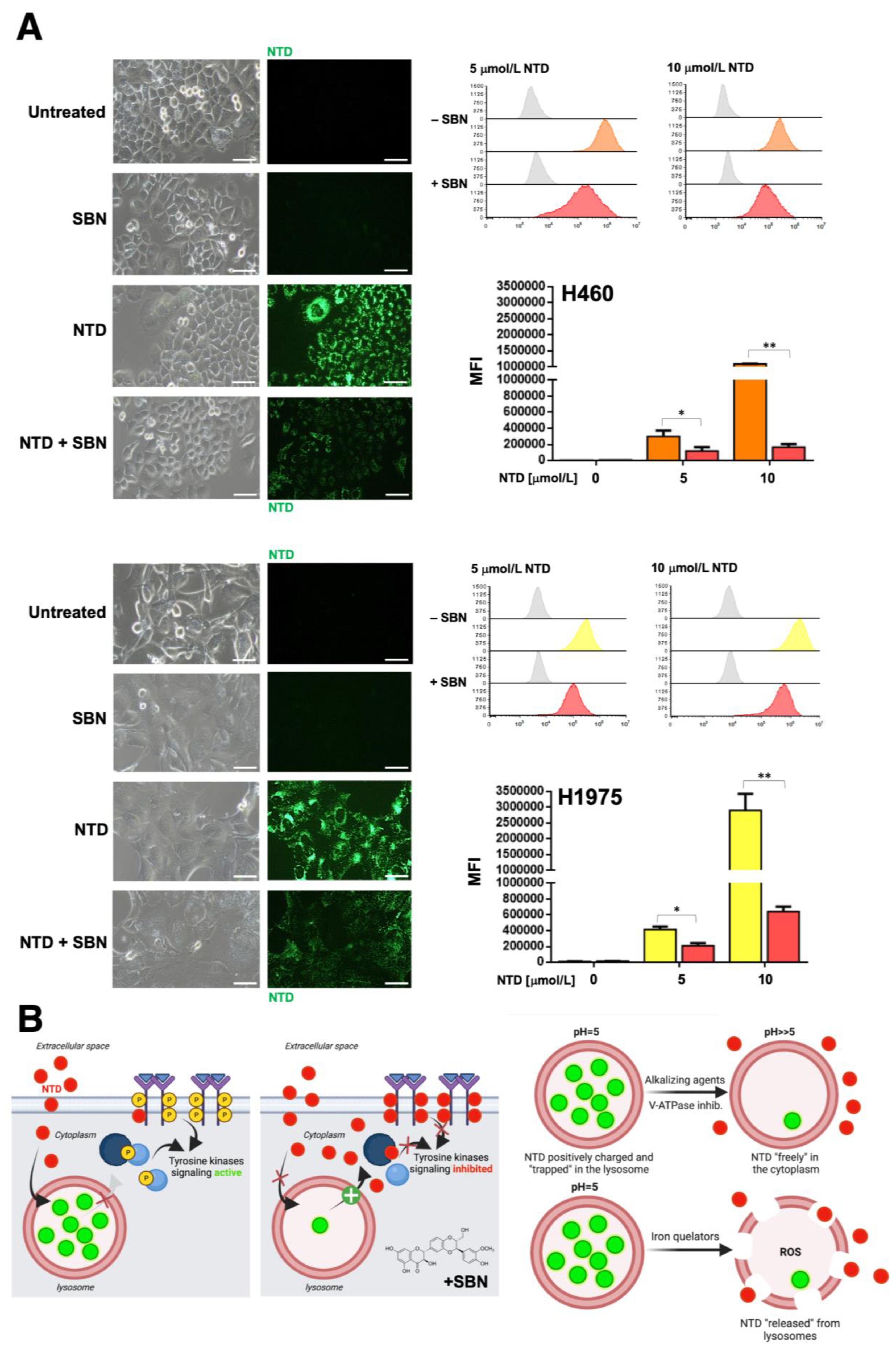
2.6. Silibinin Prevents Lysosome Sequestration of Nintedanib in NSCLC Cells
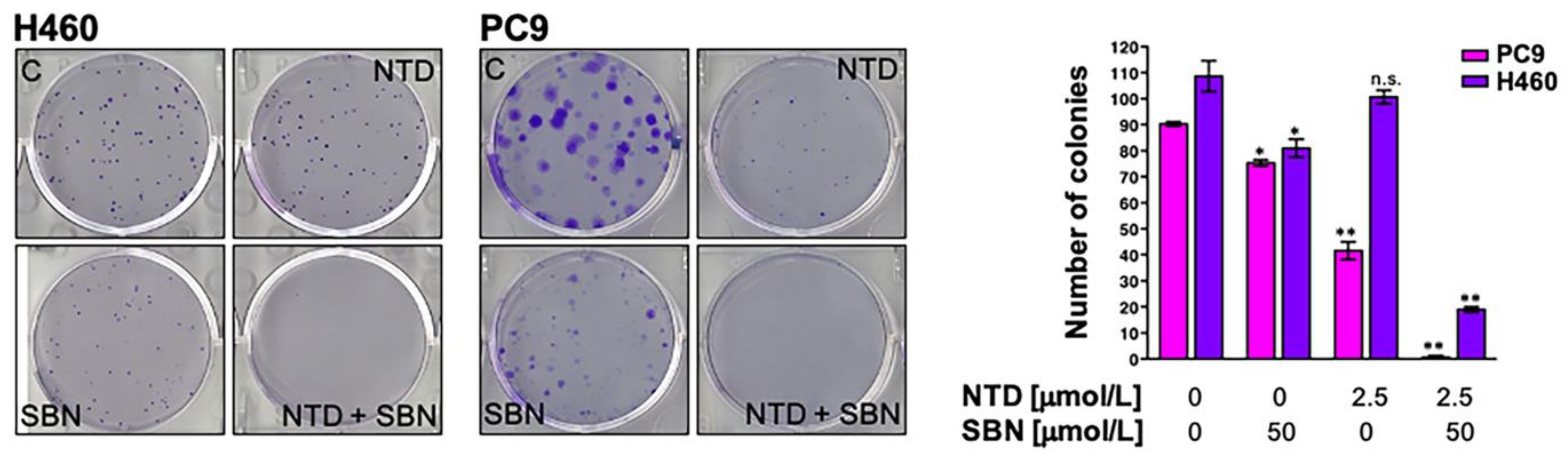
2.7. Clonogenic Assays and Real-Time Monitoring of Cell Proliferation Confirms the Synergistic Interaction between Silibinin and Nintedanib in NSCLC Cells
2.8. The Nutraceutical Use of Silibinin Enhances the Clinical Response to Nintedanib/Docetaxel in Advanced Lung Adenocarcinoma
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Reagents
4.3. MTT-Based Cell Viability Assays
4.4. Combination Index
4.5. Immunoblotting
4.6. Subcellular Fractionation
4.7. Immunofluorescence Microscopy
4.8. Nintedanib Fluorescence
4.8.1. Live Cell Fluorescence Microscopy
4.8.2. Flow Cytometry
4.9. Real-Time Cell Growth Rate
4.10. Colony Formation Assays
4.11. Patients
4.11.1. Silibinin Regimen
4.11.2. Outcomes Definitions
4.12. Statistical Analysis
4.12.1. Cell-Based Assays
4.12.2. Patients
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hilberg, F.; Roth, G.J.; Krssak, M.; Kautschitsch, S.; Sommergruber, W.; Tontsch-Grunt, U.; Garin-Chesa, P.; Bader, G.; Zoephel, A.; Quant, J.; et al. BIBF 1120: Triple Angiokinase Inhibitor with Sustained Receptor Blockade and Good Antitumor Efficacy. Cancer Res. 2008, 68, 4774–4782. [Google Scholar] [CrossRef]
- Reck, M.; Kaiser, R.; Mellemgaard, A.; Douillard, J.-Y.; Orlov, S.; Krzakowski, M.; von Pawel, J.; Gottfried, M.; Bondarenko, I.; Liao, M.; et al. Docetaxel plus nintedanib versus docetaxel plus placebo in patients with previously treated non-small-cell lung cancer (LUME-Lung 1): A phase 3, double-blind, randomised controlled trial. Lancet Oncol. 2014, 15, 143–155. [Google Scholar] [CrossRef]
- Corral, J.; Majem, M.; Rodríguez-Abreu, D.; Carcereny, E.; Cortes, Á.A.; Llorente, M.; Picazo, J.M.L.; Garcia, Y.G.; Domine, M.; Criado, M.P.L. Efficacy of nintedanib and docetaxel in patients with advanced lung adenocarcinoma treated with first-line chemotherapy and second-line immunotherapy in the nintedanib NPU program. Clin. Transl. Oncol. 2019, 21, 1270–1279. [Google Scholar] [CrossRef]
- Grohé, C.; Gleiber, W.; Haas, S.; Losem, C.; Mueller-Huesmann, H.; Schulze, M.; Franke, C.; Basara, N.; Atz, J.; Kaiser, R. Nintedanib plus docetaxel after progression on immune checkpoint inhibitor therapy: Insights from VARGADO, a prospective study in patients with lung adenocarcinoma. Future Oncol. 2019, 15, 2699–2706. [Google Scholar] [CrossRef] [PubMed]
- Navarro, P.; Bueno, M.J.; Zagorac, I.; Mondejar, T.; Sanchez, J.; Mourón, S.; Munoz, J.; López, G.G.; Jimenez-Renard, V.; Mulero, F.; et al. Targeting Tumor Mitochondrial Metabolism Overcomes Resistance to Antiangiogenics. Cell Rep. 2016, 15, 2705–2718. [Google Scholar] [CrossRef]
- Quintela-Fandino, M. Normoxic or hypoxic adaptation in response to antiangiogenic therapy: Clinical implications. Mol. Cell. Oncol. 2016, 3, e1217368. [Google Scholar] [CrossRef][Green Version]
- Bueno, M.J.; Mouron, S.; Quintela-Fandino, M. Personalising and targeting antiangiogenic resistance: A complex and multifactorial approach. Br. J. Cancer 2017, 116, 1119–1125. [Google Scholar] [CrossRef] [PubMed]
- Sonveaux, P.; Vegran, F.; Schroeder, T.; Wergin, M.C.; Verrax, J.; Rabbani, Z.N.; De Saedeleer, C.J.; Kennedy, K.M.; Diepart, C.; Jordan, B.F.; et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J. Clin. Investig. 2008, 118, 3930–3942. [Google Scholar] [CrossRef] [PubMed]
- Porporato, P.E.; Dhup, S.; Dadhich, R.K.; Copetti, T.; Sonveaux, P. Anticancer Targets in the Glycolytic Metabolism of Tumors: A Comprehensive Review. Front. Pharmacol. 2011, 2, 49. [Google Scholar] [CrossRef] [PubMed]
- Pisarsky, L.; Bill, R.; Fagiani, E.; Dimeloe, S.; Goosen, R.W.; Hagmann, J.; Hess, C.; Christofori, G. Targeting Metabolic Symbiosis to Overcome Resistance to Anti-angiogenic Therapy. Cell Rep. 2016, 15, 1161–1174. [Google Scholar] [CrossRef]
- Jiménez-Valerio, G.; Martínez-Lozano, M.; Bassani, N.; Vidal, A.; Ochoa-De-Olza, M.; Suárez, C.; García-Del-Muro, X.; Carles, J.; Viñals, F.; Graupera, M.; et al. Resistance to Antiangiogenic Therapies by Metabolic Symbiosis in Renal Cell Carcinoma PDX Models and Patients. Cell Rep. 2016, 15, 1134–1143. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.; Miéville, P.; Warren, C.M.; Saghafinia, S.; Li, L.; Peng, M.-W.; Hanahan, D. Metabolic Symbiosis Enables Adaptive Resistance to Anti-angiogenic Therapy that Is Dependent on mTOR Signaling. Cell Rep. 2016, 15, 1144–1160. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Valerio, G.; Casanovas, O. Angiogenesis and Metabolism: Entwined for Therapy Resistance. Trends Cancer 2017, 3, 10–18. [Google Scholar] [CrossRef]
- Jiménez-Valerio, G.; Casanovas, O. Antiangiogenic resistance via metabolic symbiosis. Mol. Cell. Oncol. 2016, 3, e1211979. [Google Scholar] [CrossRef][Green Version]
- Quintela-Fandino, M.; Lluch, A.; Manso, L.; Calvo, I.; Cortes, J.; García-Saenz, J.A.; Gil-Gil, M.; Martinez-Jánez, N.; Gonzalez-Martin, A.; Adrover, E.; et al. (18)F-fluoromisonidazole PET and Activity of Neoadjuvant Nintedanib in Early HER2-Negative Breast Cancer: A Window-of-Opportunity Randomized Trial. Clin. Cancer Res. 2017, 23, 1432–1441. [Google Scholar] [CrossRef] [PubMed]
- Richeldi, L.; Costabel, U.; Selman, M.; Kim, D.S.; Hansell, D.M.; Nicholson, A.G.; Brown, K.K.; Flaherty, K.R.; Noble, P.W.; Raghu, G.; et al. Efficacy of a Tyrosine Kinase Inhibitor in Idiopathic Pulmonary Fibrosis. N. Engl. J. Med. 2011, 365, 1079–1087. [Google Scholar] [CrossRef]
- Hostettler, K.E.; Zhong, J.; Papakonstantinou, E.; Karakiulakis, G.; Tamm, M.; Seidel, P.; Sun, Q.; Mandal, J.; Lardinois, D.; Lambers, C.; et al. Anti-fibrotic effects of nintedanib in lung fibroblasts derived from patients with idiopathic pulmonary fibrosis. Respir. Res. 2014, 15, 1–9. [Google Scholar] [CrossRef]
- Atanelishvili, I.; Akter, T.; Noguchi, A.; Vuyiv, O.; Wollin, L.; Silver, R.M.; Bogatkevich, G.S. Antifibrotic efficacy of nintedanib in a cellular model of systemic sclerosis-associated interstitial lung disease. Clin. Exp. Rheumatol. 2019, 37 (Suppl. S119), 115–124. [Google Scholar]
- Li, X.; Liu, X.; Deng, R.; Gao, S.; Yu, H.; Huang, K.; Jiang, Q.; Liu, R.; Li, X.; Zhang, L.; et al. Nintedanib Inhibits Wnt3a-Induced Myofibroblast Activation by Suppressing the Src/β-Catenin Pathway. Front. Pharmacol. 2020, 11, 310. [Google Scholar] [CrossRef]
- Glass, D.S.; Grossfeld, D.; Renna, H.A.; Agarwala, P.; Spiegler, P.; Kasselman, L.J.; Glass, A.D.; DeLeon, J.; Reiss, A.B. Idiopathic pulmonary fibrosis: Molecular mechanisms and potential treatment approaches. Respir. Investig. 2020, 58, 320–335. [Google Scholar] [CrossRef]
- Awasthi, N.; Hinz, S.; Brekken, R.A.; Schwarz, M.A.; Schwarz, R.E. Nintedanib, a triple angiokinase inhibitor, enhances cytotoxic therapy response in pancreatic cancer. Cancer Lett. 2015, 358, 59–66. [Google Scholar] [CrossRef]
- Hibi, M.; Kaneda, H.; Tanizaki, J.; Sakai, K.; Togashi, Y.; Terashima, M.; De Velasco, M.; Fujita, Y.; Banno, E.; Nakamura, Y.; et al. FGFR gene alterations in lung squamous cell carcinoma are potential targets for the multikinase inhibitor nintedanib. Cancer Sci. 2016, 107, 1667–1676. [Google Scholar] [CrossRef]
- Hilberg, F.; Tontsch-Grunt, U.; Baum, A.; Le, A.T.; Doebele, R.C.; Lieb, S.; Gianni, D.; Voss, T.; Garin-Chesa, P.; Haslinger, C.; et al. Triple Angiokinase Inhibitor Nintedanib Directly Inhibits Tumor Cell Growth and Induces Tumor Shrinkage via Blocking Oncogenic Receptor Tyrosine Kinases. J. Pharmacol. Exp. Ther. 2018, 364, 494–503. [Google Scholar] [CrossRef] [PubMed]
- Tai, W.-T.; Shiau, C.-W.; Li, Y.-S.; Chang, C.-W.; Huang, J.-W.; Hsueh, T.-T.; Yu, H.-C.; Chen, K.-F. Nintedanib (BIBF-1120) inhibits hepatocellular carcinoma growth independent of angiokinase activity. J. Hepatol. 2014, 61, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Vignais, M.L.; Sadowski, H.B.; Watling, D.; Rogers, N.; Gilman, M. Platelet-derived growth factor induces phosphorylation of multiple JAK family kinases and STAT proteins. Mol. Cell. Biol. 1996, 16, 1759–1769. [Google Scholar] [CrossRef] [PubMed]
- Vignais, M.L.; Gilman, M. Distinct Mechanisms of Activation of Stat1 and Stat3 by Platelet-Derived Growth Factor Receptor in a Cell-Free System. Mol. Cell. Biol. 1999, 19, 3727–3735. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.-F.; Huang, W.-J.; Zhao, J.-F.; Fu, H.-Y.; Zhang, G.-Y.; Huang, X.-J.; Lv, B.-D. The platelet-derived growth factor receptor/STAT3 signaling pathway regulates the phenotypic transition of corpus cavernosum smooth muscle in rats. PLoS ONE 2017, 12, e0172191. [Google Scholar] [CrossRef]
- Bohrer, L.R.; Chuntova, P.; Bade, L.K.; Beadnell, T.C.; Leon, R.P.; Brady, N.; Ryu, Y.; Goldberg, J.E.; Schmechel, S.C.; Koopmeiners, J.S.; et al. Activation of the FGFR—STAT3 Pathway in Breast Cancer Cells Induces a Hyaluronan-Rich Microenvironment That Licenses Tumor Formation. Cancer Res. 2014, 74, 374–386. [Google Scholar] [CrossRef]
- Li, P.; Huang, T.; Zou, Q.; Liu, D.; Wang, Y.; Tan, X.; Wei, Y.; Qiu, H. FGFR2 Promotes Expression of PD-L1 in Colorectal Cancer via the JAK/STAT3 Signaling Pathway. J. Immunol. 2019, 202, 3065–3075. [Google Scholar] [CrossRef]
- Garcia, R.; Bowman, T.L.; Niu, G.; Yu, H.; Minton, S.; A Muro-Cacho, C.; E Cox, C.; Falcone, R.; Fairclough, R.; Parsons, S.; et al. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene 2001, 20, 2499–2513. [Google Scholar] [CrossRef]
- Byers, L.A.; Sen, B.; Saigal, B.; Diao, L.; Wang, J.; Nanjundan, M.; Cascone, T.; Mills, G.B.; Heymach, J.V.; Johnson, F.M. Reciprocal Regulation of c-Src and STAT3 in Non-Small Cell Lung Cancer. Clin. Cancer Res. 2009, 15, 6852–6861. [Google Scholar] [CrossRef] [PubMed]
- Demaria, M.; Giorgi, C.; Lebiedzińska-Arciszewska, M.; Esposito, G.; D’Angeli, L.; Bartoli, A.; Gough, D.; Turkson, J.; Levy, D.; Watson, C.J.; et al. A STAT3-mediated metabolic switch is involved in tumour transformation and STAT3 addiction. Aging 2010, 2, 823–842. [Google Scholar] [CrossRef] [PubMed]
- Camporeale, A.; Demaria, M.; Monteleone, E.; Giorgi, C.; Wieckowski, M.R.; Pinton, P.; Poli, V. STAT3 Activities and Energy Metabolism: Dangerous Liaisons. Cancers 2014, 6, 1579–1596. [Google Scholar] [CrossRef]
- Demaria, M.; Camporeale, A.; Poli, V. STAT3 and metabolism: How many ways to use a single molecule? Int. J. Cancer 2014, 135, 1997–2003. [Google Scholar] [CrossRef]
- Poli, V.; Camporeale, A. STAT3-Mediated Metabolic Reprograming in Cellular Transformation and Implications for Drug Resistance. Front. Oncol. 2015, 5, 121. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Hirpara, J.L.; Eu, J.-Q.; Sethi, G.; Wang, L.; Goh, B.-C.; Wong, A.L. Targeting STAT3 and oxidative phosphorylation in oncogene-addicted tumors. Redox Biol. 2019, 25, 101073. [Google Scholar] [CrossRef]
- Furtek, S.L.; Backos, D.; Matheson, C.J.; Reigan, P. Strategies and Approaches of Targeting STAT3 for Cancer Treatment. ACS Chem. Biol. 2016, 11, 308–318. [Google Scholar] [CrossRef]
- Siveen, K.S.; Sikka, S.; Surana, R.; Dai, X.; Zhang, J.; Kumar, A.P.; Tan, B.K.; Sethi, G.; Bishayee, A. Targeting the STAT3 signaling pathway in cancer: Role of synthetic and natural inhibitors. Biochim. Biophys. Acta (BBA) Bioenerg. 2014, 1845, 136–154. [Google Scholar] [CrossRef]
- Zhao, M.; Jiang, B.; Gao, F.-H. Small molecule inhibitors of STAT3 for cancer therapy. Curr. Med. Chem. 2011, 18, 4012–4018. [Google Scholar] [CrossRef]
- Masciocchi, D.; Gelain, A.; Villa, S.; Meneghetti, F.; Barlocco, D. Signal transducer and activator of transcription 3 (STAT3): A promising target for anticancer therapy. Futur. Med. Chem. 2011, 3, 567–597. [Google Scholar] [CrossRef]
- Shih, P.-C. Revisiting the development of small molecular inhibitors that directly target the signal transducer and activator of transcription 3 (STAT3) domains. Life Sci. 2020, 242, 117241. [Google Scholar] [CrossRef] [PubMed]
- Gelain, A.; Mori, M.; Meneghetti, F.; Villa, S. Signal Transducer and Activator of Transcription Protein 3 (STAT3): An Update on its Direct Inhibitors as Promising Anticancer Agents. Curr. Med. Chem. 2019, 26, 5165–5206. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.-C.; Graf, T.N.; Sparacino, C.M.; Wani, M.C.; Wall, M.E. Complete isolation and characterization of silybins and isosilybins from milk thistle (Silybum marianum). Org. Biomol. Chem. 2003, 1, 1684–1689. [Google Scholar] [CrossRef]
- Gazák, R.; Walterova, D.; Kren, V. Silybin and Silymarin—New and Emerging Applications in Medicine. Curr. Med. Chem. 2007, 14, 315–338. [Google Scholar] [CrossRef]
- Bijak, M. Silybin, a Major Bioactive Component of Milk Thistle (Silybum marianum L. Gaernt.)—Chemistry, Bioavailability, and Metabolism. Molecules 2017, 22, 1942. [Google Scholar] [CrossRef]
- Abenavoli, L.; Izzo, A.A.; Milić, N.; Cicala, C.; Santini, A.; Capasso, R. Milk thistle (Silybum marianum): A concise overview on its chemistry, pharmacological, and nutraceutical uses in liver diseases. Phytother. Res. 2018, 32, 2202–2213. [Google Scholar] [CrossRef]
- Verdura, S.; Cuyàs, E.; Llorach-Pares, L.; Pérez-Sánchez, A.; Micol, V.; Nonell-Canals, A.; Joven, J.; Valiente, M.; Sanchez-Martinez, M.; Bosch-Barrera, J.; et al. Silibinin is a direct inhibitor of STAT3. Food Chem. Toxicol. 2018, 116, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Sánchez, A.; Cuyàs, E.; Ruiz-Torres, V.; Agulló-Chazarra, L.; Verdura, S.; González-Álvarez, I.; Bermejo, M.; Joven, J.; Micol, V.; Bosch-Barrera, J.; et al. Intestinal Permeability Study of Clinically Relevant Formulations of Silibinin in Caco-2 Cell Monolayers. Int. J. Mol. Sci. 2019, 20, 1606. [Google Scholar] [CrossRef]
- Bosch-Barrera, J.; Sais, E.; Cañete, N.; Marruecos, J.; Cuyàs, E.; Izquierdo, A.; Porta, R.; Haro, M.; Brunet, J.; Pedraza, S.; et al. Response of brain metastasis from lung cancer patients to an oral nutraceutical product containing silibinin. Oncotarget 2016, 7, 32006–32014. [Google Scholar] [CrossRef]
- Priego, N.; Zhu, L.; Monteiro, C.; Mulders, M.; Wasilewski, D.; Bindeman, W.; Doglio, L.; Martínez, L.; Martínez-Saez, E.; Ramón YCajal, S.; et al. STAT3 labels a subpopulation of reactive astrocytes required for brain metastasis. Nat. Med. 2018, 24, 1024–1035. [Google Scholar] [CrossRef]
- Bosch-Barrera, J.; Menendez, J. Silibinin and STAT3: A natural way of targeting transcription factors for cancer therapy. Cancer Treat. Rev. 2015, 41, 540–546. [Google Scholar] [CrossRef]
- Bosch-Barrera, J.; Queralt, B.; Menendez, J. Targeting STAT3 with silibinin to improve cancer therapeutics. Cancer Treat. Rev. 2017, 58, 61–69. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Lucena-Araujo, A.R.; Nakayama, S.; Pontes, L.L.D.F.; Gonzalez, D.A.; Yasuda, H.; Kobayashi, S.; Costa, D.B. Dual ALK and EGFR inhibition targets a mechanism of acquired resistance to the tyrosine kinase inhibitor crizotinib in ALK rearranged lung cancer. Lung Cancer 2014, 83, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.R.; Kim, W.S.; Choi, Y.J.; Choi, C.M.; Rho, J.K.; Lee, J.C. Epithelial-mesenchymal transition leads to crizotinib resistance in H2228 lung cancer cells with EML4-ALK translocation. Mol. Oncol. 2013, 7, 1093–1102. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Martin, A.; Cufí, S.; Oliveras-Ferraros, C.; Torres-Garcia, V.Z.; Corominas-Faja, B.; Cuyàs, E.; Bonavia, R.; Visa, J.; Martín-Castillo, B.; Barrajón-Catalán, E.; et al. IGF-1R/epithelial-to-mesenchymal transition (EMT) crosstalk suppresses the erlotinib-sensitizing effect of EGFR exon 19 deletion mutations. Sci. Rep. 2013, 3, srep02560. [Google Scholar] [CrossRef] [PubMed]
- Corominas-Faja, B.; Oliveras-Ferraros, C.; Cuyàs, E.; Segura-Carretero, A.; Joven, J.; Martin-Castillo, B.; Barrajón-Catalán, E.; Micol, V.; Bosch-Barrera, J.; Menendez, J.A. Stem cell-like ALDH(bright) cellular states in EGFR-mutant non-small cell lung cancer: A novel mechanism of acquired resistance to erlotinib targetable with the natural polyphenol silibinin. Cell Cycle 2013, 12, 3390–3404. [Google Scholar] [CrossRef]
- Chou, T.C. Drug Combination Studies and Their Synergy Quantification Using the Chou-Talalay Method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef]
- Chou, T.-C. Theoretical Basis, Experimental Design, and Computerized Simulation of Synergism and Antagonism in Drug Combination Studies. Pharmacol. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef]
- Englinger, B.; Kallus, S.; Senkiv, J.; Heilos, D.; Gabler, L.; Van Schoonhoven, S.; Terenzi, A.; Moser, P.; Pirker, C.; Timelthaler, G.; et al. Intrinsic fluorescence of the clinically approved multikinase inhibitor nintedanib reveals lysosomal sequestration as resistance mechanism in FGFR-driven lung cancer. J. Exp. Clin. Cancer Res. 2017, 36, 122. [Google Scholar] [CrossRef]
- Geisslinger, F.; Müller, M.; Vollmar, A.M.; Bartel, K. Targeting Lysosomes in Cancer as Promising Strategy to Overcome Chemoresistance—A Mini Review. Front. Oncol. 2020, 10, 1156. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J.; Zhuang, G.; Cao, Y.; Du, P.; Kim, H.-J.; Settleman, J. Drug Resistance via Feedback Activation of Stat3 in Oncogene-Addicted Cancer Cells. Cancer Cell 2014, 26, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, S.; Cormerais, Y.; Dufies, M.; Grépin, R.; Colosetti, P.; Belaid, A.; Parola, J.; Martin, A.; Lacas-Gervais, S.; Mazure, N.M.; et al. Resistance to sunitinib in renal clear cell carcinoma results from sequestration in lysosomes and inhibition of the autophagic flux. Autophagy 2015, 11, 1891–1904. [Google Scholar] [CrossRef] [PubMed]
- Llanos, S.; Megias, D.; Blanco-Aparicio, C.; Hernández-Encinas, E.; Rovira, M.; Pietrocola, F.; Serrano, M. Lysosomal trapping of palbociclib and its functional implications. Oncogene 2019, 38, 3886–3902. [Google Scholar] [CrossRef]
- Fassl, A.; Brain, C.; Abu-Remaileh, M.; Stukan, I.; Butter, D.; Stepien, P.; Feit, A.S.; Bergholz, J.; Michowski, W.; Otto, T.; et al. Increased lysosomal biomass is responsible for the resistance of triple-negative breast cancers to CDK4/6 inhibition. Sci. Adv. 2020, 6, eabb2210. [Google Scholar] [CrossRef] [PubMed]
- Halaby, R. Influence of lysosomal sequestration on multidrug resistance in cancer cells. Cancer Drug Resist. 2019, 2, 31–42. [Google Scholar] [CrossRef]
- Hraběta, J.; Belhajová, M.; Šubrtová, H.; Rodrigo, M.A.M.; Heger, Z.; Eckschlager, T. Drug Sequestration in Lysosomes as One of the Mechanisms of Chemoresistance of Cancer Cells and the Possibilities of Its Inhibition. Int. J. Mol. Sci. 2020, 21, 4392. [Google Scholar] [CrossRef]
- Simon, T.; Coquerel, B.; Petit, A.; Kassim, Y.; Demange, E.; Le Cerf, D.; Perrot, V.; Vannier, J.-P. Direct Effect of Bevacizumab on Glioblastoma Cell Lines In Vitro. NeuroMolecular Med. 2014, 16, 752–771. [Google Scholar] [CrossRef]
- Simon, T.; Gagliano, T.; Giamas, G. Direct Effects of Anti-Angiogenic Therapies on Tumor Cells: VEGF Signaling. Trends Mol. Med. 2017, 23, 282–292. [Google Scholar] [CrossRef]
- Tomida, C.; Yamagishi, N.; Nagano, H.; Uchida, T.; Ohno, A.; Hirasaka, K.; Nikawa, T.; Teshima-Kondo, S. VEGF pathway-targeting drugs induce evasive adaptation by activation of neuropilin-1/cMet in colon cancer cells. Int. J. Oncol. 2018, 52, 1350–1362. [Google Scholar] [CrossRef]
- Dudka, A.A.; Sweet, S.M.M.; Heath, J.K. Signal Transducers and Activators of Transcription-3 Binding to the Fibroblast Growth Factor Receptor Is Activated by Receptor Amplification. Cancer Res. 2010, 70, 3391–3401. [Google Scholar] [CrossRef]
- Zhitomirsky, B.; Assaraf, Y.G. Lysosomes as mediators of drug resistance in cancer. Drug Resist. Updates 2016, 24, 23–33. [Google Scholar] [CrossRef]
- Zhitomirsky, B.; Assaraf, Y.G. Lysosomal accumulation of anticancer drugs triggers lysosomal exocytosis. Oncotarget 2017, 8, 45117–45132. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, M.; Ghaffari, S.H.; Zekri, A.; Hassani, S.; Alimoghaddam, K.; Ghavamzadeh, A. Silibinin induces apoptosis and inhibits proliferation of estrogen receptor (ER)-negative breast carcinoma cells through suppression of nuclear factor kappa B activation. Arch. Iran. Med. 2014, 17, 366–371. [Google Scholar]
- Borsari, M.; Gabbi, C.; Ghelfi, F.; Grandi, R.; Saladini, M.; Severi, S.; Borella, F. Silybin, a new iron-chelating agent. J. Inorg. Biochem. 2001, 85, 123–129. [Google Scholar] [CrossRef]
- Kovacevic, Z.; Kalinowski, D.S.; Lovejoy, D.B.; Yu, Y.; Rahmanto, Y.S.; Sharpe, P.C.; Bernhardt, P.V.; Richardson, D.R. The medicinal chemistry of novel iron chelators for the treatment of cancer. Curr. Top. Med. Chem. 2011, 11, 483–499. [Google Scholar] [CrossRef] [PubMed]
- Gharagozloo, M.; Khoshdel, Z.; Amirghofran, Z. The effect of an iron (III) chelator, silybin, on the proliferation and cell cycle of Jurkat cells: A comparison with desferrioxamine. Eur. J. Pharmacol. 2008, 589, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, C.; Bomford, A.; A Geissler, C. The iron-chelating potential of silybin in patients with hereditary haemochromatosis. Eur. J. Clin. Nutr. 2010, 64, 1239–1241. [Google Scholar] [CrossRef] [PubMed]
- Moayedi Esfahani, B.A.; Reisi, N.; Mirmoghtadaei, M. Evaluating the safety and efficacy of silymarin in β-thalassemia patients: A review. Hemoglobin 2015, 39, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Ravikumar Reddy, D.; Khurana, A.; Bale, S.; Ravirala, R.; Samba Siva Reddy, V.; Mohankumar, M.; Godugu, C. Natural flavonoids silymarin and quercetin improve the brain distribution of co-administered P-gp substrate drugs. SpringerPlus 2016, 5, 1618. [Google Scholar] [CrossRef]
- Abdallah, H.; Al-Abd, A.; El-Dine, R.S.; El-Halawany, A.M. P-glycoprotein inhibitors of natural origin as potential tumor chemo-sensitizers: A review. J. Adv. Res. 2015, 6, 45–62. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Lewis, B.; Krueger, C.C.; Sargeant, T.; D’Angelo, M.; Deery, M.J.; Feret, R.; Howard, J.A.; Lilley, K.S.; Watson, C.J. Stat3-mediated alterations in lysosomal membrane protein composition. J. Biol. Chem. 2018, 293, 4244–4261. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Palmfeldt, J.; Lin, L.; Colaco, A.; Clemmensen, K.K.B.; Huang, J.; Xu, F.; Liu, X.; Maeda, K.; Luo, Y.; et al. STAT3 associates with vacuolar H+-ATPase and regulates cytosolic and lysosomal pH. Cell Res. 2018, 28, 996–1012. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.L.; Hirpara, J.L.; Pervaiz, S.; Eu, J.-Q.; Sethi, G.; Goh, B.-C. Do STAT3 inhibitors have potential in the future for cancer therapy? Expert Opin. Investig. Drugs 2017, 26, 883–887. [Google Scholar] [CrossRef]
- Chai, E.Z.P.; Shanmugam, M.K.; Arfuso, F.; Dharmarajan, A.; Wang, C.; Kumar, A.P.; Samy, R.P.; Lim, L.; Wang, L.; Goh, B.C.; et al. Targeting transcription factor STAT3 for cancer prevention and therapy. Pharmacol. Ther. 2016, 162, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Syrigos, L.; Miliasukas, S.; Zöchbauer-Müller, S.; Buchner, H.; Kitzing, T.; Kerr, K.M. 80P- Nintedanib (N) + docetaxel (D) after immunotherapy in adenocarcinoma non-small cell lung cancer (NSCLC): First results from the non-interventional LUME-BioNIS study. Ann. Oncol. 2019, 30, xi28–xi29. [Google Scholar] [CrossRef]


| Cell Line | Mutational Features |
|---|---|
| A549 | EGFR WT; KRAS mutant (G12S) |
| H460 | EGFR WT; KRAS mutant (Q61H) |
| H1993 | EGFR WT; MET amplification |
| H3122 | EGFR WT; EML4-ALK variant 1 |
| H2228 | EGFR WT; EML4-ALK variant 3 |
| H1975 | EGFR mutant (L858R/T790M); KRAS WT |
| PC9 | EGFR mutant (DelE746-A750) |
| Study Cohort | Control Cohort | ||
|---|---|---|---|
| (n = 18) | (n = 41) | p-Value | |
| Sex | 0.187 | ||
| Male | 12 (67%) | 34 (83%) | |
| Female | 6 (33%) | 7 (17%) | |
| Age | 0.490 | ||
| Mean ± SD (range) | 61.2 ± 10.3 (43–79) | 59.5 ± 7.3 (45–71) | |
| Line of treatment | 1.000 | ||
| 2nd line | 14 (78%) | 32 (78%) | |
| ≥3rd line | 4 (22%) | 9 (22%) | |
| TTF 1st line (months) | 0.439 | ||
| Median (p25,p75) | 4.7 (4.1, 6.5) | 4.4 (1.9, 7.6) | |
| Response to 1st line | 0.188 | ||
| Response (PR) | 5 (27.8%) | 13 (31.7%) | |
| Stable disease | 9 (50.0%) | 11 (26.8%) | |
| Progression disease | 4 (22.2%) | 17 (41.5%) | |
| Initial docetaxel dose | 0.001 | ||
| 75 mg/m2 | 2 (11%) | 23 (56%) | |
| <75 mg/m2 | 16 (88.9%) | 18 (44%) | |
| ECOG PS | 0.144 | ||
| 0 | 5 (27.8%) | 3 (7.3%) | |
| 1 | 13 (72.2%) | 36 (87.8%) | |
| 2 | 0 (0%) | 2 (4.9%) | |
| EGFR status | 0.546 | ||
| EGFR-mutant | 0 (0%) | 3 (7.3%) | |
| EGFR-wild-type | 18 (100%) | 38 (92.7%) | |
| KRAS status | 0.099 | ||
| KRAS-mutant | 4 (22.2%) | 12 (29.3%) | |
| KRAS-wild-type | 4 (22.2%) | 18 (43.9%) | |
| KRAS-not evaluable | 10 (55.6%) | 11 (26.8%) | |
| PD-L1 status | 0.222 | ||
| 0% | 3 (16.7%) | 11 (26.8%) | |
| 1–49% | 1 (5.5%) | 5 (12.2%) | |
| ≥50% | 3 (16.7%) | 1 (2.4%) | |
| Not evaluable | 11 (61.1%) | 24 (58.6%) |
| Univariate Analysis | Multivariate Analysis | |||||
|---|---|---|---|---|---|---|
| HR | (95%CI) | p-Value | HR | (95%CI) | p-Value | |
| Age (years) | ||||||
| ≤65 | 1 | (referent) | 1 | (referent) | ||
| >65 | 0.60 | (0.33–1.10) | 0.096 | 0.74 | (0.39–1.37) | 0.335 |
| Sex | ||||||
| Female | 1 | (referent) | 1 | (referent) | ||
| Male | 1.06 | (0.57–1.98) | 0.847 | 0.72 | (0.47–1.70) | 0.890 |
| Docetaxel | ||||||
| 75 mg/m2 | 1 | (referent) | 1 | (referent) | ||
| <75 mg/m2 | 2.23 | (1.18–4.22) | 0.013 * | 3.30 | (1.69–6.45) | <0.001 * |
| Legasil® | ||||||
| No | 1 | (referent) | 1 | (referent) | ||
| Yes | 0.74 | (0.42–1.31) | 0.302 | 0.43 | (0.22–0.84) | 0.013 * |
| Univariate Analysis | Multivariate Analysis | |||||
|---|---|---|---|---|---|---|
| HR | (95% CI) | p-Value | HR | (95% CI) | p-Value | |
| Age (years) | ||||||
| ≤65 | 1 | (referent) | 1 | (referent) | ||
| >65 | 1.05 | (0.56–1.95) | 0.886 | 1.07 | (0.58–2.00) | 0.823 |
| Sex | ||||||
| Female | 1 | (referent) | 1 | (referent) | ||
| Male | 1.26 | (0.65–2.45) | 0.503 | 1.19 | (0.60–2.32) | 0.621 |
| Docetaxel | ||||||
| 75 mg/m2 | 1 | (referent) | 1 | (referent) | ||
| <75 mg/m2 | 1.80 | (1.02–3.15) | 0.041 * | 2.22 | (1.18–4.17) | 0.014 * |
| Legasil® | ||||||
| No | 1 | (referent) | 1 | (referent) | ||
| Yes | 0.95 | (0.52–1.75) | 0.878 | 0.63 | (0.32–1.25) | 0.190 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bosch-Barrera, J.; Verdura, S.; Ruffinelli, J.C.; Carcereny, E.; Sais, E.; Cuyàs, E.; Palmero, R.; Lopez-Bonet, E.; Hernández-Martínez, A.; Oliveras, G.; et al. Silibinin Suppresses Tumor Cell-Intrinsic Resistance to Nintedanib and Enhances Its Clinical Activity in Lung Cancer. Cancers 2021, 13, 4168. https://doi.org/10.3390/cancers13164168
Bosch-Barrera J, Verdura S, Ruffinelli JC, Carcereny E, Sais E, Cuyàs E, Palmero R, Lopez-Bonet E, Hernández-Martínez A, Oliveras G, et al. Silibinin Suppresses Tumor Cell-Intrinsic Resistance to Nintedanib and Enhances Its Clinical Activity in Lung Cancer. Cancers. 2021; 13(16):4168. https://doi.org/10.3390/cancers13164168
Chicago/Turabian StyleBosch-Barrera, Joaquim, Sara Verdura, José Carlos Ruffinelli, Enric Carcereny, Elia Sais, Elisabet Cuyàs, Ramon Palmero, Eugeni Lopez-Bonet, Alejandro Hernández-Martínez, Gloria Oliveras, and et al. 2021. "Silibinin Suppresses Tumor Cell-Intrinsic Resistance to Nintedanib and Enhances Its Clinical Activity in Lung Cancer" Cancers 13, no. 16: 4168. https://doi.org/10.3390/cancers13164168
APA StyleBosch-Barrera, J., Verdura, S., Ruffinelli, J. C., Carcereny, E., Sais, E., Cuyàs, E., Palmero, R., Lopez-Bonet, E., Hernández-Martínez, A., Oliveras, G., Buxó, M., Izquierdo, A., Morán, T., Nadal, E., & Menendez, J. A. (2021). Silibinin Suppresses Tumor Cell-Intrinsic Resistance to Nintedanib and Enhances Its Clinical Activity in Lung Cancer. Cancers, 13(16), 4168. https://doi.org/10.3390/cancers13164168








