Simple Summary
Cancer is often characterized by profound changes in metabolism, some of which are appropriate targets for therapeutic intervention. Many cancers overexpress the two-electron reductase NQO1, which can bioactivate the drug β-lapachone, inducing a futile redox cycle that liberates large amounts of reactive oxygen species and results in subsequent cell death. However, β-lapachone has off-target toxicities in red blood cells, which makes minimal dosing for chemotherapeutic response desirable. Here, we show that magnetic resonance-based detection of [2H7]glucose metabolism provides a robust metric of NQO1 activation, as the redox perturbation causes downregulation of glycolytic flux that is detectable in the HDO and lactate signals. Imaging of either metabolic product could provide constraints for a continual reassessment model for controlling therapeutic dosing levels.
Abstract
Treatment of cancers with β-lapachone causes NAD(P)H: quinone oxidoreductase 1 (NQO1) to generate an unstable hydroquinone that regenerates itself in a futile cycle while producing reactive oxygen species (ROS) in the form of superoxide and subsequently hydrogen peroxide. Rapid accumulation of ROS damages DNA, hyperactivates poly-ADP-ribose polymerase-I, causes massive depletion of NAD+/ATP, and hampers glycolysis. Cells overexpressing NQO1 subsequently die rapidly through an NAD+-keresis mechanism. Assessing changes in glycolytic rates caused by NQO1 bioactivation would provide a means of assessing treatment efficacy, potentially lowering the chemotherapeutic dosage, and reducing off-target toxicities. NQO1-mediated changes in glycolytic flux were readily detected in A549 (lung), MiaPaCa2 (pancreatic), and HCT-116 (colon) cancer cell lines by 2H-NMR after administration of [2H7]glucose. The deuterated metabolic products 2H-lactate and HDO were quantified, and linear relationships with glucose consumption for both products were observed. The higher concentration of HDO compared to 2H-lactate allows for more sensitive measurement of the glycolytic flux in cancer. Gas chromatography-mass spectrometry analysis agreed with the NMR results and confirmed downregulated energy metabolism in NQO1+ cells after β-lapachone treatment. The demonstrated method is ideal for measuring glycolytic rates, the effects of chemotherapeutics that target glycolysis, and has the potential for in vivo translation.
1. Introduction
Metabolic reprogramming is one of the key hallmarks of cancer cell metabolism [1,2]. Multiple cancers display an increased glucose uptake under normoxic conditions and favor lactate production as opposed to aerobic respiration (i.e., the Warburg effect) [3]. These metabolic defects have been studied extensively as a way to differentiate cancer cells from healthy cells, both for diagnosis and treatment purposes [4,5]. Among many therapeutic targets, NAD(P)H: quinone oxidoreductase 1 (NQO1) is an enzyme involved in cellular detoxifying reactions which has been recognized as a potential target for cancer treatment [6,7,8]. β-lapachone is a bioactivatable chemotherapeutic agent that directly targets cancer cell lines over-expressing NQO1, leading to the interruption of glycolytic metabolism and changes in the redox state of the cell [9]. NQO1 quinone detoxification is highly dependent on the stability of the hydroquinone generated from the two-electron reduction it catalyzes [10]. β-lapachone treatment takes advantage of the NQO1 catalyzed reaction through the formation of an unstable hydroquinone that regenerates itself while producing peroxides in a futile cycle [11]. This process causes the rapid accumulation of reactive oxygen species (ROS), leading to DNA damage, followed by hyperactivation of poly(ADP-ribose) polymerase-1 (PARP1), leading to NAD+ and ATP loss, and cell death by a mechanism known as NAD+-keresis [12]. β-lapachone-induced cytotoxicity directly involves the inhibition of the NAD+-dependent glyceraldehyde 3-phosphate dehydrogenase (GAPDH), which suppresses overall glycolytic flux [6,12]. While β-lapachone has shown significant promise in Phase-1 clinical trials, off-target, dose-dependent toxicity in red blood cells due to methemoglobinemia has caused significant safety concerns [13]. The drug β-lapachone causes a profound interruption of glycolytic metabolism, which makes this pathway a prime target for metabolic imaging. A means of assessing NQO1 bioactivation in vivo could empower adaptive dosing strategies [14] that could reduce the amount of agent needed while maintaining its anti-cancer benefits. The glucose analog [18F]2-fluoro-2-deoxy-glucose (18FDG) is widely used clinically with positron emission tomography (FDG-PET) to produce images of glucose uptake in tumors, but it does not report on downstream metabolism, and causes radioactive exposure that carries a risk for secondary cancer [15,16]. Without the ability to monitor not only glucose uptake, but also its downstream metabolism, FDG-PET is an unsuitable avenue for imaging NQO1 activation. Recently, we showed that HDO production from [2H7]glucose could be utilized as a marker to quantitatively identify cancer metabolism [17]. We utilized hepatocellular carcinoma HUH-7 cells and normal AML-12 hepatocytes, and showed that HDO production significantly increased in HUH-7 cells compared to normal AML-12 cells [17]. While lactate production has been used as an indicator of upregulated glycolysis, the previous study showed HDO is a sensitive marker that can be applied as a safer alternative to radiotracer techniques [17]. We also demonstrated this approach is suitable for brain imaging in vivo in a rat model [18]. With the recent demonstration of 2H imaging as an effective means for detecting cancer in vivo in humans [19], extension of this method to detect the action of chemotherapeutics is logical.
Here, we utilized A549 lung carcinoma and MiaPaCa2 pancreatic cancer cells, which express high levels of NQO1, and HCT-116 colon cancer cells with low levels of NQO1, to test the hypothesis that [2H7]glucose could be used as a metabolic contrast agent sensitive to a β-lapachone-mediated reduction in glycolytic flux. NQO1 expression levels in these cancer cells were established previously by Western blot analysis [11,20], and shown to be sustained in cancer cells after β-lapachone treatment [9,21]. Additionally, we confirmed that these cell lines do not express the C609T polymorphism, which affects NQO1 activity [22,23,24]. Experimentally, we quantitatively measured decreased glycolytic rates in β-lapachone treated cancer cells using HDO, 2H-lactate, and residual [2H7]glucose in cell culture media. The estimates of HDO production rate combined with the production of the 2H-lactate results in a comprehensive accounting of the [2H7]glucose metabolism in control and β-lapachone treated cells. HDO production is linear with lactate production, but is characterized by a much larger change in absolute signal intensity, i.e., greater dynamic range. Our previous work demonstrated this method in normal versus cancerous cells. Here, HDO detection is extended as a method for monitoring chemotherapeutic inhibition of glycolysis.
2. Materials and Methods
2.1. Cell Lines, Chemicals, and Media
A549 (lung), MiaPaCa2 (pancreatic), and HCT-116 (colon) cancer cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Dulbecco’s Modified Eagle Medium (DMEM), Phosphate Buffered Saline (PBS) and Dimethyl Sulfoxide (DMSO) were purchased from Thermo Scientific (Waltham, MA, USA). Fetal Bovine Serum (FBS) was purchased from Atlas Biological (Fort Collins, CO, USA). Deuterated pyrazine (Pyrazine-D4), D-glucose, [1,2,3,4,5,6,6-2H7]-D-glucose ([2H7]glucose), and 2,2-dimethyl-3,4-dihydrobenzo[h]chromene-5,6-dione (β-lapachone) were purchased from Sigma Aldrich, (St. Louis, MO, USA). Methoxyamine hydrochloride in pyridine (MOX reagent) and N-methyl-N-(tert-butyldimethylsilyl)-trifluoroacetamide + 1% tertbutyldimethylchlorosilane (MTBSTFA + TBDMS) were purchased from Thermo Scientific, (Waltham, MA, USA).
2.2. Cell Culture
A549, MiaPaCa2, and HCT-116 cell lines were maintained in a complete growth medium composed of Dulbecco’s Modified Eagle Medium (DMEM) with 10% v/v FBS, 50 µg/mL penicillin, 50 µg/mL streptomycin, 10 µg/mL neomycin. Cell lines were cultured at 37 °C in a 95% air and 5% CO2 atmosphere in an air-jacketed incubator (Heracell Vios 160i, Thermo Scientific, Waltham, MA, USA). Every 3 days growth media was replenished, and once at 80% confluence cells were subcultured 1:10 into six 100 mm OD (56.7 cm2 culture area) cell culture plates. All cell lines were grown to 70–80% confluency (~10 million total cells) and washed once with warm PBS and incubated with 4 mL each of either DMEM with 5.5 mM [2H7]glucose and 6 µM β-lapachone (dissolved in DMSO) or DMEM with 5.5 mM [2H7]glucose and DMSO vehicle for 2 h. During the 2 h treatment, 100 μL aliquots were collected at 0 min, 15 min, 60 min, and 2 h. After the 2 h time point, all DMEM was collected and cells were washed once with warm PBS and incubated with 4 mL each of DMEM with 5.5 mM [2H7]glucose for 6 h, withdrawing 100 µL aliquots at 0 min, 30 min, 2 h, and 6 h. After the 6 h time point cells were trypsinized and collected for O2 consumption measurements (Supplementary Figure S8). An Oxygraph+ System from Hansatech Instruments (Pentley, KL, UK) was used to measure O2 consumption by analyzing 1.5 mL of the harvested cell suspension in a 37 °C sealed water chamber connected to an S1 Oxygen Electrode Disc that measured voltage differences in the suspension based on the amount of O2 present in solution.
2.3. NMR Sample Preparation
Cell media samples were prepared without an extraction. Each cell media sample of A549, MiaPaCa2, and HCT-116 cells was spiked with a 25 mM pyrazine-D4 internal standard stock solution to attain a final concentration of 2.5 mM. Addition of pyrazine-D4 internal standard allowed for quantification of HDO, 2H-lactate, and unconsumed [2H7]glucose from 2H-NMR data. A total of 50 μL of cell media sample for each time point was transferred into 1.7 mm NMR sample tubes for NMR data acquisition.
2.4. 2H-NMR Spectroscopy
A Bruker 14.1 T magnet system equipped with an Avance Neo Console (Bruker Biospin) and 1.7 mm CryoProbe was used to acquire 2H-NMR data of the deuterium-labeled metabolites in the cell media samples. The deuterium lock channel was used to acquire the 2H-NMR spectra at 92.12 MHz resonant frequency. An acquisition time (AQ) of 1 s and a relaxation delay (d1) of 2 s (total, 3 s of repetition time) with a 90° pulse was used to acquire all 2H-NMR spectra. A total of 4096 complex data points were digitized with the spectral width of 11 ppm using 256 scans for each of the 12 FIDs (3072 scans) for each of the samples. All NMR data were collected at room temperature (25 °C).
2.5. 2H-NMR Data Processing and Quantification of HDO, 2H-Lactate, and Residual [2H7]Glucose
TopSpin 4.0.3 was used for data acquisition. 2H-NMR spectra were transferred to MestReNova v14.0.1-23284 (Mestrelab Research S.L.) for further processing. The 2H-NMR spectra were refined by adjusting the exponential window function to 0.5 Hz and increasing the zero-filling of the free induction decay (FID) to 8192 data points before the Fourier Transform (FT). Manual phase corrections were made for each spectrum, as well as automatic spline baseline corrections. A total of 12 FIDs were acquired for each sample and all 12 2H-NMR spectra were aligned with respect to the pyrazine-D4 peak and combined to account for peak shifting due to magnetic field drift over the course of the experiment. Concentrations of HDO, 2H-lactate and residual [2H7]glucose in the cell media were calculated using peak areas of the internal standard pyrazine-D4. The concentration of pyrazine-D4 (2.5 mM) was used to quantify the concentration of HDO and residual [2H7]glucose, normalized to the number of deuterons responsible for corresponding resonances. 2H-lactate concentration was corrected for mono- and di-deuterated lactate isotopomers using the fractional enrichments of lactate m+1 and m+2 mass isotopologues. The time-series data were imported into GraphPad Prism and linear regression statistical analysis was performed. Simultaneously, the significant difference between the slopes of control and treatment groups was determined. All linear regression analysis was performed in Graphpad Prism (GraphPad Software, La Jolla California USA, www.graphpad.com).
2.6. GC-MS Sample Preparation
Media (20 µL) samples were dried in the Reacti-vial reaction vials under nitrogen gas airflow. Cells were extracted with 1 mL of Acetonitrile:Isopropanol:Water (3:3:2, v:v:v), then centrifuged for 15 min at 10,000× g at 4 °C. The supernatant was transferred into a new centrifuge tube and dried in a speedvac. Dried cell extract was resuspended in 0.5 mL of Acetonitrile:Water (1:1, v:v) and centrifuged for 5 min at 10,000× g. The supernatant was transferred into Reacti-vial reaction vials and dried down under nitrogen gas airflow. DL-Norleucine was added as an internal standard. Fifty microliters of 2% MOX reagent were added to the vial and vortex for 5 s followed by incubation at 30 °C for 90 min on a heating block with a magnetic stirrer for faster reaction and smooth mixing of the samples. Fifty microliters of MTBSTFA +TBDMS were added to each vial and incubated at 60 °C for 1 h. Reacti-vials were centrifuged at 8000× g for 10 min and the supernatant was transferred into GC vials.
2.7. GC-MS Analysis
GC-MS analysis was performed with Thermo Scientific ISQ Single Quadrupole Mass Spectrometer and Trace 1310 Gas Chromatograph (GC-MS, Thermo Scientific, USA) equipped with 30 m long Restek 95% dimethyl/5% diphenyl polysiloxane RTX-5MS column, 0.25 mm internal diameter, 0.25 µm film, with 10 m empty guard column (Restek, Bellefonte, PA, USA). The initial oven temperature was 60 °C for 60 s and the ramp oven temperature increased to 325 °C at 10 °C/min with 5 min final hold time at the end. Ion source temperature was maintained at 230 °C with electron ionization energy at 70 eV and a quadrupole mass analyzer was utilized for separation of ions. Helium was used as carrier gas at a flow rate of 1 mL/min. The Mass Isotopomer Distribution (MID) of the lactate and citrate was obtained by integration of extracted ion chromatograms from each of the isotopologues. The MID of lactate and citrate were corrected for natural isotope abundance using the Isotopomer Network Compartmental Analysis (INCA) software [25]. Extracellular and intracellular metabolite’s total intensities were normalized to the intensity of DL-Norleucine internal standard and compared between control and treated groups. Significant differences for metabolites between control and treatment groups were established using Student’s t-test and FDR correction utilizing RStudio (RStudio Version 1.4.1717, PBC, Boston Massachusetts, USA, www.rstudio.com (accessed on 17 August 2021)).
3. Results
3.1. 2H-NMR Spectra of Cell Culture Media
2H-NMR spectra were recorded as a function of time for cell media samples during treatment and post-treatment periods for A549, MiaPaCa2 and HCT-116 cancer cell lines. HDO and 2H-lactate signal intensity increase with incubation time while [2H7]glucose signal decreases for DMSO control A549 cells (Figure 1A). Signal intensities of HDO and 2H-lactate increase slowly with incubation time, which matches a slow decrease of [2H7]glucose signal intensity in β-lapachone treated A549 cells (Figure 1B).
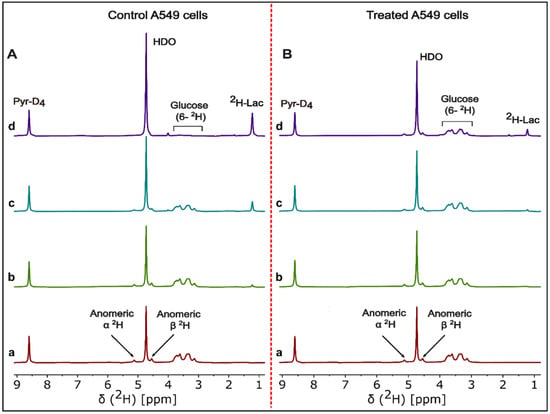
Figure 1.
Representative 2H-NMR spectra of cell media time course analysis. Stacked plot of 2H-NMR spectra of DMSO control (left, panel A) and β-lapachone treated (right, panel B) of the A549 cell line incubated with 5.5 mM [2H7]glucose. Cell media samples were withdrawn at (a) 0 min, (b) 30 min, (c) 2 h, and (d) 6 h incubation time points of the β-lapachone post-treatment cultures of A549 cancer cells. Labeling in the (a and d) spectra of panel A,B shows the signals for pyrazine-D4, HDO, residual [2H7]glucose, and 2H-lactate.
3.2. Quantification of the 2H-Lactate, HDO and [2H7]Glucose in Cell Media
[2H7]glucose consumption was significantly higher in control cells at the 120 min time point for both treatment and post-treatment periods (Figure 2 and Figure S1A,B, Tables S1 and S2). For the post-treatment period, [2H7]glucose consumption rates for control and treated A549 cells were 0.014 ± 0.0004 and 0.003 ± 0.0006 μmol/106 cells/min (Figure 2B and Figure 3), respectively, with a significantly higher rate in control cells (p = 0.0001). Linearity for 2H-lactate and HDO production from A549 cells into the culture medium is excellent with an R2 of 0.99. HDO production was corrected for the natural abundance concentration in each cell media sample. The production rates of HDO for A549 cells were 0.050 ± 0.005 and 0.037 ± 0.002 μmol/106 cells/min for control and treated cells, respectively, and were significant (p = 0.02) between both groups in the treatment period (Figure 2C). In the post-treatment period, HDO production rates were significantly higher in A549 control cells (0.053 ± 0.002 and 0.017 ± 0.003 μmol/106 cells/min, respectively, p = 0.0001) (Figure 2D). The production rates of 2H-lactate in control and treated cells during treatment were also significantly different (0.02 ± 0.0005 and 0.007 ± 0.004 μmol/106 cells/min, respectively, p = 0.0001) (Figure 2E and Figure 3). The post-treatment period rates were 0.02 ± 0.001 and 0.005 ± 0.001 μmol/106 cells/min, respectively, (p = 0.0001) (Figure 2F). Similar results were shown in the MiaPaCa2 cells (Figure 3, Tables S1 and S2).
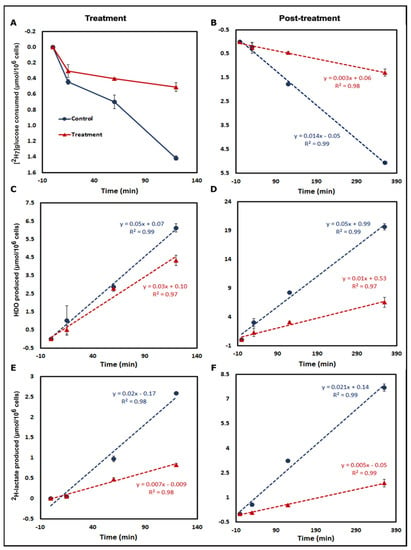
Figure 2.
Time course analysis of [2H7]glucose metabolism. Kinetic plots of treatment (1st column) and post-treatment (2nd column) [2H7]glucose metabolism by control and treated A549 cells incubated in DMEM with 5.5 mM [2H7]glucose. Analysis of [2H7]glucose consumption for both periods is shown in (A,B). The kinetics of HDO production are displayed in panels (C,D). Similarly, kinetic data for 2H-lactate production are shown in panels (E,F). Results were calculated from 2H-NMR data of each of the cell media samples including a blank withdrawn at the 0 min time point for each time period. (Note: [2H7]glucose consumption and HDO and 2H-lactate production were measured in the cell media samples withdrawn at 0 min, 15 min, 1 h, and 2 h for treatment and 0 min, 30 min, 2 h, and 6 h time points of the post-treatment period. Three biological replicates were used for each group and data are represented as mean ± standard error of mean (SEM)).
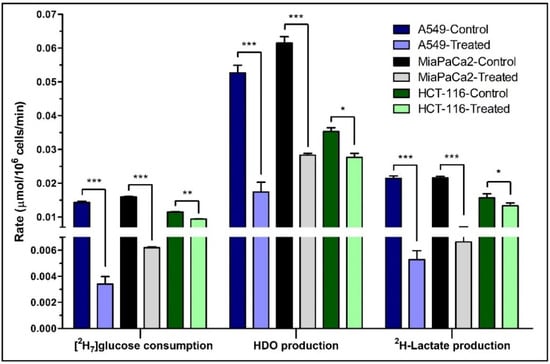
Figure 3.
The rates of [2H7]glucose consumption and HDO/2H-lactate production. The consumption rate of [2H7]glucose and production rates of HDO and 2H-lactate are shown for the post-treatment period. Rates were calculated from the kinetic data of control and β-lapachone treated A549, MiaPaCa2 and HCT-116 cancer cells. Biological triplicate data are represented as mean ± SEM. Significance levels between the slopes of control and treatment groups were determined using the statistical analysis in Graphpad Prism and shown as: “*” if p ≤ 0.05, “**” if p ≤ 0.01, and “***” if p ≤ 0.001.
HCT-116 cells express less NQO1, and we hypothesized lower significant differences between control and treated cells for glucose consumption and production of HDO and 2H-lactate after β-lapachone exposure (Figure 3 and Table S1). Despite lower levels of NQO1, HCT-116 cells still showed significant differences on treatment, though less robustly than the A549 and MiaPaca2 cell lines. In general, higher NQO1 expression [11,19] correlates with greater differences in glycolytic rates between control versus treatment as expected in A549 and MiaPaCa2 cell lines (Figure 3, Tables S1 and S2).
3.3. Correlation between [2H7]Glucose Consumption and 2H-Lactate, HDO Production
Correlation plots were made for the post-treatment period to determine relationships between glucose consumption and the production of the downstream metabolites. In Figure 4, panel A and D clearly show the linearity between [2H7]glucose consumption and HDO or 2H-lactate production for A549 cells, respectively. This behavior is preserved even in β-lapachone treated cells. The concentration of produced HDO was higher than 2H-lactate, producing a metric of glycolysis with a greater dynamic range than lactate (Figure S1 and Table S2). The sensitivity of HDO production over 2H-lactate production with respect to the [2H7]glucose consumption is also evidenced by the slopes in Figure 4. In HCT-116 cells, 0.32 [2H7]glucose units are required to produce an HDO and 0.76 to produce lactate. Similar trends were found in the case of A549 and MiaPaCa2 cells (Figure 4). Additionally, the linear trends between [2H7]glucose consumption and HDO/2H-lactate production led us to calculate the 2H mass balance of consumed [2H7]glucose and its incorporation into 2H-lactate and HDO at the 6 h time point. In the case of A549 cells, mass balance accountability is lower (84.1%) than treated cells (92.4%) whereas, in the case of HCT-116 cells, where NQO1 is less abundant, it was 68.4 and 70.8% for control and treated cells, respectively (Table 1).
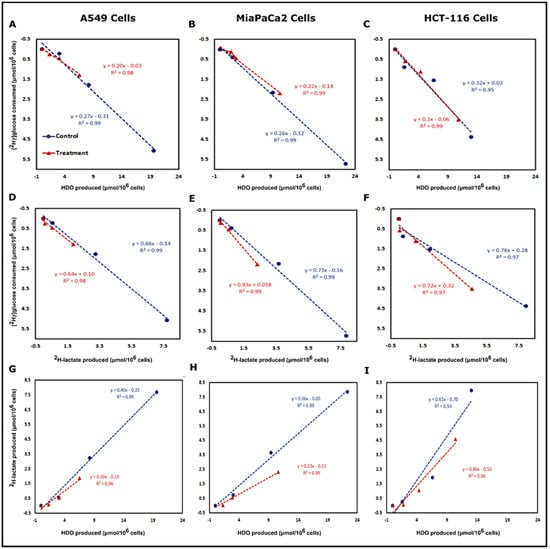
Figure 4.
HDO and 2H-lactate production are directly related to [2H7]glucose consumption. Correlation plots of [2H7]glucose consumption vs. HDO production (top row) and 2H-lactate production (middle row) and correlation between 2H-lactate vs. HDO production (bottom row) are shown for the post-treatment period of A549 (Panels A,D and G), MiaPaCa2 (Panels B,E and H) and HCT-116 (Panels C,F and I). Correlations in all conditions were linear with R2 values > 0.96. Correlation plots were plotted from the calculated concentration of consumed [2H7]glucose and produced 2H-lactate and HDO by control and treated cells. The excellent linearity between 2H-lactate and HDO production and higher production of HDO demonstrates that the HDO could be a much more sensitive marker than lactate for monitoring NQO1 bioactivation.

Table 1.
2H mass balance between consumed [2H7]glucose and produced HDO and 2H-lactate.
3.4. Correlation between 2H-Lactate and HDO Production
Correlation plots between HDO and 2H-lactate production for all three cancer cell lines demonstrated linearity between control and treated cells in the post-treatment phase (Figure 4). In A549 cells, 0.40 2H-lactate was produced per HDO, which drops to 0.30 lactate/HDO in the β-lapachone treatment group. However, HDO concentration is more than 2× than that of the 2H-lactate (Figure 4G). Similarly, MiaPaCa2 and HCT-116 cells also demonstrated a linear correlation between HDO and 2H-lactate production in the control and treated cells post-treatment (Figure 4H,I). The linear correlation between lactate and HDO production was excellent with R2 of 0.99 for control A549 and MiaPaCa2 cells, whereas treated A549 and MiaPaCa2 cells showed linearity with R2 of 0.96 and 0.99, respectively (Figure 4G,H). The linear correlation was slightly lower for HCT-116 cells with R2 of 0.93 and 0.96 for control and treated cells, respectively (Figure 4I).
3.5. Fractional Enrichment of Lactate and Citrate Isotopologues
Natural isotope abundance corrected fractional enrichments of lactate and citrate isotopologues at 360 min time points were analyzed for assessing glycolytic and TCA cycle activity. A549 cells showed significant differences in all lactate isotopologue patterns (Figure 5A). The citrate m+1 isotopologues were significantly higher in control A549 cells (Figure 5B). Lactate m+2 and citrate m+1 isotopologues were significantly higher in control MiaPaCa2 cells (Figure 5D,E). HCT-116 cells showed insignificant differences for lactate and citrate (m+1 and m+2) isotopologues whereas lactate m+0 isotopologues were significantly lower in control HCT-116 cells (Figure 5G,H).
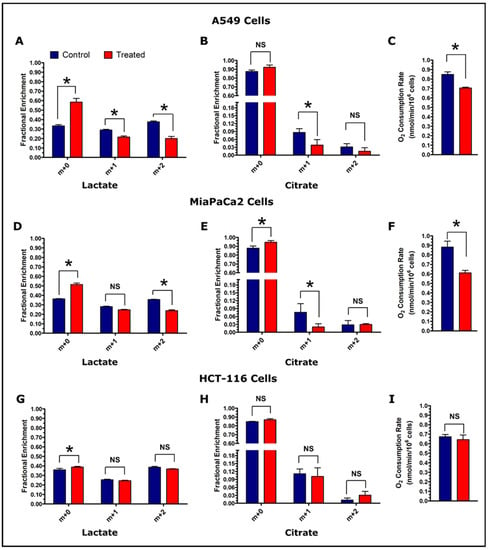
Figure 5.
Assessment of glycolytic and TCA cycle activity in control and treated cells. Fractional enrichment of lactate and citrate, and O2 consumption rate are shown for A549 cells (Panels, A–C), MiaPaCa2 cells (Panels, D–F), and HCT-116 cells (Panels, G–I). Enrichment results were calculated from the GC-MS data of cell media samples collected at the 360 min post-treatment time point. The GC-MS data were corrected for natural isotope abundance using the Isotopomer Network Compartmental Analysis (INCA) software. (Note: N = 3: biological replicate data are represented as mean ± SEM. Significant levels were calculated using the Student’s t-test between control and treated groups and are displayed as: non-significant “NS” if p > 0.05 and “*” if p ≤ 0.05. Oxygen consumption was measured at the end of tracer experiment (total time period of 8 h) and O2 consumption rates were calculated for the number of cells used for the measurements and normalized per million cells).
3.6. Oxygen Consumption Rate of the Cancer Cell Lines
Oxygen consumption is a general index of metabolic turnover, and was measured after the completion of the tracer experiment. The O2 consumption rates of A549 and MiaPaCa2 control cells were found significantly higher in treatment cells (Figure 5C,F). On the other hand, HCT-116 cells did not show a significant difference in O2 consumption at the conclusion of the experiment (Figure 5I).
3.7. Metabolic Profiling of Control and β-Lapachone Treated Cancer Cell Lines
To assess the consumption and production of extracellular metabolites, 6 h cell media samples were analyzed by GC-MS. Eighteen key metabolites were identified and compared between control and treatment groups across all three cancer cell lines. More extracellular metabolites were found to be significantly different among treatment and control groups for NQO1+ cells, A549 and MiaPaCa2 (Figures S4 and S5), than for NQO1- HCT-116 cells (Figure S6). Notably, lactate levels were significantly lower in treatment group media for both A549 (p = 0.03) and MiaPaCa2 (p = 0.01) but showed the opposite non-significant trend in HCT-116 cell media (Figures S4–S6). Analysis of cell media showed higher levels of glycine, tyrosine, glutamine, serine, isoleucine, leucine, and valine in treated cells. To assess intracellular metabolite levels, GC-MS analysis was conducted on A549 and HCT-116 cell extracts. More metabolites with greater significant differences were detected between the control and treatment groups of A549 cells compared to HCT-116 cells. The intracellular levels of lactate agree with the analysis of lactate production shown in Figure 3. Treated A549 cells contained ~10 times less lactate than control cells (p = 0.003) (Figure 6), whereas treated HCT-116 cells contained ~1.75 times less lactate than control cells (p = 0.004) (Figure S7). Treated A549 cells also showed significantly lower levels of some TCA cycle intermediates, polar amino acids, and branched-chain amino acids (Figure 6). Both A549 and HCT-116 cells showed significantly lower levels of glutamine in treated cells (Figure 6 and Figure S7). Most of the intracellular metabolites showed insignificant differences between control and treated HCT-116 cells.
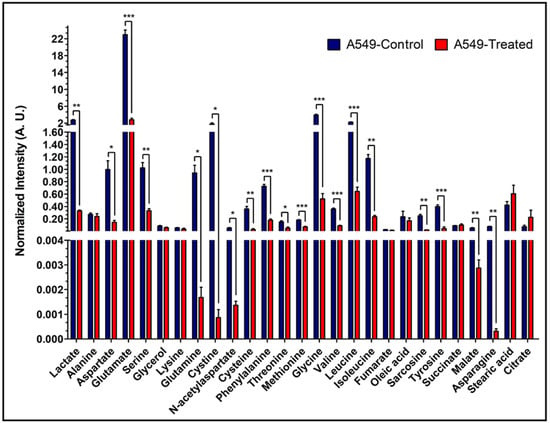
Figure 6.
Intracellular levels of the metabolites in A549 cells. Bar plots of GC-MS analysis-based levels of different intracellular metabolites from A549 cells. Shown are the normalized intensities of the differential levels of metabolites between control and treated A549 cells. (Note: N = 3: biological replicate data are represented as mean ± SEM. Student’s t-test was used to calculate statistical significance followed by false discovery rate (FDR) performed in RStudio, to adjust p values and represented as: “*” if p ≤ 0.05, “**” if p ≤ 0.01, and “***” if p ≤ 0.001).
4. Discussion
Deuterium NMR was used sporadically in the late 1980s, [26,27] but has re-emerged as a method for measuring turnover in highly metabolic systems. Recently, deuterium (2H) magnetic resonance imaging (DMI) was used to assess cancer metabolism, as it provides requisite chemical and temporal resolution [19,28]. The low natural abundance (0.0115%) of deuterium atoms [29] maximizes the specificity of deuterated tracers for metabolic flux measurements in cell culture and in vivo studies. Deuterium is a non-radioactive nucleus, making perdeuterated glucose (or [2H7]glucose) a safe tracer for in vitro and in vivo studies. It has been demonstrated that the total glycolytic rate is not affected by perdeuteration in glucose [30], suggesting negligible effects on the total glycolytic rate in these experiments. With this experimental design, we sought to determine if [2H7]glucose could be used to measure changes in glycolysis after administration of β-lapachone, which is known to deplete NAD+ stores in NQO1+ cells [11]. While β-lapachone has demonstrated powerful effects in a variety of cancer types, it also has known off-target effects on the red blood cell [13]. The common side-effect of hemolytic anemia limits drug dosing and has caused some clinical trials to be terminated early. The adaptive dose-finding approach to maximize efficacy while minimizing toxicities can be significantly amplified with stronger parametrization of favorable results [31]. Because of the strong, logical connection between β-lapachone action and decreased glycolytic flux, the analytical methods we demonstrate here suggest themselves as potential biomarkers for input into an adaptive dosing paradigm. Using this specific model, we demonstrate a global accounting of perturbations in glycolysis by measuring HDO and lactate production. This method produces a more satisfactory accounting of glucose metabolism than observing lactate alone, which in the case of β-lapachone action is doubly inhibited by the loss of the NAD+ cofactor at GAPDH and LDH.
The NMR data (Figure 1) have a high signal-to-noise ratio, allowing very exact estimates of concentration. Time series data showed a quantitative estimate of increased glucose consumption in control cancer cells based on increased HDO and 2H-lactate and decreased residual [2H7]glucose signals. Figure 1B showed reduced signal intensities for HDO and 2H-lactate compared to control cells (Figure 1A), indicating the effects of NAD+ depletion on glycolytic flux [6,11].
Apart from 2H-lactate, HDO also acts as a sensitive marker for glycolytic rate because the loss of 2H from [2H7]glucose occurs in many enzymatic processes during glycolysis, and therefore may provide a more global view of glucose utilization. Loss of 2H atoms from C6 of [6,6-2H2]glucose is mainly due to the catalytic action of pyruvate kinase (PK) whereas loss of 2H from C1 of [1-2H]glucose is due to both pyruvate kinase (PK) and phosphomannose isomerase (PMI) [32]. The presence of a 2H kinetic isotope effect at triose phosphate isomerase (TPI), results in more 2H liberation from dihydroxyacetone phosphate (DHAP) [33,34]. HDO is produced by TPI from the C2 position of GA3P and DHAP, within the course of direct metabolism of one equivalent of [2H7]glucose to pyruvate [17]. HDO production from the C1 and C2 positions can occur through the interconversion of fructose-6-phosphate (F6P) and mannose-6-phosphate (M6P) via PMI [35]. Enolase enzymatic activity on phospho-enol pyruvate (PEP) and keto-enol tautomerization in pyruvate are also prominent sites of HDO production during glycolysis [19,36,37,38,39,40]. Additionally, deuterium atoms could be lost during the conversion of pyruvate into alanine and lactate via alanine aminotransferase (ALT) and lactate dehydrogenase (LDH) reactions, respectively [41]. Figure 7 summarizes the reactions that generate 2H-lactate and HDO from [2H7]glucose during glycolysis. Apart from glycolysis, deuterium could also be lost to solvent water in the form of HDO in several reactions in the tricarboxylic acid (TCA) cycle [28,42].
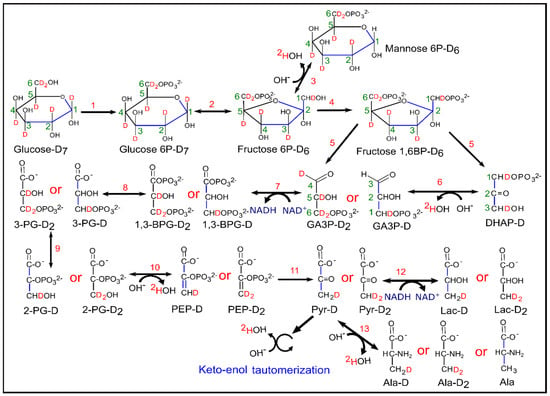
Figure 7.
Schematic diagram of glycolysis and evolution of HDO from [2H7]glucose. Enzymatic reactions indicated by numbers in the figure were catalyzed by (1) hexokinase, (2) phosphoglucose isomerase, (3) phosphomannose isomerase, (4) phosphofructokinase, (5) aldolase, (6) triose phosphate isomerase, (7) glyceraldehyde 3-phosphate dehydrogenase, (8) phosphoglycerate kinase, (9) phosphoglyceromutase, (10) enolase, (11) pyruvate kinase, (12) lactate dehydrogenase and (13) alanine amino transferase. (Note: number of deuterons, which ends up in the methyl group of lactate isotopomers were written with the name of lactate precursors from GA3P-D and GA3P-D2 onwards).
Treatment of NQO1+ A549 cells with β-lapachone showed clear differences in [2H7]glucose consumption, 2H-lactate and HDO production between control and treatment groups (Figure 2). Both during treatment and post-treatment, HDO and 2H-lactate production is linear. NQO1/β-lapachone induced futile cycling consumes NADPH while producing large amounts of hydrogen peroxide that attack DNA and overwhelm repair mechanisms causing the hyperactivation of poly-(ribose-AMP) polymerase (PARP1), leading to a significant loss NAD+/ATP pools [20,24]. Thus, glycolytic enzymes that depend on NAD+/NADH like GAPDH and LDH are highly affected by β-lapachone treatment, leading to a reduction in glycolytic rate [11]. In NQO1+ cells, we observed that 2H-lactate production is drastically reduced. LDH catalytic dependence on NAD+/NADH suggests that the production of 2H-lactate does not reflect glucose utilization because it is doubly reduced by β-lapachone-dependent NAD+/NADH depletion; once by upstream GAPDH inhibition and secondly by an insufficient pool of NAD+/NADH cofactor for LDH. On the other hand, HDO serves as a better indicator of overall drug-dependent disruptions of glycolysis because multiple glycolytic enzymes are responsible for HDO production. Additionally, measurement of HDO production can denote distinguishable differences in drug-treated and non-treated cells with robust signals corresponding to high concentrations of HDO detected in extracellular media, exemplified in A549 and MiaPaCa2 cells (Figure 2 and Figure S2). The drug-dependent effects on glycolytic rate by β-lapachone treatment are verified by the response of NQO1- HCT-116 cells showing lower differences in [2H7]glucose consumption and 2H-lactate/HDO production between treatment and control cells (Figure S3).
Correlation plots demonstrated an excellent linear slope for HDO production for both control and treatment cells, with a much higher concentration of HDO compared to 2H-lactate. The linear slopes show that much less [2H7]glucose is required to show the differences in glycolytic rates between control and treated cells using HDO as a marker (Figure 4). This reinforces the idea that HDO production can serve as a biomarker of β-lapachone action. Congruently, we observed a significant reduction in 2H-lactate production, which coincides with the implications of the direct effects of NAD+/NADH depletion on LDH. Theoretically, NADPH depletion might increase the flux of glucose-derived carbon through the pentose phosphate pathway (PPP). Previously, we showed the possible fate of [2H7]glucose for HDO/2H-lactate production, and the loss of 2H atoms from glycolysis, sugar isomerization, and potentially the PPP [17]. A recent publication using flux balance analysis demonstrated that the malic enzyme and isocitrate dehydrogenase are the most likely sources of NADPH that would be involved with the β-lapachone-mediated futile cycle [43]. Therefore, we do not expect the PPP to be a major source of HDO in our experimental conditions. The mass balance between [2H7]glucose consumption and HDO/2H-lactate production is close to 85–90%, so it is conceivable that some of the 2H-label is lost in other metabolic pathways. For example, 2H-labeling in metabolites involved in multiple pathways downstream of glucose is shown in Supplementary Figure S9. As these metabolites were not quantitated by standard addition, we cannot include them in the mass balance analysis.
The excellent linearity between 2H-lactate and HDO production shown in the correlation plots of Figure 4 demonstrate that glycolytic rate can be measured by HDO with high sensitivity even in the presence of interrupted metabolism—in this case, the loss of the NAD+ cofactor for LDH and GAPDH. Mass balance calculations were in good agreement for the more glycolytic A549 and MiaPaCa2 cell lines, but a significant undercounting of deuterium-labeled metabolites in the HCT-116 cells suggest that other pathways than glycolysis may be more active. Mass isotopologue analysis further confirmed reduced glycolysis and tricarboxylic acid (TCA) cycle metabolism in β-lapachone treated cells, as indicated by significantly lower labeling of deuterium in lactate and citrate isotopologues than control cells at the 6 h time point. Lower deuterium enrichment in downstream glucose metabolites is concomitantly supported by O2 consumption (Figure 5). Consistent with phenotypic glucose metabolism and β-lapachone drug action, treated NQO1+ cells consumed significantly lower O2 than control cells at the end of the [2H7]glucose tracer experiment (total time 8 h) whereas NQO1- HCT-116 cells showed insignificantly lower O2 consumption in treated cells (Figure 5C,F,I). Previous work measured excess O2 consumption during the treatment period, but did not measure O2 consumption in a post-treatment setting [24]. This work indicates that a significant decrease in TCA cycle turnover is a durable effect of NQO1 bioactivation in the post-treatment phase.
To confirm the metabolic effects of β-lapachone treatment, a comparative analysis of 18 metabolites between control and treated cells in media was performed by GC-MS. The detected lactate level was significantly lower in β-lapachone treated cancer cells (Figure 6), consistent with interruption of glycolysis due to β-lapachone treatment [11]. Additionally, the metabolites panel showed disruptions in the production and consumption of key metabolites mediated by β-lapachone treatment with significant differences observed in NQO1+ cell lines and non-significant trends seen in NQO1− HCT-116 cells. The lactate/alanine ratio reflects the NAD+/NADH equilibrium in the cytosol [44]. A549 cells have a control to treatment lactate/alanine ratio of ~7 while HCT-116 cells have a ratio of ~2; indicating a loss of reducing equivalents in A549 (NQO1+) β-lapachone treated cells. The intracellular metabolic panel also indicated that β-lapachone treatment perturbs the TCA cycle and amino acid metabolism, including branched-chain amino acids (BCAA) (Figure 6). As a whole, extracellular and intracellular metabolomic analysis alludes to potential avenues for β-lapachone combinatorial treatments targeting additional NAD+/NADH-dependent reactions. Significantly higher levels of intracellular BCAAs (valine, isoleucine, and leucine) in control NQO1+ cells (Figure 6), in contrast to extracellular levels (Figure S4), indicates that there is a possibility that β-lapachone action is inhibiting BCAA consumption by limiting NAD+/NADH availability of branched chained α-ketoacid dehydrogenase (BCKDH); a primary mediator of BCAA catabolism [45]. Similarly, significant differential levels of intracellular malate, aspartate, and asparagine observed between control and treated NQO1+ cells in contrast to non-significant extracellular differences suggest inhibition of the NADH dependent malate-aspartate shuttle [46]. BCAA catabolism or the malate-aspartate shuttle are logical targets for possible synergies with β-lapachone treatment.
5. Conclusions
Here, we report that HDO and 2H-lactate production can be used as a sensitive marker to measure differences in glycolytic metabolism caused by β-lapachone drug action. Recent results using [6,6-2H2]glucose as a tracer indicated that HDO production did not correlate well with glycolytic activity in an in vivo cancer model [47]. However, the amplified HDO production associated with the perdeuterated glucose might be sufficient to overcome the HDO signal associated with peripheral metabolism to yield an imaging biomarker for β-lapachone-mediated interruption of glycolysis. In vivo investigations of this hypothesis are underway. Deuterated lactate is an alternative biomarker for further investigation. It should be noted that the perdeuterated glucose tracer used here should produce more deuterated lactate, as both the C6 and C1 positions of glucose will label lacate-C3. Either of these markers could be used as inputs for an adaptive dosing model to reduce β-lapachone-associated toxicity that limits its use in human cancers at this time.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13164165/s1, Figure S1: Comparative analysis shows reduced glycolytic flux in NQO1+ β-lapachone treated cells, Figure S2: Kinetic analysis of glycolysis in MiaPaCa2 cells, Figure S3: Time course measurement of glycolytic rate in HCT-116 cells, Figure S4: Extracellular metabolic panel shows differential level of metabolites between control and treated A549 cells, Figure S5: Extracellular differential level of metabolites in MiaPaCa2 cells, Figure S6: Analysis of the extracellular metabolites in HCT-116 cells, Figure S7: Intracellular levels of metabolites in HCT-116 cells, Figure S8: Schematic diagram representing the experimental timeline, Figure S9: The 2H-labeling in the metabolites of control and treated cells, Table S1: Kinetic analysis during β-lapachone treatment, Table S2: Kinetic analysis data of post-treatment periods.
Author Contributions
Conceptualization, R.M., M.C.C. and M.E.M.; Data curation, R.M. and M.C.C.; Funding acquisition, M.E.M.; Investigation, R.M. and M.C.C.; Methodology, R.M. and M.C.C.; Supervision, M.E.M.; Writing—original draft, R.M. and M.C.C.; Writing—review & editing, M.E.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Institute of Health grants: NIH P41-122698, 5U2CDK119889, and NIH R01-105346. Funding was also provided by NSF DMR-1644779.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in this article and Supplementary Materials and will also be available from the corresponding author upon request.
Acknowledgments
A portion of this work was performed at the National High Magnetic Field Laboratory, which is supported by National Science Foundation Cooperative Agreement number DMR-1644779, & the State of Florida. The authors gratefully acknowledge the teaching and leadership of the late David Boothman, who inspired this work with his relentless enthusiasm.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [PubMed]
- Phan, L.M.; Yeung, S.-C.J.; Lee, M.-H. Cancer Metabolic Reprogramming: Importance, Main Features, and Potentials for Precise Targeted Anti-Cancer Therapies. Cancer Biol. Med. 2014, 11, 1–19. [Google Scholar] [CrossRef]
- Warburg, O. On Respiratory Impairment in Cancer Cells. Science 1956, 124, 269–270. [Google Scholar] [PubMed]
- Gatenby, R.A.; Gillies, R.J. Why Do Cancers Have High Aerobic Glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; DeBerardinis, R.J. Understanding the Intersections between Metabolism and Cancer Biology. Cell 2017, 168, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, G.; Silvers, M.A.; Ilcheva, M.; Liu, Y.; Moore, Z.R.; Luo, X.; Gao, J.; Anderson, G.; Liu, L.; Sarode, V.; et al. Tumor-Selective Use of DNA Base Excision Repair Inhibition in Pancreatic Cancer Using the NQO1 Bioactivatable Drug, β-Lapachone. Sci. Rep. 2015, 5, 17066. [Google Scholar] [CrossRef] [PubMed]
- Belinsky, M.; Jaiswal, A.K. NAD(P)H:Quinone Oxidoreductase1 (DT-Diaphorase) Expression in Normal and Tumor Tissues. Cancer Metastasis Rev. 1993, 12, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Joseph, P.; Xie, T.; Xu, Y.; Jaiswal, A.K. NAD(P)H:Quinone Oxidoreductase1 (DT-Diaphorase): Expression, Regulation, and Role in Cancer. Oncol. Res. 1994, 6, 525–532. [Google Scholar]
- Pink, J.J.; Planchon, S.M.; Tagliarino, C.; Varnes, M.E.; Siegel, D.; Boothman, D.A. NAD(P)H:Quinone Oxidoreductase Activity Is the Principal Determinant of β-Lapachone Cytotoxicity. J. Biol. Chem. 2000, 275, 5416–5424. [Google Scholar] [CrossRef]
- Ross, D.; Siegel, D. Functions of NQO1 in Cellular Protection and CoQ10 Metabolism and Its Potential Role as a Redox Sensitive Molecular Switch. Front. Physiol. 2017, 8, 595. [Google Scholar] [CrossRef]
- Silvers, M.A.; Deja, S.; Singh, N.; Egnatchik, R.A.; Sudderth, J.; Luo, X.; Beg, M.S.; Burgess, S.C.; DeBerardinis, R.J.; Boothman, D.A.; et al. The NQO1 Bioactivatable Drug, β-Lapachone, Alters the Redox State of NQO1+ Pancreatic Cancer Cells, Causing Perturbation in Central Carbon Metabolism. J. Biol. Chem. 2017, 292, 18203–18216. [Google Scholar] [CrossRef]
- Moore, Z.; Chakrabarti, G.; Luo, X.; Ali, A.; Hu, Z.; Fattah, F.J.; Vemireddy, R.; DeBerardinis, R.J.; Brekken, R.A.; Boothman, D.A. NAMPT Inhibition Sensitizes Pancreatic Adenocarcinoma Cells to Tumor-Selective, PAR-Independent Metabolic Catastrophe and Cell Death Induced by β-Lapachone. Cell Death Dis. 2015, 6, e1599. [Google Scholar] [CrossRef]
- Gerber, D.E.; Beg, M.S.; Fattah, F.; Frankel, A.E.; Fatunde, O.; Arriaga, Y.; Dowell, J.E.; Bisen, A.; Leff, R.D.; Meek, C.C.; et al. Phase 1 Study of ARQ 761, a β-Lapachone Analogue That Promotes NQO1-Mediated Programmed Cancer Cell Necrosis. Br. J. Cancer 2018, 119, 928–936. [Google Scholar] [CrossRef]
- Iasonos, A.; O’Quigley, J. Adaptive Dose-Finding Studies: A Review of Model-Guided Phase I Clinical Trials. J. Clin. Oncol. 2014, 32, 2505–2511. [Google Scholar] [CrossRef]
- Linet, M.S.; Slovis, T.L.; Miller, D.L.; Kleinerman, R.; Lee, C.; Rajaraman, P.; Gonzalez, D.A.B. Cancer Risks Associated With External Radiation From Diagnostic Imaging Procedures. CA Cancer J. Clin. 2012, 62, 75–100. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, A.; Jaimini, A.; Tripathi, M.; Souza, M.D.; Sharma, R.; Mondal, A. Estimation of Radiation Dose to Patients from 18 FDG Whole Body PET / CT Investigations Using Dynamic PET Scan Protocol. Indian J. Med. Res. 2015, 721–731. [Google Scholar] [CrossRef]
- Mahar, R.; Donabedian, P.L.; Merritt, M.E. HDO Production from [2H7]Glucose Quantitatively Identifies Warburg Metabolism. Sci. Rep. 2020, 10, 8885. [Google Scholar] [CrossRef] [PubMed]
- Mahar, R.; Zeng, H.; Giacalone, A.; Ragavan, M.; Mareci, T.H.; Merritt, M.E. Deuterated Water Imaging of the Rat Brain Following Metabolism of [2H7]Glucose. Magn. Reson. Med. 2021, mrm.28700. [Google Scholar] [CrossRef]
- De Feyter, H.M.; Behar, K.L.; Corbin, Z.A.; Fulbright, R.K.; Brown, P.B.; McIntyre, S.; Nixon, T.W.; Rothman, D.L.; de Graaf, R.A. Deuterium Metabolic Imaging (DMI) for MRI-Based 3D Mapping of Metabolism in Vivo. Sci. Adv. 2018, 4, eaat7314. [Google Scholar] [CrossRef] [PubMed]
- Bey, E.A.; Bentle, M.S.; Reinicke, K.E.; Dong, Y.; Yang, C.-R.; Girard, L.; Minna, J.D.; Bornmann, W.G.; Gao, J.; Boothman, D.A. An NQO1- and PARP-1-Mediated Cell Death Pathway Induced in Non-Small-Cell Lung Cancer Cells by β-Lapachone. Proc. Natl. Acad. Sci. USA 2007, 104, 11832–11837. [Google Scholar] [CrossRef]
- Kim, D.; Cho, J.-Y. NQO1 Is Required for β-Lapachone-Mediated Downregulation of Breast-Cancer Stem-Cell Activity. Int. J. Mol. Sci. 2018, 19, 3813. [Google Scholar] [CrossRef] [PubMed]
- Lajin, B.; Alachkar, A. The NQO1 Polymorphism C609T (Pro187Ser) and Cancer Susceptibility: A Comprehensive Meta-Analysis. Br. J. Cancer 2013, 109, 1325–1337. [Google Scholar] [CrossRef] [PubMed]
- Anwar, A.; Dehn, D.; Siegel, D.; Kepa, J.K.; Tang, L.J.; Pietenpol, J.A.; Ross, D. Interaction of Human NAD(P)H:Quinone Oxidoreductase 1 (NQO1) with the Tumor Suppressor Protein P53 in Cells and Cell-Free Systems. J. Biol. Chem. 2003, 278, 10368–10373. [Google Scholar] [CrossRef]
- Huang, X.; Motea, E.A.; Moore, Z.R.; Yao, J.; Dong, Y.; Chakrabarti, G.; Kilgore, J.A.; Silvers, M.A.; Patidar, P.L.; Cholka, A.; et al. Leveraging an NQO1 Bioactivatable Drug for Tumor-Selective Use of Poly(ADP-Ribose) Polymerase Inhibitors. Cancer Cell 2016, 30, 940–952. [Google Scholar] [CrossRef]
- Young, J.D. INCA: A Computational Platform for Isotopically Non-Stationary Metabolic Flux Analysis. Bioinformatics 2014, 30, 1333–1335. [Google Scholar] [CrossRef]
- Ackerman, J.J.H.; Ewy, C.S.; Becker, N.N.; Shalwitz, R.A. Deuterium Nuclear Magnetic Resonance Measurements of Blood Flow and Tissue Perfusion Employing 2H2O as a Freely Diffusible Tracer. Proc. Natl. Acad. Sci. USA 1987, 84, 4099–4102. [Google Scholar] [CrossRef]
- Ewy, C.S.; Ackerman, J.J.H.; Balaban, R.S. Deuterium NMR Cerebral Imaging in Situ. Magn. Reson. Med. 1988, 8, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Zhu, X.-H.; Zhang, Y.; Mateescu, G.; Chen, W. Quantitative Assessment of Brain Glucose Metabolic Rates Using in Vivo Deuterium Magnetic Resonance Spectroscopy. J. Cereb. Blood Flow Metab. 2017, 37, 3518–3530. [Google Scholar] [CrossRef]
- Klein, P.D.; Klein, E.R. Stable Isotopes: Origins and Safety. J. Clin. Pharmacol. 1986, 26, 378–382. [Google Scholar] [CrossRef]
- Funk, A.M.; Anderson, B.L.; Wen, X.; Hever, T.; Khemtong, C.; Kovacs, Z.; Sherry, A.D.; Malloy, C.R. The Rate of Lactate Production from Glucose in Hearts Is Not Altered by Per-Deuteration of Glucose. J. Magn. Reson. 2017, 284, 86–93. [Google Scholar] [CrossRef]
- Zhang, W.; Sargent, D.J.; Mandrekar, S. An Adaptive Dose-Finding Design Incorporating Both Toxicity and Efficacy. Statist. Med. 2006, 25, 2365–2383. [Google Scholar] [CrossRef]
- Ben-Yoseph, O.; Kingsley, P.B.; Ross, B.D. Metabolic Loss of Deuterium from Isotopically Labeled Glucose. Magn. Reson. Med. 1994, 32, 405–409. [Google Scholar] [CrossRef]
- Browning, J.D.; Burgess, S.C. Use of 2H2O for Estimating Rates of Gluconeogenesis: Determination and Correction of Error Due to Transaldolase Exchange. Am. J. Physiol. Endocrinol. Metab. 2012, 303, E1304–E1312. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Leadlay, P.F.; Albery, W.J.; Knowles, J.R. Energetics of Triosephosphate Isomerase: Deuterium Isotope Effects in the Enzyme-Catalyzed Reaction. Biochemistry 1976, 15, 5617–5620. [Google Scholar] [CrossRef] [PubMed]
- Chandramouli, V.; Ekberg, K.; Schumann, W.C.; Wahren, J.; Landau, B.R. Origins of the Hydrogen Bound to Carbon 1 of Glucose in Fasting: Significance in Gluconeogenesis Quantitation. Am. J. Physiol. Endocrinol. Metab. 1999, 277, E717–E723. [Google Scholar] [CrossRef]
- Read, J.; Pearce, J.; Li, X.; Muirhead, H.; Chirgwin, J.; Davies, C. The Crystal Structure of Human Phosphoglucose Isomerase at 1.6 Å Resolution: Implications for Catalytic Mechanism, Cytokine Activity and Haemolytic Anaemia. J. Mol. Biol. 2001, 309, 447–463. [Google Scholar] [CrossRef]
- Lorentzen, E.; Siebers, B.; Hensel, R.; Pohl, E. Mechanism of the Schiff Base Forming Fructose-1,6-Bisphosphate Aldolase: Structural Analysis of Reaction Intermediates. Biochemistry 2005, 44, 4222–4229. [Google Scholar] [CrossRef] [PubMed]
- Reis, M.; Alves, C.N.; Lameira, J.; Tuñón, I.; Martí, S.; Moliner, V. The Catalytic Mechanism of Glyceraldehyde 3-Phosphate Dehydrogenase from Trypanosoma Cruzi Elucidated via the QM/MM Approach. Phys. Chem. Chem. Phys. 2013, 15, 3772–3785. [Google Scholar] [CrossRef][Green Version]
- Lebioda, L.; Stec, B. Mechanism of Enolase: The Crystal Structure of Enolase-Mg2+-2-Phosphoglycerate/Phosphoenolpyruvate Complex at 2.2-Å Resolution. Biochemistry 1991, 30, 2817–2822. [Google Scholar] [CrossRef]
- Hanau, S.; Montin, K.; Cervellati, C.; Magnani, M.; Dallocchio, F. 6-Phosphogluconate Dehydrogenase Mechanism. J. Biol. Chem. 2010, 285, 21366–21371. [Google Scholar] [CrossRef] [PubMed]
- Funk, A.M.; Wen, X.; Hever, T.; Maptue, N.R.; Khemtong, C.; Sherry, A.D.; Malloy, C.R. Effects of Deuteration on Transamination and Oxidation of Hyperpolarized 13C-Pyruvate in the Isolated Heart. J. Magn. Reson. 2019, 301, 102–108. [Google Scholar] [CrossRef]
- Aguayo, J.B.; McLennan, I.J.; Graham, C.; Cheng, H.-M. Dynamic Monitoring of Corneal Carbohydrate Metabolism Using High-Resolution Deuterium NMR Spectroscopy. Exp. Eye Res. 1988, 47, 337–343. [Google Scholar] [CrossRef]
- Lewis, J.E.; Costantini, F.; Mims, J.; Chen, X.; Furdui, C.M.; Boothman, D.A.; Kemp, M.L. Genome-Scale Modeling of NADPH-Driven β-Lapachone Sensitization in Head and Neck Squamous Cell Carcinoma. Antioxid. Redox Signal. 2018, 29, 937–952. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, J.M.; Kudej, R.K.; LaNoue, K.F.; Vatner, S.F.; Lewandowski, E.D. Limited Transfer of Cytosolic NADH into Mitochondria at High Cardiac Workload. Am. J. Physiol. Heart Circ. Physiol. 2004, 286, H2237–H2242. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.A.; Kobayashi, R.; Murakami, T.; Shimomura, Y. Regulation of Branched-Chain α-Keto Acid Dehydrogenase Kinase Expression in Rat Liver. J. Nutr. 2001, 131, 841S–845S. [Google Scholar] [CrossRef]
- McKenna, M.C.; Dienel, G.A.; Sonnewald, U.; Waagepetersen, H.S.; Schousboe, A. Energy Metabolism of the Brain. In Basic Neurochemistry; Elsevier: Amsterdam, The Netherlands, 2012; pp. 200–231. ISBN 978-0-12-374947-5. [Google Scholar]
- Kreis, F.; Wright, A.J.; Hesse, F.; Fala, M.; Hu, D.; Brindle, K.M. Measuring Tumor Glycolytic Flux in Vivo by Using Fast Deuterium MRI. Radiology 2020, 294, 289–296. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).