USP19 and RPL23 as Candidate Prognostic Markers for Advanced-Stage High-Grade Serous Ovarian Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Specimens
2.2. Library Preparation and mRNA Sequencing
2.3. Transcriptome Analysis
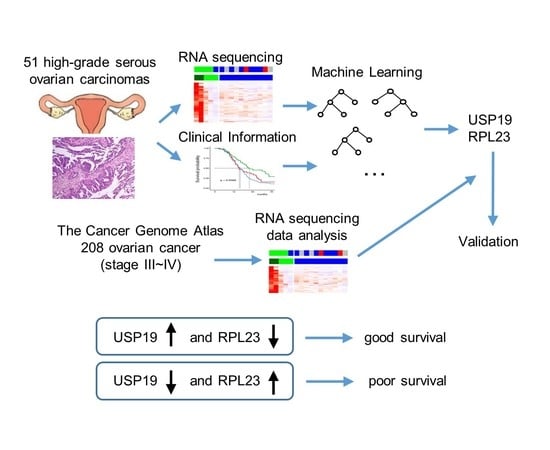
2.4. Identification of Candidate Prognostic Biomarker Genes
2.5. Model Validation
2.6. Validation of the Prognostic Value of USP19 and RPL23 Expression
2.7. Statistical Analysis
3. Results
3.1. Clinical Characteristics of Patients with HGSC
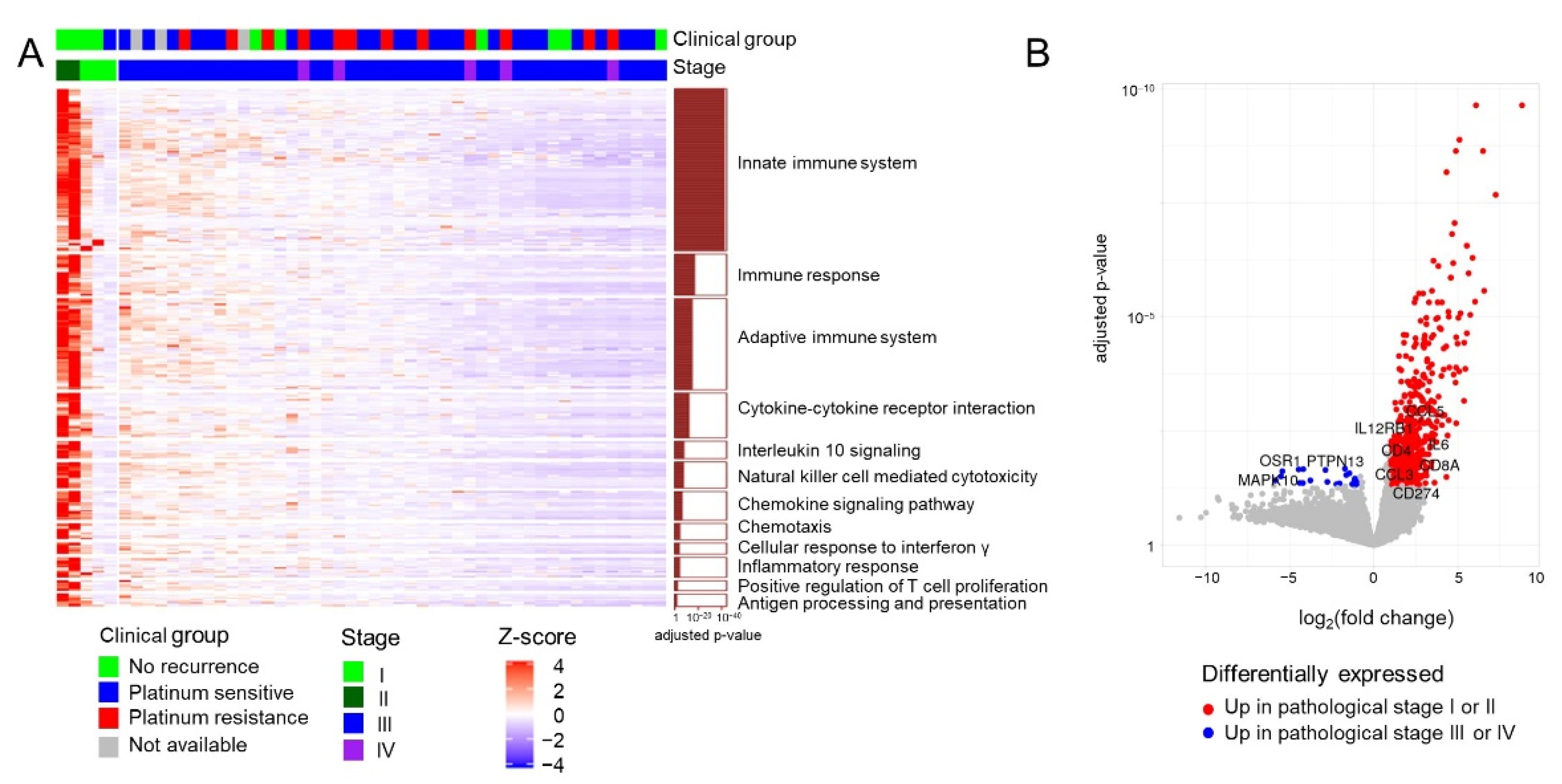
3.2. Gene Expression Profiles in 51 Patients with HGSC
3.3. Identification of Candidate Prognostic Biomarker Genes
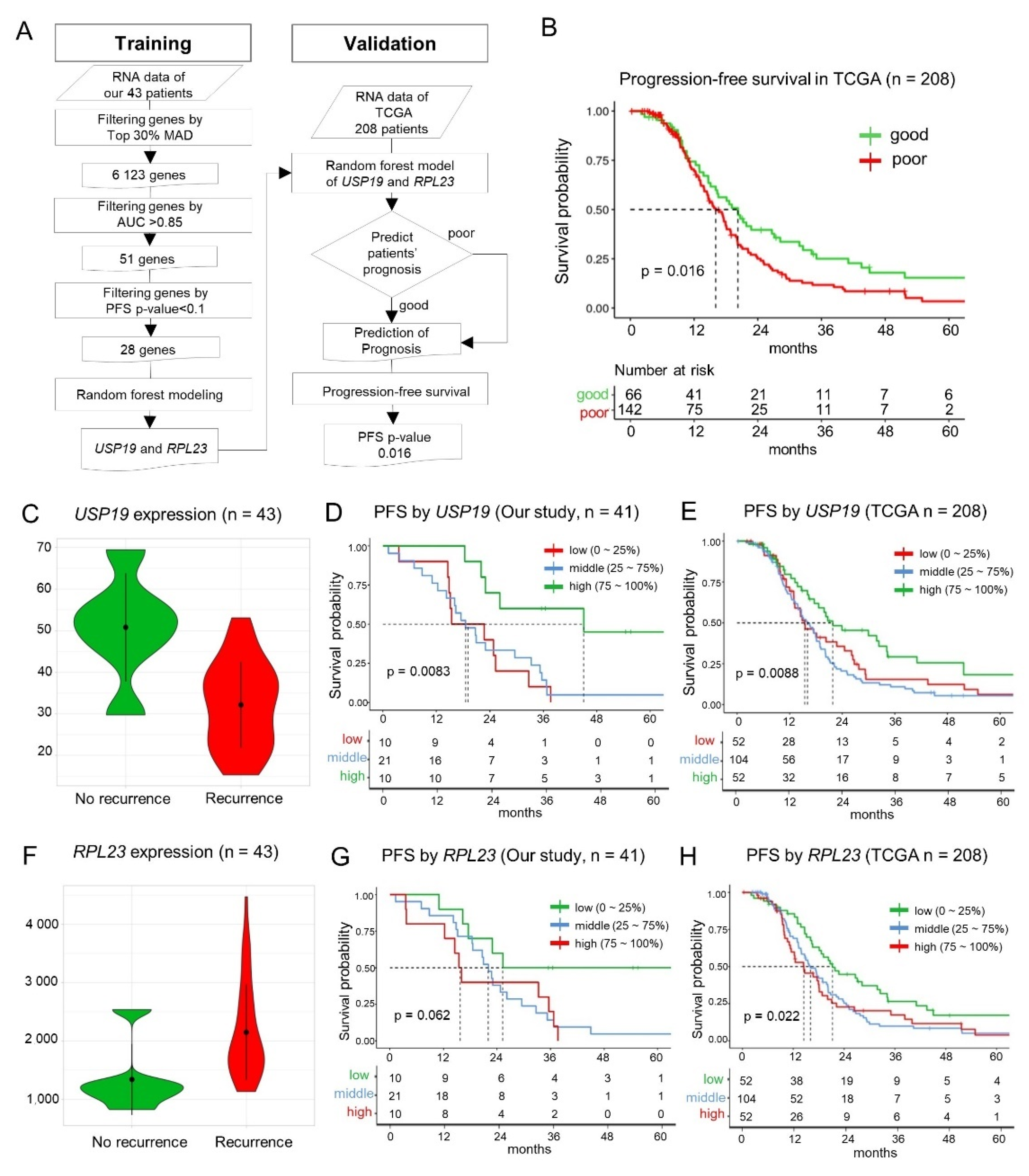
3.4. Machine Learning—Based Identification of Key Prognostic Genes
3.5. Validation of the Random Forest Model Employing USP19 and RPL23 Expression
3.6. Prognostic Value of USP19 and RPL23 Expression—Lower USP19 and Higher RPL23 Expression in Patients with Recurrence
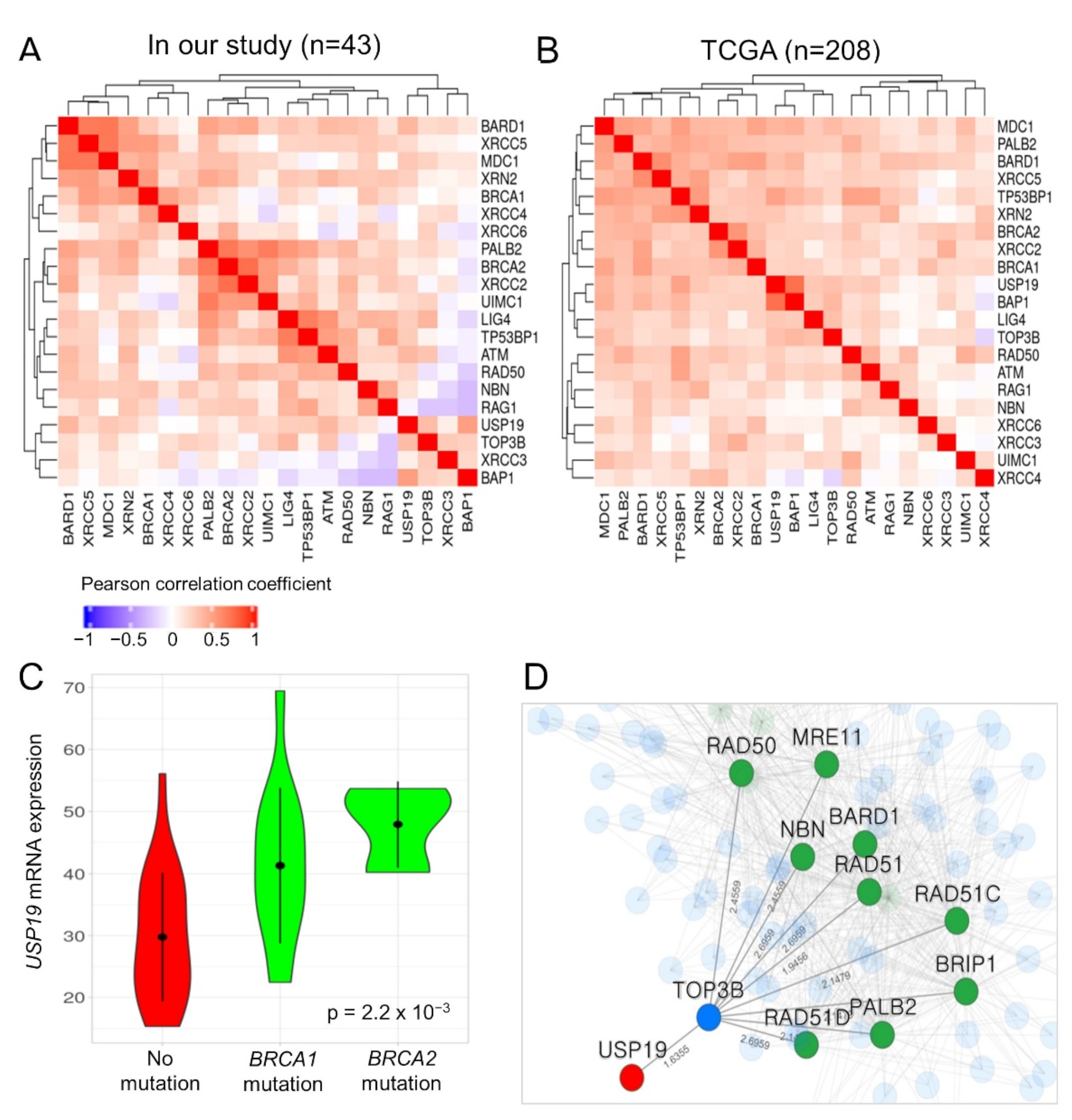
3.7. Functional Association between USP19 and DNA Double-Strand Break (DSB) Repair Genes such as BRCA1/2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reid, B.M.; Permuth, J.B.; Sellers, T.A. Epidemiology of ovarian cancer: A review. Cancer Biol. Med. 2017, 14, 9–32. [Google Scholar] [CrossRef] [PubMed]
- Webb, P.M.; Jordan, S.J. Epidemiology of epithelial ovarian cancer. Best Pract. Res. Clin. Obstet. Gynaecol. 2017, 41, 3–14. [Google Scholar] [CrossRef]
- Lisio, M.-A.; Fu, L.; Goyeneche, A.; Gao, Z.-H.; Telleria, C. High-Grade Serous Ovarian Cancer: Basic Sciences, Clinical and Therapeutic Standpoints. Int. J. Mol. Sci. 2019, 20, 952. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.M.; Kannan, L.; Geistlinger, L.; Kofia, V.; Safikhani, Z.; Gendoo, D.M.; Parmigiani, G.; Birrer, M.J.; Haibe-Kains, B.; Waldron, L. Consensus on Molecular Subtypes of High-Grade Serous Ovarian Carcinoma. Clin. Cancer Res. 2018, 24, 5037–5047. [Google Scholar] [CrossRef] [PubMed]
- Verhaak, R.G.; Tamayo, P.; Yang, J.-Y.; Hubbard, D.; Zhang, H.; Creighton, C.J.; Fereday, S.; Lawrence, M.; Carter, S.L.; Mermel, C.; et al. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J. Clin. Investig. 2012, 123, 517–525. [Google Scholar] [CrossRef]
- Helland, A.; Anglesio, M.S.; George, J.; Cowin, P.A.; Johnstone, C.N.; House, C.M.; Sheppard, K.; Etemadmoghadam, D.; Melnyk, N.; Rustgi, A.K.; et al. Deregulation of MYCN, LIN28B and LET7 in a Molecular Subtype of Aggressive High-Grade Serous Ovarian Cancers. PLoS ONE 2011, 6, e18064. [Google Scholar] [CrossRef]
- Tothill, R.; Tinker, A.V.; George, J.; Brown, R.; Fox, S.; Lade, S.; Johnson, D.S.; Trivett, M.K.; Etemadmoghadam, D.; Locandro, B.; et al. Novel Molecular Subtypes of Serous and Endometrioid Ovarian Cancer Linked to Clinical Outcome. Clin. Cancer Res. 2008, 14, 5198–5208. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- McCarthy, D.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Ducie, J.; Dao, F.; Considine, M.; Olvera, N.; Shaw, P.A.; Kurman, R.J.; Shih, I.-M.; Soslow, R.A.; Cope, L.; Levine, D.A. Molecular analysis of high-grade serous ovarian carcinoma with and without associated serous tubal intra-epithelial carcinoma. Nat. Commun. 2017, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Sing, T.; Sander, O.; Beerenwinkel, N.; Lengauer, T. ROCR: Visualizing classifier performance in R. Bioinformatics 2005, 21, 3940–3941. [Google Scholar] [CrossRef]
- Therneau, T.M.; Grambsch, P.M. Modeling Survival Data, Extending the Cox Model; Springer: New York, NY, USA, 2000. [Google Scholar]
- Choi, M.C.; Hwang, S.; Kim, S.; Jung, S.G.; Park, H.; Joo, W.D.; Song, S.H.; Lee, C.; Kim, T.-H.; Kang, H.; et al. Clinical Impact of Somatic Variants in Homologous Recombination Repair-Related Genes in Ovarian High-Grade Serous Carcinoma. Cancer Res. Treat. 2020, 52, 634–644. [Google Scholar] [CrossRef]
- Shahriyari, L.; Abdel-Rahman, M.; Cebulla, C. BAP1 expression is prognostic in breast and uveal melanoma but not colon cancer and is highly positively correlated with RBM15B and USP19. PLoS ONE 2019, 14, e0211507. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Kim, C.Y.; Yang, S.; Kim, E.; Hart, T.; Marcotte, E.; Lee, I. HumanNet v2: Human gene networks for disease research. Nucleic Acids Res. 2019, 47, D573–D580. [Google Scholar] [CrossRef]
- James, F.R.; Jiminez-Linan, M.; Alsop, J.; Mack, M.; Song, H.; Brenton, J.D.; Pharoah, P.D.P.; Ali, H.R. Association between tumour infiltrating lymphocytes, histotype and clinical outcome in epithelial ovarian cancer. BMC Cancer 2017, 17, 657. [Google Scholar] [CrossRef]
- Qi, Y.; Li, X.; Chang, C.-K.; Xu, F.; He, Q.; Zhao, Y.; Wu, L. Ribosomal protein L23 negatively regulates cellular apoptosis via the RPL23/Miz-1/c-Myc circuit in higher-risk myelodysplastic syndrome. Sci. Rep. 2017, 7, 2323. [Google Scholar] [CrossRef]
- Shi, Y.; Zhai, H.; Wang, X.; Han, Z.; Liu, C.; Lan, M.; Du, J.; Guo, C.; Zhang, Y.; Wu, K.; et al. Ribosomal proteins S13 and L23 promote multidrug resistance in gastric cancer cells by suppressing drug-induced apoptosis. Exp. Cell Res. 2004, 296, 337–346. [Google Scholar] [CrossRef]
- Newton, T.R.; Parsons, P.G.; Lincoln, D.J.; Cummings, M.C.; Wyld, D.K.; Webb, P.M.; Green, A.C.; Boyle, G.M. Expression profiling correlates with treatment response in women with advanced serous epithelial ovarian cancer. Int. J. Cancer 2006, 119, 875–883. [Google Scholar] [CrossRef]
- Hassink, G.C.; Zhao, B.; Sompallae, R.; Altun, M.; Gastaldello, S.; Zinin, N.V.; Masucci, M.G.; Lindsten, K. The ER-resident ubiquitin-specific protease 19 participates in the UPR and rescues ERAD substrates. EMBO Rep. 2009, 10, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Tian, S.; Chen, Y.; Zhang, C.; Xie, W.; Xia, X.; Cui, J.; Wang, R. USP 19 modulates autophagy and antiviral immune responses by deubiquitinating Beclin-1. EMBO J. 2016, 35, 866–880. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.-H.; Choi, J.-H.; Park, J.-H.; Cho, H.-J.; Park, J.-J.; Lee, E.-J.; Li, L.; Choi, Y.-K.; Baek, K.-H. Ubiquitin specific protease 19 involved in transcriptional repression of retinoic acid receptor by stabilizing CORO2A. Oncotarget 2016, 7, 34759–34772. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kumari, N.; Jaynes, P.W.; Saei, A.; Iyengar, P.; Richard, J.L.C.; Eichhorn, P.J.A. The roles of ubiquitin modifying enzymes in neoplastic disease. Biochim. Biophys. Acta Rev. Cancer 2017, 1868, 456–483. [Google Scholar] [CrossRef]
- Lai, K.P.; Chen, J.; Tse, W.K.F. Role of Deubiquitinases in Human Cancers: Potential Targeted Therapy. Int. J. Mol. Sci. 2020, 21, 2548. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Guo, J.; North, B.J.; Wang, B.; Cui, C.-P.; Li, H.; Tao, K.; Zhang, L.; Wei, W. Functional analysis of deubiquitylating enzymes in tumorigenesis and development. Biochim. Biophys. Acta Rev. Cancer 2019, 1872, 188312. [Google Scholar] [CrossRef]
- Wu, M.; Tu, H.-Q.; Chang, Y.; Tan, B.; Wang, G.; Zhou, J.; Wang, L.; Mu, R.; Zhang, W.-N. USP19 deubiquitinates HDAC1/2 to regulate DNA damage repair and control chromosomal stability. Oncotarget 2017, 8, 2197–2208. [Google Scholar] [CrossRef]
- Hu, W.; Su, Y.; Fei, X.; Wang, X.; Zhang, G.; Su, C.; Du, T.; Yang, T.; Wang, G.; Tang, Z.; et al. Ubiquitin specific peptidase 19 is a prognostic biomarker and affect the proliferation and migration of clear cell renal cell carcinoma. Oncol. Rep. 2020, 43, 1964–1974. [Google Scholar] [CrossRef] [PubMed]
- He, W.-T.; Zheng, X.-M.; Zhang, Y.-H.; Gao, Y.; Song, A.-X.; van der Goot, G.; Hu, H.-Y. Cytoplasmic Ubiquitin-Specific Protease 19 (USP19) Modulates Aggregation of Polyglutamine-Expanded Ataxin-3 and Huntingtin through the HSP90 Chaperone. PLoS ONE 2016, 11, e0147515. [Google Scholar] [CrossRef]
- Zhang, T.; Wallis, M.; Petrovic, V.; Challis, J.; Kalitsis, P.; Hudson, D.F. Loss of TOP3B leads to increased R-loop formation and genome instability. Open Biol. 2019, 9, 190222. [Google Scholar] [CrossRef]
- Morales, J.C.; Richard, P.; Patidar, P.L.; Motea, E.A.; Dang, T.; Manley, J.; Boothman, D.A. XRN2 Links Transcription Termination to DNA Damage and Replication Stress. PLoS Genet. 2016, 12, e1006107. [Google Scholar] [CrossRef] [PubMed]
- Incorvaia, L.; Passiglia, F.; Rizzo, S.; Galvano, A.; Listì, A.; Barraco, N.; Maragliano, R.; Calò, V.; Natoli, C.; Ciaccio, M.; et al. “Back to a false normality”: New intriguing mechanisms of resistance to PARP inhibitors. Oncotarget 2016, 8, 23891–23904. [Google Scholar] [CrossRef]
- Garvin, A.J. Beyond reversal: Ubiquitin and ubiquitin-like proteases and the orchestration of the DNA double strand break repair response. Biochem. Soc. Trans. 2019, 47, 1881–1893. [Google Scholar] [CrossRef] [PubMed]
| Characteristic | Overall (n = 51) | % | |
|---|---|---|---|
| Age (Years), Median (Range) | 55 (36–77) | ||
| Cancer site | |||
| Ovary | 49 | 96.0 | |
| Fallopian tube | 1 | 2.0 | |
| Peritoneum | 1 | 2.0 | |
| Stage | |||
| I | 3 | 5.9 | |
| II | 2 | 3.9 | |
| III | 41 | 80.4 | |
| IV | 5 | 9.8 | |
| Platinum sensitivity | |||
| No recurrence | 10 | 19.6 | |
| Recurrence | |||
| Platinum sensitive | 26 | 51.0 | |
| Platinum resistant | 12 | 23.5 | |
| Lost to follow up | 3 | 5.9 | |
| Characteristic | Our Study (Training Set) | TCGA (Validation Set) | p-Value | |
|---|---|---|---|---|
| Number of patients | 43 | 208 | - | |
| Age (mean years, range) | 56 (37–77) | 59 (31–87) | 0.073 | |
| Progression-free survival (median months, 95% confidence interval) | 22.8 (18.4–32.8) | 17.5 (14.9–19.9) | 0.089 | |
| Overall survival (median months, 95% confidence interval) | Not reached | 45.4 (41.9–53.9) | 0.0012 | |
| Stage | III | 38 (88.4%) | 183 (88.0%) | 1.0 |
| IV | 5 (11.6%) | 25 (12.0%) | ||
| BRCA1/2 mutation | Germline | 14 (32.6%) | 1 (0.5%) | 9.5 × 10−6 |
| Somatic | 3 (7.0%) | 9 (4.3%) | ||
| Germline + somatic | 0 | 2 (1.0%) | ||
| No mutation | 25 (58.1%) | 122 (58.7%) | ||
| Not available | 1 (2.3%) | 74 (35.6%) | ||
| Cohort | Model | Variable | Coefficient | Exp (coef.) = Hazard Ratio | SE (coef.) | p-Value |
|---|---|---|---|---|---|---|
| This study (n = 42) | USP19 exp. | USP19 | −0.040 | 0.96 | 0.014 | 0.0048 * |
| BRCA mutation | BRCA | −0.15 | 0.86 | 0.35 | 0.67 | |
| USP19 exp. + BRCA mutation | USP19 # | −0.056 | 0.95 | 0.018 | 0.0018 * | |
| BRCA | 0.73 | 2.07 | 0.45 | 0.10 | ||
| TCGA (n = 134) | USP19 exp. | USP19 | −0.014 | 0.99 | 0.0085 | 0.091 |
| BRCA mutation | BRCA | −0.62 | 0.54 | 0.35 | 0.081 | |
| USP19 exp. + BRCA mutation | USP19 # | −0.023 | 0.98 | 0.0097 | 0.017 * | |
| BRCA | −0.89 | 0.41 | 0.38 | 0.018 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, H.; Choi, M.C.; Kim, S.; Jeong, J.-Y.; Kwon, A.-Y.; Kim, T.-H.; Kim, G.; Joo, W.D.; Park, H.; Lee, C.; et al. USP19 and RPL23 as Candidate Prognostic Markers for Advanced-Stage High-Grade Serous Ovarian Carcinoma. Cancers 2021, 13, 3976. https://doi.org/10.3390/cancers13163976
Kang H, Choi MC, Kim S, Jeong J-Y, Kwon A-Y, Kim T-H, Kim G, Joo WD, Park H, Lee C, et al. USP19 and RPL23 as Candidate Prognostic Markers for Advanced-Stage High-Grade Serous Ovarian Carcinoma. Cancers. 2021; 13(16):3976. https://doi.org/10.3390/cancers13163976
Chicago/Turabian StyleKang, Haeyoun, Min Chul Choi, Sewha Kim, Ju-Yeon Jeong, Ah-Young Kwon, Tae-Hoen Kim, Gwangil Kim, Won Duk Joo, Hyun Park, Chan Lee, and et al. 2021. "USP19 and RPL23 as Candidate Prognostic Markers for Advanced-Stage High-Grade Serous Ovarian Carcinoma" Cancers 13, no. 16: 3976. https://doi.org/10.3390/cancers13163976
APA StyleKang, H., Choi, M. C., Kim, S., Jeong, J.-Y., Kwon, A.-Y., Kim, T.-H., Kim, G., Joo, W. D., Park, H., Lee, C., Song, S. H., Jung, S. G., Hwang, S., & An, H. J. (2021). USP19 and RPL23 as Candidate Prognostic Markers for Advanced-Stage High-Grade Serous Ovarian Carcinoma. Cancers, 13(16), 3976. https://doi.org/10.3390/cancers13163976