Plasma Based Protein Signatures Associated with Small Cell Lung Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Specimen
2.2. SCLC Cell Line-Derived Conditioned Media
2.3. Mass Spectrometry Analyses of Human Plasmas
2.4. Mass Spectrometry Analyses of SCLC Cell Line Conditioned Media
2.5. Ingenuity Pathway Enrichment Analysis
2.6. Statistical Analysis
3. Results
3.1. Proteomic Profiling Reveals Signatures Associated with Oncogenic Drivers Manifest in Plasmas at Early Stages of SCLC
3.2. Intersection of SCLC-Associated Protein Signatures between Human and SCLC Cell Line-Derived Conditioned Medium
3.3. Proteomic Signatures in Plasmas Collected within One Year Prior to Diagnosis of SCLC
3.4. Proteomic Findings in Plasmas Collected More Than One Year Prior to DIAGNOSIS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Small-Cell Lung Cancer Cell Lines | |||
|---|---|---|---|
| Cell Line | Type | Neuroendocrine Status | Subtype ‡ |
| HCC4002 | SCLC | NE | SCLC-N |
| H209 | SCLC | NE | SCLC-A |
| H2195 | SCLC | NE | SCLC-A |
| H345 | SCLC | NE | SCLC-A |
| H524 | SCLC | NE | SCLC-N |
| H69 | SCLC | NE | SCLC-A |
| HCC4001 | SCLC | NE | SCLC-N |
| HCC4003 | SCLC | NE | SCLC-A |
| HCC4004 | SCLC | NE | SCLC-N |
| HCC4005 | SCLC | NE | SCLC-N |
| H1607 | SCLC | Non-NE | SCLC-Y |
| H211 | SCLC | Non-NE | SCLC-P |
| H2679 | SCLC | Non-NE | SCLC-P |
| H526 | SCLC | Non-NE | SCLC-Y |
| H69AD | SCLC | Non-NE | SCLC-Y |
| H82 | SCLC | Non-NE | SCLC-P |
| H1048 | SCLC | Non-NE | SCLC-P |
| MYC-Associated Target | Reference(s) Linking to EMT |
|---|---|
| ANKHD1 | [53] |
| ANPEP | [54,55] |
| CDH12 | [56] |
| CDH13 | [57] |
| CIT | [58] |
| CLASP2 | [59] |
| DIAPH3 | [60] |
| EGFL7 | [61] |
| FBXO11 | [62] |
| FOXP2 | [63] |
| IQGAP3 | [64] |
| PAX1 | [65] |
| PLEKHA1 | [66] |
| THBS1 | [45,46] |
| TPM1 | [67] |
| TPX2 | [68] |
| YWHAZ | [44] |
| MDACC Early Stage SCLC Cohort | ||
| Top 5 Canonical Pathways | ||
| Pathway | p-Value | # Molecules |
| Actin Cytoskeleton Signaling | 4.01 × 109 | 9 |
| Epithelial Adherens Junction Signaling | 7.30 × 108 | 7 |
| Germ Cell-Sertoli Cell Junction Signaling | 2.38 × 106 | 6 |
| ILK Signaling | 5.67 × 106 | 6 |
| FAK Signaling | 7.47 × 106 | 5 |
| SCLC Pre-Diagnostic (0–1 year] | ||
| Top 5 Canonical Pathways | ||
| Pathway | p-Value | # Molecules |
| Epithelial Adherens Junction Signaling | 9.10 × 105 | 5 |
| Dilated Cardiomyopathy Signaling Pathway | 8.66 × 104 | 4 |
| Phagosome Maturation | 9.34 × 104 | 4 |
| Remodeling of Epithelial Adherens Junctions | 9.42 × 104 | 3 |
| Germ Cell-Sertoli Cell Junction Signaling | 1.39 × 103 | 4 |
| Top 5 Canonical Pathways | ||
|---|---|---|
| Pathway | p-Value | # Molecules |
| Integrin Signaling | 6.97 × 106 | 5 |
| VEGF Signaling | 8.01 × 106 | 4 |
| Paxillin Signaling | 1.15 × 105 | 4 |
| Actin Cytoskeleton Signaling | 1.50 × 105 | 5 |
| FAK Signaling | 1.65 × 105 | 4 |
References
- Park, K.S.; Liang, M.C.; Raiser, D.M.; Zamponi, R.; Roach, R.R.; Curtis, S.J.; Walton, Z.; Schaffer, B.E.; Roake, C.M.; Zmoos, A.F.; et al. Characterization of the cell of origin for small cell lung cancer. Cell Cycle 2011, 10, 2806–2815. [Google Scholar] [CrossRef]
- Van Meerbeeck, J.P.; Fennell, D.A.; De Ruysscher, D.K. Small-cell lung cancer. Lancet 2011, 378, 1741–1755. [Google Scholar] [CrossRef]
- Lee, C.B.; Morris, D.E.; Fried, D.B.; Socinski, M.A. Current and evolving treatment options for limited stage small cell lung cancer. Curr. Opin. Oncol. 2006, 18, 162–172. [Google Scholar] [CrossRef]
- Aberle, D.R.; Adams, A.M.; Berg, C.D.; Black, W.C.; Clapp, J.D.; Fagerstrom, R.M.; Gareen, I.F.; Gatsonis, C.; Marcus, P.M.; Sicks, J.D. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [CrossRef] [PubMed]
- de Koning, H.J.; van der Aalst, C.M.; de Jong, P.A.; Scholten, E.T.; Nackaerts, K.; Heuvelmans, M.A.; Lammers, J.J.; Weenink, C.; Yousaf-Khan, U.; Horeweg, N.; et al. Reduced Lung-Cancer Mortality with Volume CT Screening in a Randomized Trial. N. Engl. J. Med. 2020, 382, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Pasquinelli, M.M.; Kovitz, K.L.; Koshy, M.; Menchaca, M.G.; Liu, L.; Winn, R.; Feldman, L.E. Outcomes from a Minority-Based Lung Cancer Screening Program vs the National Lung Screening Trial. JAMA Oncol. 2018, 4, 1291–1293. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.; Pattanayak, P.; Szabo, E.; Pinsky, P. Characteristics and Outcomes of Small Cell Lung Cancer Detected by CT Screening. Chest 2018, 154, 1284–1290. [Google Scholar] [CrossRef]
- Guida, F.; Sun, N.; Bantis, L.E.; Muller, D.C.; Li, P.; Taguchi, A.; Dhillon, D.; Kundnani, D.L.; Patel, N.J.; Yan, Q.; et al. Assessment of Lung Cancer Risk on the Basis of a Biomarker Panel of Circulating Proteins. JAMA Oncol. 2018, 4, e182078. [Google Scholar] [CrossRef]
- Fahrmann, J.F.; Vykoukal, J.; Fleury, A.; Tripathi, S.; Dennison, J.B.; Murage, E.; Wang, P.; Yu, C.Y.; Capello, M.; Creighton, C.J.; et al. Association between plasma diacetylspermine and tumor spermine synthase with outcome in triple negative breast cancer. J. Natl. Cancer Inst. 2019, 112, 607–616. [Google Scholar] [CrossRef]
- Fahrmann, J.F.; Bantis, L.E.; Capello, M.; Scelo, G.; Dennison, J.B.; Patel, N.; Murage, E.; Vykoukal, J.; Kundnani, D.L.; Foretova, L.; et al. A Plasma-Derived Protein-Metabolite Multiplexed Panel for Early-Stage Pancreatic Cancer. J. Natl. Cancer Inst. 2019, 111, 372–379. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Taguchi, A.; Politi, K.; Pitteri, S.J.; Lockwood, W.W.; Faca, V.M.; Kelly-Spratt, K.; Wong, C.H.; Zhang, Q.; Chin, A.; Park, K.S.; et al. Lung cancer signatures in plasma based on proteome profiling of mouse tumor models. Cancer Cell 2011, 20, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Bork, E.; Hansen, M.; Urdal, P.; Paus, E.; Holst, J.J.; Schifter, S.; Fenger, M.; Engbaek, F. Early detection of response in small cell bronchogenic carcinoma by changes in serum concentrations of creatine kinase, neuron specific enolase, calcitonin, ACTH, serotonin and gastrin releasing peptide. Eur. J. Cancer Clin. Oncol. 1988, 24, 1033–1038. [Google Scholar] [CrossRef]
- Molina, R.; Auge, J.M.; Escudero, J.M.; Marrades, R.; Vinolas, N.; Carcereny, E.; Ramirez, J.; Filella, X. Mucins CA 125, CA 19.9, CA 15.3 and TAG-72.3 as tumor markers in patients with lung cancer: Comparison with CYFRA 21-1, CEA, SCC and NSE. Tumour Biol. 2008, 29, 371–380. [Google Scholar] [CrossRef]
- Pujol, J.L.; Quantin, X.; Jacot, W.; Boher, J.M.; Grenier, J.; Lamy, P.J. Neuroendocrine and cytokeratin serum markers as prognostic determinants of small cell lung cancer. Lung Cancer 2003, 39, 131–138. [Google Scholar] [CrossRef]
- Shibayama, T.; Ueoka, H.; Nishii, K.; Kiura, K.; Tabata, M.; Miyatake, K.; Kitajima, T.; Harada, M. Complementary roles of pro-gastrin-releasing peptide (ProGRP) and neuron specific enolase (NSE) in diagnosis and prognosis of small-cell lung cancer (SCLC). Lung Cancer 2001, 32, 61–69. [Google Scholar] [CrossRef]
- Borglum, T.; Rehfeld, J.F.; Drivsholm, L.B.; Hilsted, L. Processing-independent quantitation of chromogranin a in plasma from patients with neuroendocrine tumors and small-cell lung carcinomas. Clin. Chem. 2007, 53, 438–446. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Holdenrieder, S.; von Pawel, J.; Dankelmann, E.; Duell, T.; Faderl, B.; Markus, A.; Siakavara, M.; Wagner, H.; Feldmann, K.; Hoffmann, H.; et al. Nucleosomes, ProGRP, NSE, CYFRA 21-1, and CEA in monitoring first-line chemotherapy of small cell lung cancer. Clin. Cancer Res. 2008, 14, 7813–7821. [Google Scholar] [CrossRef] [PubMed]
- Giovanella, L.; Piantanida, R.; Ceriani, L.; Bandera, M.; Novario, R.; Bianchi, L.; Roncari, G. Immunoassay of neuron-specific enolase (NSE) and serum fragments of cytokeratin 19 (CYFRA 21.1) as tumor markers in small cell lung cancer: Clinical evaluation and biological hypothesis. Int. J. Biol. Markers 1997, 12, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Mollaoglu, G.; Guthrie, M.R.; Böhm, S.; Brägelmann, J.; Can, I.; Ballieu, P.M.; Marx, A.; George, J.; Heinen, C.; Chalishazar, M.D.; et al. MYC Drives Progression of Small Cell Lung Cancer to a Variant Neuroendocrine Subtype with Vulnerability to Aurora Kinase Inhibition. Cancer Cell 2017, 31, 270–285. [Google Scholar] [CrossRef]
- Pedersen, N.; Mortensen, S.; Sørensen, S.B.; Pedersen, M.W.; Rieneck, K.; Bovin, L.F.; Poulsen, H.S. Transcriptional gene expression profiling of small cell lung cancer cells. Cancer Res. 2003, 63, 1943–1953. [Google Scholar] [PubMed]
- Peifer, M.; Fernandez-Cuesta, L.; Sos, M.L.; George, J.; Seidel, D.; Kasper, L.H.; Plenker, D.; Leenders, F.; Sun, R.; Zander, T.; et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat. Genet. 2012, 44, 1104–1110. [Google Scholar] [CrossRef] [PubMed]
- Umemura, S.; Mimaki, S.; Makinoshima, H.; Tada, S.; Ishii, G.; Ohmatsu, H.; Niho, S.; Yoh, K.; Matsumoto, S.; Takahashi, A.; et al. Therapeutic priority of the PI3K/AKT/mTOR pathway in small cell lung cancers as revealed by a comprehensive genomic analysis. J. Thorac. Oncol. 2014, 9, 1324–1331. [Google Scholar] [CrossRef] [PubMed]
- Rudin, C.M.; Poirier, J.T.; Byers, L.A.; Dive, C.; Dowlati, A.; George, J.; Heymach, J.V.; Johnson, J.E.; Lehman, J.M.; MacPherson, D.; et al. Molecular subtypes of small cell lung cancer: A synthesis of human and mouse model data. Nat. Rev. Cancer 2019, 19, 289–297. [Google Scholar] [CrossRef]
- Wilson, D.O.; Weissfeld, J.L.; Fuhrman, C.R.; Fisher, S.N.; Balogh, P.; Landreneau, R.J.; Luketich, J.D.; Siegfried, J.M. The Pittsburgh Lung Screening Study (PLuSS): Outcomes within 3 years of a first computed tomography scan. Am. J. Respir Crit Care Med. 2008, 178, 956–961. [Google Scholar] [CrossRef]
- Hankin, J.H.; Stram, D.O.; Arakawa, K.; Park, S.; Low, S.H.; Lee, H.P.; Yu, M.C. Singapore Chinese Health Study: Development, validation, and calibration of the quantitative food frequency questionnaire. Nutr. Cancer 2001, 39, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.-E.; Mann, M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 2006, 1, 2650–2660. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Rouillard, A.D.; Gundersen, G.W.; Fernandez, N.F.; Wang, Z.; Monteiro, C.D.; McDermott, M.G.; Ma’ayan, A. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, 2016, baw100. [Google Scholar] [CrossRef]
- Lachmann, A.; Xu, H.; Krishnan, J.; Berger, S.I.; Mazloom, A.R.; Ma’ayan, A. ChEA: Transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 2010, 26, 2438–2444. [Google Scholar] [CrossRef]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Pobbati, A.V.; Hong, W. Emerging roles of TEAD transcription factors and its coactivators in cancers. Cancer Biol. Ther. 2013, 14, 390–398. [Google Scholar] [CrossRef]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in mechanotransduction. Nature 2011, 474, 179–183. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, Y.; Tang, P.M.K.; Cheng, A.S.L.; Yu, J.; To, K.F.; Kang, W. Mechanotransduction and Cytoskeleton Remodeling Shaping YAP1 in Gastric Tumorigenesis. Int. J. Mol. Sci. 2019, 20, 1576. [Google Scholar] [CrossRef] [PubMed]
- Aseervatham, J. Cytoskeletal Remodeling in Cancer. Biology 2020, 9, 385. [Google Scholar] [CrossRef]
- Walker, C.; Mojares, E.; Del Río Hernández, A. Role of Extracellular Matrix in Development and Cancer Progression. Int. J. Mol. Sci. 2018, 19, 3028. [Google Scholar] [CrossRef]
- Lambert, A.W.; Weinberg, R.A. Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat. Rev. Cancer 2021, 21, 325–338. [Google Scholar] [CrossRef]
- Tanaka, S.; Jiang, Y.; Martinez, G.J.; Tanaka, K.; Yan, X.; Kurosaki, T.; Kaartinen, V.; Feng, X.H.; Tian, Q.; Wang, X.; et al. Trim33 mediates the proinflammatory function of Th17 cells. J. Exp. Med. 2018, 215, 1853–1868. [Google Scholar] [CrossRef] [PubMed]
- Prabhala, P.; Wright, D.B.; Robbe, P.; Bitter, C.; Pera, T.; Ten Hacken, N.H.T.; van den Berge, M.; Timens, W.; Meurs, H.; Dekkers, B.G.J. Laminin α4 contributes to airway remodeling and inflammation in asthma. Am. J. Physiol. Lung Cell Mol. Physiol. 2019, 317, L768–L777. [Google Scholar] [CrossRef] [PubMed]
- Ireland, A.S.; Micinski, A.M.; Kastner, D.W.; Guo, B.; Wait, S.J.; Spainhower, K.B.; Conley, C.C.; Chen, O.S.; Guthrie, M.R.; Soltero, D.; et al. MYC Drives Temporal Evolution of Small Cell Lung Cancer Subtypes by Reprogramming Neuroendocrine Fate. Cancer Cell 2020, 38, 60–78. [Google Scholar] [CrossRef]
- Patel, A.S.; Yoo, S.; Kong, R.; Sato, T.; Sinha, A.; Karam, S.; Bao, L.; Fridrikh, M.; Emoto, K.; Nudelman, G.; et al. Prototypical oncogene family Myc defines unappreciated distinct lineage states of small cell lung cancer. Sci. Adv. 2021, 7, eabc2578. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Girard, L.; Zhang, Y.A.; Haruki, T.; Papari-Zareei, M.; Stastny, V.; Ghayee, H.K.; Pacak, K.; Oliver, T.G.; Minna, J.D.; et al. Small cell lung cancer tumors and preclinical models display heterogeneity of neuroendocrine phenotypes. Transl. Lung Cancer Res. 2018, 7, 32–49. [Google Scholar] [CrossRef]
- Calvo, F.; Ege, N.; Grande-Garcia, A.; Hooper, S.; Jenkins, R.P.; Chaudhry, S.I.; Harrington, K.; Williamson, P.; Moeendarbary, E.; Charras, G.; et al. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat. Cell Biol. 2013, 15, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Chuang, S.M.; Yang, M.F.; Liao, J.W.; Yu, S.L.; Chen, J.J. A novel function of YWHAZ/β-catenin axis in promoting epithelial-mesenchymal transition and lung cancer metastasis. Mol. Cancer Res. 2012, 10, 1319–1331. [Google Scholar] [CrossRef] [PubMed]
- Jayachandran, A.; Anaka, M.; Prithviraj, P.; Hudson, C.; McKeown, S.J.; Lo, P.H.; Vella, L.J.; Goding, C.R.; Cebon, J.; Behren, A. Thrombospondin 1 promotes an aggressive phenotype through epithelial-to-mesenchymal transition in human melanoma. Oncotarget 2014, 5, 5782–5797. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xu, D.; Liu, Z.; Li, Y.; Zhang, C.; Gong, Y.; Jiang, Y.; Xing, B. THBS1 facilitates colorectal liver metastasis through enhancing epithelial–mesenchymal transition. Clin. Transl. Oncol. 2020, 22, 1730–1740. [Google Scholar] [CrossRef] [PubMed]
- Allen, A.; Gau, D.; Francoeur, P.; Sturm, J.; Wang, Y.; Martin, R.; Maranchie, J.; Duensing, A.; Kaczorowski, A.; Duensing, S.; et al. Actin-binding protein profilin1 promotes aggressiveness of clear-cell renal cell carcinoma cells. J. Biol. Chem. 2020, 295, 15636–15649. [Google Scholar] [CrossRef]
- de Semir, D.; Bezrookove, V.; Nosrati, M.; Scanlon, K.R.; Singer, E.; Judkins, J.; Rieken, C.; Wu, C.; Shen, J.; Schmudermayer, C.; et al. PHIP drives glioblastoma motility and invasion by regulating the focal adhesion complex. Proc. Natl. Acad. Sci. USA 2020, 117, 9064–9073. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Xu, H.; Gong, L.; Cao, D.; Jin, T.; Wang, Y.; Pi, J.; Yang, Y.; Yi, X.; Liao, D.; et al. Vinculin orchestrates prostate cancer progression by regulating tumor cell invasion, migration, and proliferation. Prostate 2021, 81, 347–356. [Google Scholar] [CrossRef]
- Vykoukal, J.; Sun, N.; Aguilar-Bonavides, C.; Katayama, H.; Tanaka, I.; Fahrmann, J.F.; Capello, M.; Fujimoto, J.; Aguilar, M.; Wistuba, I.I.; et al. Plasma-derived extracellular vesicle proteins as a source of biomarkers for lung adenocarcinoma. Oncotarget 2017, 8, 95466–95480. [Google Scholar] [CrossRef]
- Fahrmann, J.F.; Mao, X.; Irajizad, E.; Katayama, H.; Capello, M.; Tanaka, I.; Kato, T.; Wistuba, I.I.; Maitra, A.; Ostrin, E.J.; et al. Plasma-Derived Extracellular Vesicles Convey Protein Signatures that Reflect Pathophysiology in Lung and Pancreatic Adenocarcinomas. Cancers 2020, 12, 1147. [Google Scholar] [CrossRef]
- Ottesen, A.H.; Christensen, G.; Omland, T.; Røsjø, H. Glycosylated Chromogranin A: Potential Role in the Pathogenesis of Heart Failure. Curr. Heart Fail. Rep. 2017, 14, 478–488. [Google Scholar] [CrossRef] [PubMed]
- Yao, P.; Li, Y.; Shen, W.; Xu, X.; Zhu, W.; Yang, X.; Cao, J.; Xing, C. ANKHD1 silencing suppresses the proliferation, migration and invasion of CRC cells by inhibiting YAP1-induced activation of EMT. Am. J. Cancer Res. 2018, 8, 2311–2324. [Google Scholar]
- Kim, Y.; Han, D.; Min, H.; Jin, J.; Yi, E.C.; Kim, Y. Comparative proteomic profiling of pancreatic ductal adenocarcinoma cell lines. Mol. Cells 2014, 37, 888–898. [Google Scholar] [CrossRef] [PubMed]
- Vergara, D.; Simeone, P.; Franck, J.; Trerotola, M.; Giudetti, A.; Capobianco, L.; Tinelli, A.; Bellomo, C.; Fournier, I.; Gaballo, A.; et al. Translating epithelial mesenchymal transition markers into the clinic: Novel insights from proteomics. EuPA Open Proteom. 2016, 10, 31–41. [Google Scholar] [CrossRef]
- Ma, J.; Zhao, J.; Lu, J.; Wang, P.; Feng, H.; Zong, Y.; Ou, B.; Zheng, M.; Lu, A. Cadherin-12 enhances proliferation in colorectal cancer cells and increases progression by promoting EMT. Tumor Biol. 2016, 37, 9077–9088. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Yuan, H.; Meng, Z.; Yang, C.; Li, Z.; Li, M.; Zhang, Z.; Gan, Y.; Tu, H. Cadherin 13 Inhibits Pancreatic Cancer Progression and Epithelial-mesenchymal Transition by Wnt/β-Catenin Signaling. J. Cancer 2020, 11, 2101–2112. [Google Scholar] [CrossRef]
- Haiping, C.; Qi, X.; Dawei, L.; Qiang, W. Citron Rho-interacting serine/threonine kinase knockdown suppresses prostate cancer cell proliferation and metastasis by blocking Hippo-YAP pathway. Nan Fang Yi Ke Da Xue Xue Bao 2019, 39, 257–263. [Google Scholar] [CrossRef]
- Zhu, B.; Qi, L.; Liu, S.; Liu, W.; Ou, Z.; Chen, M.; Liu, L.; Zu, X.; Wang, J.; Li, Y. CLASP2 is involved in the EMT and early progression after transurethral resection of the bladder tumor. BMC Cancer 2017, 17, 105. [Google Scholar] [CrossRef] [PubMed]
- Morley, S.; You, S.; Pollan, S.; Choi, J.; Zhou, B.; Hager, M.H.; Steadman, K.; Spinelli, C.; Rajendran, K.; Gertych, A.; et al. Regulation of microtubule dynamics by DIAPH3 influences amoeboid tumor cell mechanics and sensitivity to taxanes. Sci. Rep. 2015, 5, 12136. [Google Scholar] [CrossRef]
- Luo, B.H.; Xiong, F.; Wang, J.P.; Li, J.H.; Zhong, M.; Liu, Q.L.; Luo, G.Q.; Yang, X.J.; Xiao, N.; Xie, B.; et al. Epidermal growth factor-like domain-containing protein 7 (EGFL7) enhances EGF receptor-AKT signaling, epithelial-mesenchymal transition, and metastasis of gastric cancer cells. PLoS ONE 2014, 9, e99922. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Lin, M.; Liu, Y.; Wang, Z.-W.; Zhu, X. Emerging role of F-box proteins in the regulation of epithelial-mesenchymal transition and stem cells in human cancers. Stem Cell Res. Ther. 2019, 10, 124. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, T.; Guo, M.; Li, Y.; Zhang, Q.; Tan, G.; Yu, L.; Tan, Y. FOXA2-Interacting FOXP2 Prevents Epithelial-Mesenchymal Transition of Breast Cancer Cells by Stimulating E-Cadherin and PHF2 Transcription. Front. Oncol. 2021, 11, 605025. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Qin, N.; Zhou, Q.; Chen, Y.; Huang, S.; Chen, B.; Shen, G.; Jia, H. Role of IQGAP3 in metastasis and epithelial–mesenchymal transition in human hepatocellular carcinoma. J. Transl. Med. 2017, 15, 176. [Google Scholar] [CrossRef]
- Siletz, A.; Schnabel, M.; Kniazeva, E.; Schumacher, A.J.; Shin, S.; Jeruss, J.S.; Shea, L.D. Dynamic transcription factor networks in epithelial-mesenchymal transition in breast cancer models. PLoS ONE 2013, 8, e57180. [Google Scholar] [CrossRef]
- Shapiro, I.M.; Cheng, A.W.; Flytzanis, N.C.; Balsamo, M.; Condeelis, J.S.; Oktay, M.H.; Burge, C.B.; Gertler, F.B. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 2011, 7, e1002218. [Google Scholar] [CrossRef] [PubMed]
- Kubo, E.; Hasanova, N.; Fatma, N.; Sasaki, H.; Singh, D.P. Elevated tropomyosin expression is associated with epithelial-mesenchymal transition of lens epithelial cells. J. Cell Mol. Med. 2013, 17, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Zheng, B.; Li, J.; Lv, X.; Zhang, H.; Yu, F.; Kong, L.; Li, Y.; Yu, M.; Fang, L.; et al. TPX2 level correlates with cholangiocarcinoma cell proliferation, apoptosis, and EMT. Biomed. Pharmacother. 2018, 107, 1286–1293. [Google Scholar] [CrossRef] [PubMed]
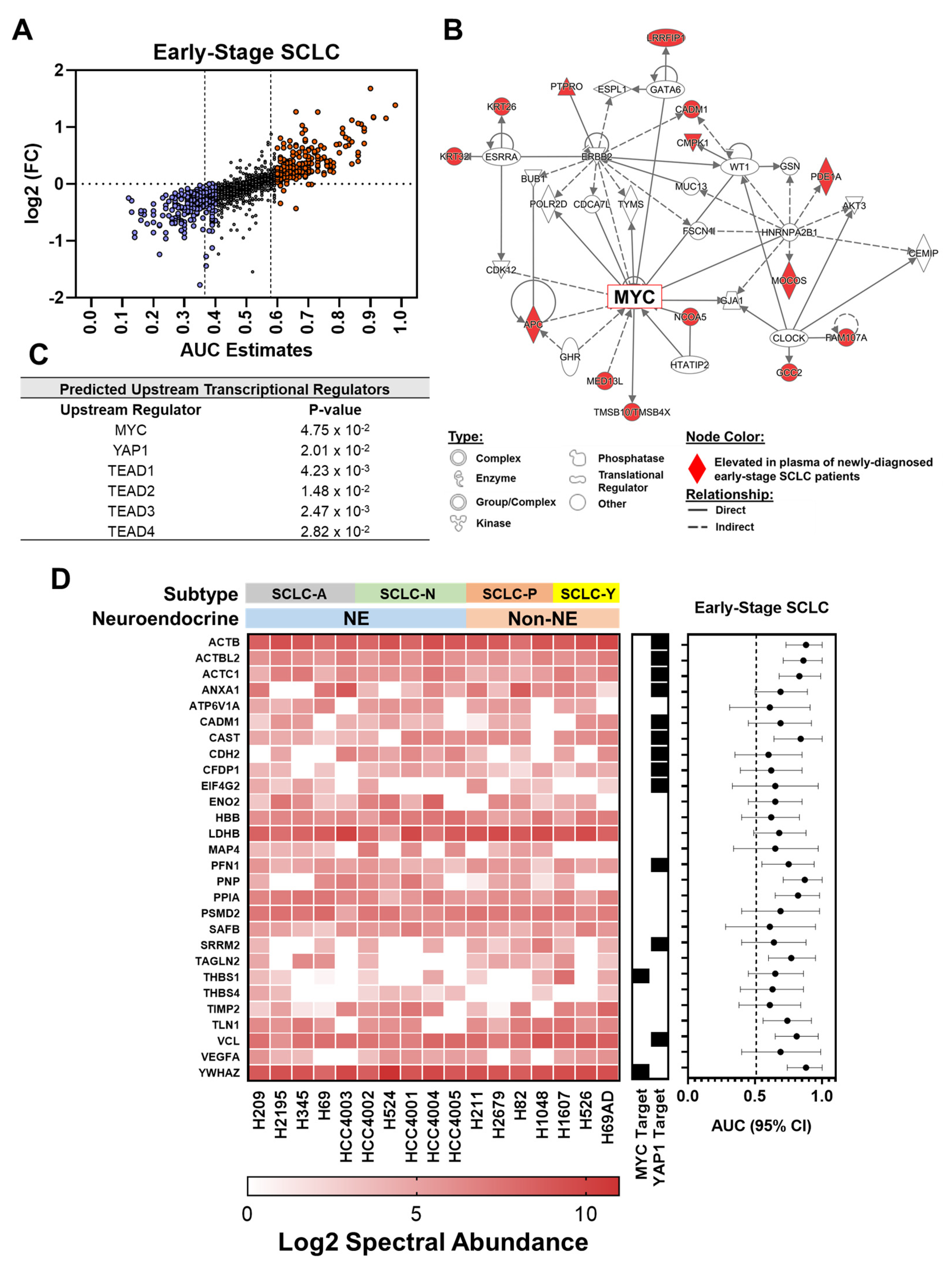
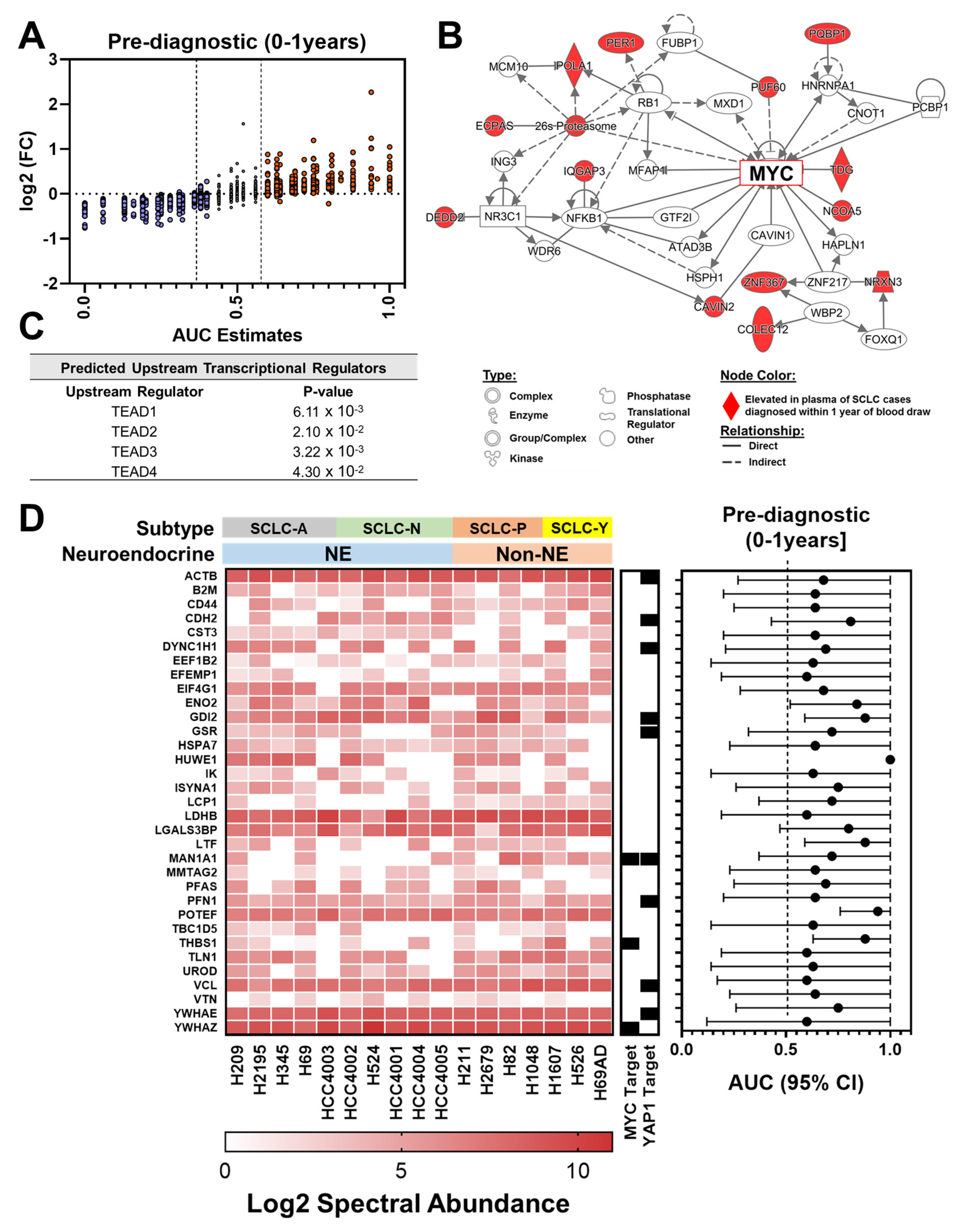
| Patient and Tumor Characteristics | Cases | Controls | p† |
|---|---|---|---|
| N | 15 | 15 | |
| Age, mean ± stdev | 67 ± 10 | 64 ± 5 | 0.330 |
| Sex, N (%) | |||
| Male | 8 (53.3%) | 8 (53.3%) | |
| Female | 7 (46.7%) | 7 (46.7%) | |
| Stage, N (%) | |||
| I | 6 (40%) | - | |
| II | 9 (60%) | - | |
| Smoking PYs, mean ± stdev | 63 ± 27 | 51 ± 18 | 0.200 |
| Patient and Tumor Characteristics | Cases | Controls |
|---|---|---|
| N | 15 | 15 |
| Age, mean ± stdev | 62.6 ± 8.7 | 62.5 ± 8.9 |
| Years from Dx, median (min/max) | 2.4 (0.7, 12.3) | - |
| Sex, N (%) | ||
| Female | 8 (53.3%) | 8 (53.3%) |
| Male | 7 (46.7%) | 7 (46.7%) |
| Smoking Status, N (%) | ||
| Former | 2 (13.3%) | 2 (13.3%) |
| Current | 12 (80.0%) | 12 (80.0%) |
| Never | 1 (6.7%) | 1 (6.7%) |
| Protein | At-Dx | Pre-Dx | Quantified in SCLC CM ‡ | MYC Downstream Target ║ | YAP1 Downstream Target ║ |
|---|---|---|---|---|---|
| Early Stage SCLC † | (0–1 Year] † | ||||
| ACTB | 0.88 (0.73–1.00) | 0.68 (0.27–1.00) | Yes | - | Yes |
| C9 | 0.60 (0.38–0.81) | 0.60 (0.19–1.00) | - | - | - |
| CA1 | 0.64 (0.43–0.85) | 0.60 (0.19–1.00) | - | - | - |
| CDH2 | 0.60 (0.35–0.85) | 0.81 (0.43–1.00) | Yes | - | Yes |
| COL6A6 | 0.61 (0.3–0.92) | 0.64 (0.23–1.00) | - | - | - |
| CPT2 | 0.78 (0.52–1.00) | 0.64 (0.20–1.00) | - | - | Yes |
| CRP | 0.68 (0.49–0.88) | 0.64 (0.25–1.00) | - | - | - |
| D2HGDH | 0.66 (0.46–0.87) | 0.63 (0.14–1.00) | - | - | - |
| ENO2 | 0.65 (0.45–0.85) | 0.84 (0.52–1.00) | Yes | - | - |
| KIF27 | 0.64 (0.34–0.94) | 0.75 (0.26–1.00) | - | - | - |
| KLHDC10 | 0.62 (0.30–0.94) | 0.63 (0.14–1.00) | - | - | Yes |
| LDHB | 0.68 (0.49–0.88) | 0.60 (0.19–1.00) | Yes | - | - |
| LONRF1 | 0.86 (0.66–1.00) | 0.94 (0.76–1.00) | - | - | - |
| MLPH | 0.75 (0.49–1.00) | 0.69 (0.21–1.00) | - | - | - |
| MYD88 | 0.70 (0.48–0.92) | 0.76 (0.41–1.00) | - | - | - |
| NCOA5 | 0.63 (0.37–0.88) | 0.60 (0.19–1.00) | - | - | - |
| OLFML2A | 0.66 (0.36–0.95) | 1.00 (1.00–1.00) | - | - | - |
| OPLAH | 0.60 (0.36–0.85) | 0.60 (0.21–0.99) | - | - | - |
| PER1 | 0.65 (0.31–1.00) | 0.64 (0.25–1.00) | - | Yes | - |
| PFN1 | 0.75 (0.55–0.94) | 0.64 (0.20–1.00) | Yes | - | Yes |
| PPBP | 0.62 (0.41–0.83) | 0.76 (0.41–1.00) | - | - | - |
| RAB17 | 0.61 (0.28–0.94) | 0.64 (0.25–1.00) | - | - | - |
| S100A12 | 0.73 (0.54–0.92) | 0.94 (0.76–1.00) | - | - | - |
| S100A8 | 0.69 (0.49–0.89) | 0.72 (0.32–1.00) | - | - | - |
| THBS1 | 0.65 (0.45–0.86) | 0.88 (0.63–1.00) | Yes | Yes | - |
| TLN1 | 0.74 (0.56–0.92) | 0.60 (0.19–1.00) | Yes | - | - |
| TRRAP | 0.66 (0.36–0.95) | 0.63 (0.14–1.00) | - | - | - |
| UNC80 | 0.65 (0.42–0.89) | 0.81 (0.43–1.00) | - | - | - |
| USP4 | 0.60 (0.38–0.81) | 0.94 (0.76–1.00) | - | - | - |
| VCL | 0.81 (0.65–0.97) | 0.60 (0.17–1.00) | Yes | - | Yes |
| YWHAZ | 0.88 (0.74–1.00) | 0.60 (0.12–1.00) | Yes | Yes | - |
| Protein | Cases (Mean ± SD) | Controls (Mean ± SD) | Odds Ratio † | 1-Sided p-Value | Elevated (AUC ≥ 0.60) in Case Plasmas ‡ |
|---|---|---|---|---|---|
| ACSL1 | 1.45 ± 0.28 | 1.3 ± 0.15 | 2.24 | 0.1000 | |
| AJM1 | 1.57 ± 0.65 | 1.27 ± 0.34 | 2.01 | 0.0880 | |
| AKAP9 | 2.2 ± 0.63 | 1.92 ± 0.35 | 2.02 | 0.1350 | |
| ALPI | 1.26 ± 0.09 | 1.17 ± 0.14 | 2.60 | 0.0630 | |
| APOD | 1.52 ± 1.39 | 1.05 ± 0.22 | 2.59 | 0.1110 | |
| C4BPA | 1.28 ± 0.17 | 1.14 ± 0.24 | 2.21 | 0.0580 | |
| C9 | 1.18 ± 0.14 | 1.05 ± 0.22 | 2.24 | 0.0550 | ✓ |
| CARD6 | 1.38 ± 1.29 | 0.93 ± 0.15 | 2.36 | 0.1430 | |
| CCDC115 | 1.12 ± 0.14 | 1.01 ± 0.06 | 9.43 | 0.0230 | |
| CDHR1 | 1.93 ± 2.24 | 1.05 ± 0.15 | 25.60 | 0.0960 | |
| CDKL1 | 1.09 ± 0.1 | 1.03 ± 0.07 | 2.18 | 0.0950 | |
| CTNND2 | 1.16 ± 0.29 | 0.98 ± 0.17 | 2.35 | 0.0800 | |
| CTPS1 | 1.57 ± 0.83 | 1.25 ± 0.24 | 2.11 | 0.1520 | |
| DDX31 | 0.88 ± 0.21 | 0.71 ± 0.08 | 10.87 | 0.0110 | |
| ERCC4 | 1.76 ± 1.11 | 1.22 ± 0.39 | 2.97 | 0.1000 | |
| FBXL8 | 1.09 ± 0.17 | 0.98 ± 0.11 | 2.37 | 0.0430 | |
| FGA | 2.07 ± 3.5 | 0.93 ± 0.21 | 4.74 | 0.1050 | |
| FGB | 1.83 ± 2.6 | 1.02 ± 0.2 | 2.32 | 0.1290 | |
| FGG | 2.25 ± 3.88 | 0.95 ± 0.2 | 13.01 | 0.0890 | |
| FUCA1 | 1.17 ± 0.23 | 1.01 ± 0.14 | 2.85 | 0.0520 | |
| HCFC2 | 1.35 ± 0.2 | 1.25 ± 0.06 | 2.72 | 0.0820 | ✓ |
| HEXD | 1.83 ± 2.23 | 0.95 ± 0.22 | 4.98 | 0.1050 | |
| HYDIN | 1.11 ± 0.26 | 0.96 ± 0.16 | 2.43 | 0.0850 | ✓ |
| IGHM | 1.94 ± 0.99 | 1.39 ± 0.41 | 2.49 | 0.0460 | ✓ |
| JMJD1C | 0.73 ± 0.13 | 0.65 ± 0.1 | 2.27 | 0.1040 | |
| KIFC2 | 2.66 ± 1.96 | 1.74 ± 0.45 | 5.02 | 0.0730 | |
| LAMA4 | 1.14 ± 0.29 | 0.97 ± 0.2 | 2.22 | 0.0910 | ✓ |
| LENG9 | 1.41 ± 0.16 | 1.28 ± 0.19 | 2.25 | 0.0860 | |
| MAP3K7 | 1.09 ± 0.18 | 1.01 ± 0.04 | 3.84 | 0.0910 | |
| MAPKAPK5-AS1 | 1.11 ± 0.5 | 0.87 ± 0.16 | 2.87 | 0.0510 | |
| MBD5 | 1 ± 0.09 | 0.93 ± 0.08 | 2.84 | 0.0480 | |
| MOV10L1 | 1.24 ± 0.16 | 1.13 ± 0.21 | 2.03 | 0.1380 | |
| MX1 | 1.23 ± 0.09 | 1.15 ± 0.12 | 2.34 | 0.0870 | |
| NACA | 1.46 ± 0.18 | 1.35 ± 0.2 | 2.00 | 0.1230 | |
| NLN | 1.21 ± 0.11 | 1.13 ± 0.12 | 2.27 | 0.0850 | |
| NUCB2 | 1.24 ± 0.12 | 1.14 ± 0.18 | 2.16 | 0.0960 | |
| OTUD6A | 0.9 ± 0.11 | 0.83 ± 0.09 | 2.31 | 0.0840 | |
| PARD3 | 1.4 ± 0.62 | 1.14 ± 0.22 | 2.26 | 0.1390 | |
| PCNT | 1.11 ± 0.28 | 0.9 ± 0.27 | 2.56 | 0.0640 | |
| PDE4DIP | 1.74 ± 1.12 | 1.28 ± 0.35 | 2.28 | 0.1230 | |
| PHC1 | 0.84 ± 0.31 | 0.7 ± 0.04 | 3.05 | 0.1110 | |
| PHRF1 | 1.03 ± 0.48 | 0.84 ± 0.17 | 2.11 | 0.1320 | |
| PIEZO1 | 1.34 ± 0.24 | 1.19 ± 0.18 | 2.27 | 0.1090 | |
| PLEC | 1.29 ± 0.24 | 1.09 ± 0.25 | 2.85 | 0.0520 | |
| PML | 1.15 ± 0.12 | 0.94 ± 0.29 | 3.36 | 0.0460 | |
| POLR1B | 3.17 ± 0.7 | 2.74 ± 0.62 | 2.23 | 0.1020 | |
| RBFA | 1.26 ± 0.37 | 1.04 ± 0.29 | 2.06 | 0.1060 | |
| RCCD1 | 1.15 ± 0.1 | 1.05 ± 0.09 | 6.71 | 0.0220 | |
| RNASE4 | 1.29 ± 0.67 | 1.05 ± 0.16 | 2.58 | 0.0970 | |
| RTKN | 1.3 ± 0.62 | 1.07 ± 0.09 | 2.41 | 0.1440 | ✓ |
| SLC4A10 | 1.51 ± 0.33 | 1.25 ± 0.2 | 3.75 | 0.0320 | |
| SLC6A15 | 1.22 ± 0.41 | 1.05 ± 0.06 | 2.75 | 0.1200 | |
| SPANXA2-OT1 | 1.78 ± 1.47 | 1.35 ± 0.15 | 2.01 | 0.1480 | |
| THRAP3 | 1.17 ± 0.19 | 1.04 ± 0.18 | 2.20 | 0.0920 | |
| TNKS1BP1 | 0.96 ± 0.07 | 0.92 ± 0.05 | 2.12 | 0.1120 | |
| TNNI3K | 2.36 ± 1.6 | 1.71 ± 0.24 | 8.87 | 0.0500 | |
| TRIM33 | 2.17 ± 4.2 | 0.73 ± 0.25 | 38.99 | 0.0720 | ✓ |
| TTC6 | 1.12 ± 0.86 | 0.79 ± 0.14 | 2.65 | 0.1280 | |
| VPS13C | 1.39 ± 0.44 | 1.21 ± 0.13 | 2.41 | 0.1330 | |
| WDR44 | 1.48 ± 1.59 | 0.92 ± 0.12 | 2.42 | 0.1440 | |
| WDR46 | 1.24 ± 0.41 | 0.95 ± 0.32 | 2.83 | 0.0570 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fahrmann, J.F.; Katayama, H.; Irajizad, E.; Chakraborty, A.; Kato, T.; Mao, X.; Park, S.; Murage, E.; Rusling, L.; Yu, C.-Y.; et al. Plasma Based Protein Signatures Associated with Small Cell Lung Cancer. Cancers 2021, 13, 3972. https://doi.org/10.3390/cancers13163972
Fahrmann JF, Katayama H, Irajizad E, Chakraborty A, Kato T, Mao X, Park S, Murage E, Rusling L, Yu C-Y, et al. Plasma Based Protein Signatures Associated with Small Cell Lung Cancer. Cancers. 2021; 13(16):3972. https://doi.org/10.3390/cancers13163972
Chicago/Turabian StyleFahrmann, Johannes F., Hiroyuki Katayama, Ehsan Irajizad, Ashish Chakraborty, Taketo Kato, Xiangying Mao, Soyoung Park, Eunice Murage, Leona Rusling, Chuan-Yih Yu, and et al. 2021. "Plasma Based Protein Signatures Associated with Small Cell Lung Cancer" Cancers 13, no. 16: 3972. https://doi.org/10.3390/cancers13163972
APA StyleFahrmann, J. F., Katayama, H., Irajizad, E., Chakraborty, A., Kato, T., Mao, X., Park, S., Murage, E., Rusling, L., Yu, C.-Y., Cai, Y., Hsiao, F. C., Dennison, J. B., Tran, H., Ostrin, E., Wilson, D. O., Yuan, J.-M., Vykoukal, J., & Hanash, S. (2021). Plasma Based Protein Signatures Associated with Small Cell Lung Cancer. Cancers, 13(16), 3972. https://doi.org/10.3390/cancers13163972









