Challenges in the Use of Artificial Intelligence for Prostate Cancer Diagnosis from Multiparametric Imaging Data
Abstract
Simple Summary
Abstract
1. Introduction
2. AI in PCa Characterization
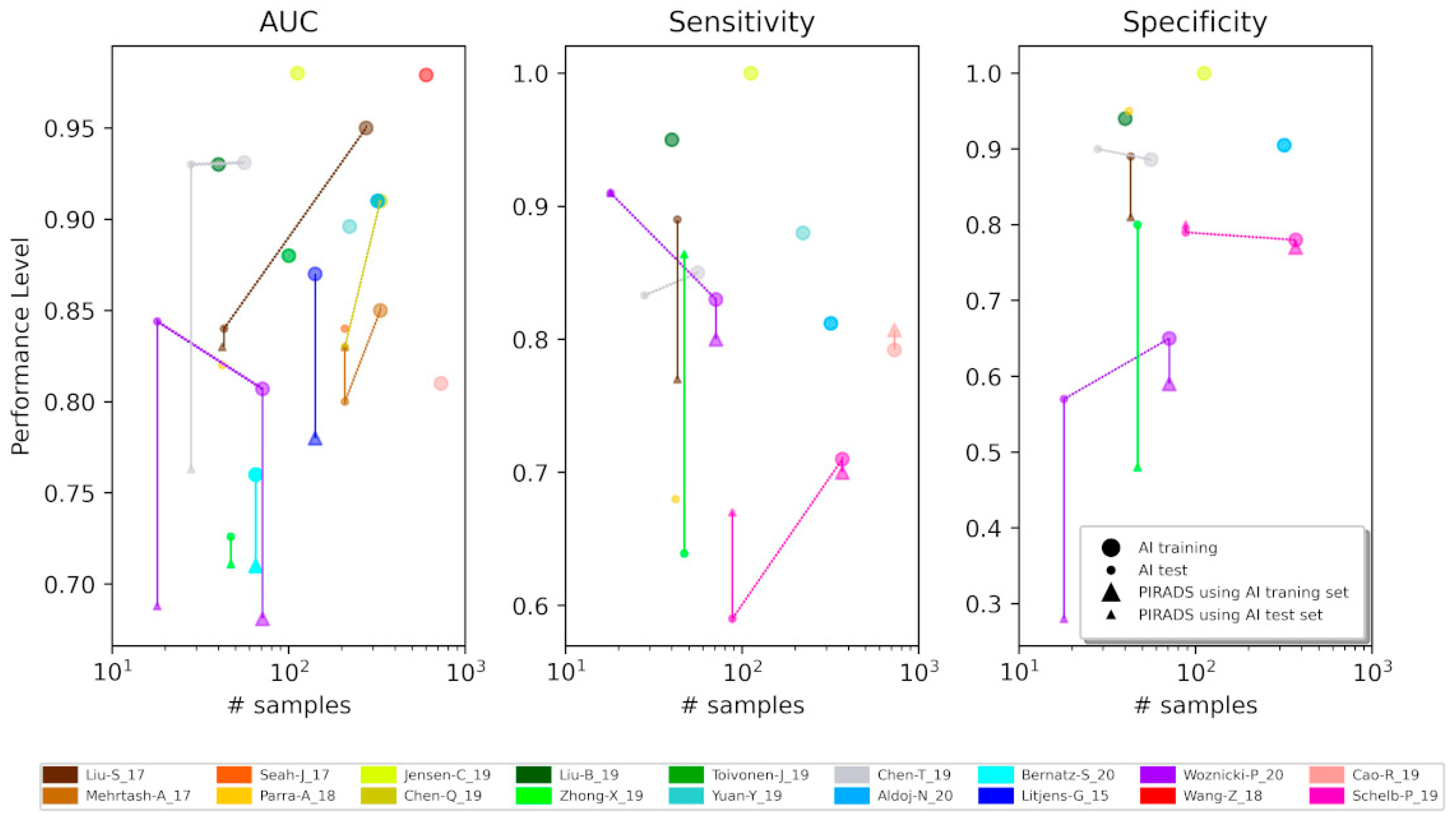
2.1. Overfitting
2.2. Standardization and Reproducibility
3. The Role of Public Databases
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rawla, P. Epidemiology of Prostate Cancer. World J. Oncol. 2019, 10, 63–89. [Google Scholar] [CrossRef]
- Caverly, T.J.; Hayward, R.A.; Reamer, E.; Zikmund-Fisher, B.J.; Connochie, D.; Heisler, M.; Fagerlin, A. Presentation of Benefits and Harms in US Cancer Screening and Prevention Guidelines: Systematic Review. J. Natl. Cancer Inst. 2016, 108, 1–8. [Google Scholar] [CrossRef]
- Patel, P.; Wang, S.; Siddiqui, M.M. The Use of Multiparametric Magnetic Resonance Imaging (mpMRI) in the Detection, Evaluation, and Surveillance of Clinically Significant Prostate Cancer (csPCa). Curr. Urol. Rep. 2019, 20, 1–9. [Google Scholar] [CrossRef]
- Borghesi, M.; Bianchi, L.; Barbaresi, U.; Vagnoni, V.; Corcioni, B.; Gaudiano, C.; Fiorentino, M.; Giunchi, F.; Chessa, F.; Garofalo, M.; et al. Diagnostic performance of MRI/TRUS fusion-guided biopsies vs. systematic prostate biopsies in biopsy-naive, previous negative biopsy patients and men undergoing active surveillance. Minarva Urol. Nephrol. 2021, 73, 357–366. [Google Scholar] [CrossRef]
- Schiavina, R.; Bianchi, L.; Borghesi, M.; Dababneh, H.; Chessa, F.; Pultrone, C.V.; Angiolini, A.; Gaudiano, C.; Porreca, A.; Fiorentino, M.; et al. MRI Displays the Prostatic Cancer Anatomy and Improves the Bundles Management before Robot-Assisted Radical Prostatectomy. J. Endourol. 2018, 32, 315–321. [Google Scholar] [CrossRef]
- Schiavina, R.; Droghetti, M.; Novara, G.; Bianchi, L.; Gaudiano, C.; Panebianco, V.; Borghesi, M.; Piazza, P.; Mineo Bianchi, F.; Guerra, M.; et al. The role of multiparametric MRI in active surveillance for low-risk prostate cancer: The ROMAS randomized controlled trial. Urol. Oncol. Semin. Orig. Investig. 2020, 39, 433.e1–433.e7. [Google Scholar] [CrossRef] [PubMed]
- Turkbey, B.; Rosenkrantz, A.B.; Haider, M.A.; Padhani, A.R.; Villeirs, G.; Macura, K.J.; Tempany, C.M.; Choyke, P.L.; Cornud, F.; Margolis, D.J.; et al. Prostate Imaging Reporting and Data System Version 2.1: 2019 Update of Prostate Imaging Reporting and Data System Version 2. Eur. Urol. 2019, 76, 340–351. [Google Scholar] [CrossRef]
- Hassanzadeh, E.; Glazer, D.I.; Dunne, R.M.; Fennessy, F.M.; Harisinghani, M.G.; Tempany, C.M. Prostate Imaging Reporting and Data System Version 2 (PI- RADS v2): A pictorial review. Abdom. Radiol. 2017, 42, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Schoots, I.G. MRI in early prostate cancer detection: How to manage indeterminate or equivocal PI-RADS 3 lesions? Transl. Androl. Urol. 2018, 7, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images are more than pictures, they are data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Chaddad, A.; Kucharczyk, M.J.; Cheddad, A.; Clarke, S.E.; Hassan, L.; Ding, S.; Rathore, S.; Zhang, M.; Katib, Y.; Bahoric, B.; et al. Magnetic Resonance Imaging Based Radiomic Models of Prostate Cancer: A Narrative Review. Cancers 2021, 13, 552. [Google Scholar] [CrossRef]
- Bardis, M.D.; Houshyar, R.; Chang, P.D.; Ushinsky, A.; Glavis-Bloom, J.; Chahine, C.; Bui, T.L.; Rupasinghe, M.; Filippi, C.G.; Chow, D.S. Applications of artificial intelligence to prostate multiparametric mri (Mpmri): Current and emerging trends. Cancers 2020, 12, 1204. [Google Scholar] [CrossRef] [PubMed]
- Bi, W.L.; Hosny, A.; Schabath, M.B.; Giger, M.L.; Birkbak, N.J.; Mehrtash, A.; Allison, T.; Arnaout, O.; Abbosh, C.; Dunn, I.F.; et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. Cancer J. Clin. 2019, 69, 127–157. [Google Scholar] [CrossRef] [PubMed]
- Schiavina, R.; Bianchi, L.; Lodi, S.; Cercenelli, L.; Chessa, F.; Bortolani, B.; Gaudiano, C.; Casablanca, C.; Droghetti, M.; Porreca, A.; et al. Real-time Augmented Reality Three-dimensional Guided Robotic Radical Prostatectomy: Preliminary Experience and Evaluation of the Impact on Surgical Planning. Eur. Urol. Focus 2020. [Google Scholar] [CrossRef] [PubMed]
- Schiavina, R.; Bianchi, L.; Chessa, F.; Barbaresi, U.; Cercenelli, L.; Lodi, S.; Gaudiano, C.; Bortolani, B.; Angiolini, A.; Bianchi, F.M.; et al. Augmented Reality to Guide Selective Clamping and Tumor Dissection During Robot-assisted Partial Nephrectomy: A Preliminary Experience. Clin. Genitourin. Cancer 2020, 19, e149–e155. [Google Scholar] [CrossRef]
- Goldenberg, S.L.; Nir, G.; Salcudean, S.E. A new era: Artificial intelligence and machine learning in prostate cancer. Nat. Rev. Urol. 2019, 16, 391–403. [Google Scholar] [CrossRef]
- Liu, S.; Zheng, H.; Feng, Y.; Li, W. Prostate Cancer Diagnosis using Deep Learning with 3D Multiparametric MRI. SPIE Med. Imaging 2017, 10134, 1–4. [Google Scholar]
- Mehrtash, A.; Sedghi, A.; Ghafoorian, M.; Taghipour, M.; Tempany, C.M.; Wells, W.M., 3rd; Kapur, T.; Mousavi, P.; Abolmaesumi, P.; Fedorov, A. Classification of clinical significance of MRI prostate findings using 3D convolutional neural networks. Proc. SPIE Int. Soc. Opt. Eng. 2017, 10134, 101342A. [Google Scholar] [CrossRef]
- Chen, T.; Li, M.; Gu, Y.; Zhang, Y.; Yang, S.; Wei, C.; Wu, J.; Li, X.; Zhao, W.; Shen, J. Prostate Cancer Differentiation and Aggressiveness: Assessment With a Radiomic-Based Model vs. PI-RADS v2. J. Magn. Reson. Imaging 2019, 49, 875–884. [Google Scholar] [CrossRef]
- Aldoj, N.; Lukas, S.; Dewey, M.; Penzkofer, T. Semi-automatic classification of prostate cancer on multi-parametric MR imaging using a multi-channel 3D convolutional neural network. Eur. Radiol. 2020, 30, 1243–1253. [Google Scholar] [CrossRef]
- Bernatz, S.; Ackermann, J.; Mandel, P.; Kaltenbach, B.; Zhdanovich, Y.; Harter, P.N.; Döring, C.; Hammerstingl, R.; Bodelle, B.; Smith, K.; et al. Comparison of machine learning algorithms to predict clinically significant prostate cancer of the peripheral zone with multiparametric MRI using clinical assessment categories and radiomic features. Eur. Radiol. 2020, 30, 6757–6769. [Google Scholar] [CrossRef]
- Litjens, G.J.S.; Barentsz, J.O.; Karssemeijer, N.; Huisman, H.J. Clinical evaluation of a computer-aided diagnosis system for determining cancer aggressiveness in prostate MRI. Eur. Radiol. 2015, 25, 3187–3199. [Google Scholar] [CrossRef]
- Woźnicki, P.; Westhoff, N.; Huber, T.; Riffel, P.; Froelich, M.F.; Gresser, E.; von Hardenberg, J.; Mühlberg, A.; Michel, M.S.; Schoenberg, S.O.; et al. Multiparametric MRI for prostate cancer characterization: Combined use of radiomics model with PI-RADS and clinical parameters. Cancers 2020, 12, 1767. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, C.; Cheng, D.; Wang, L.; Yang, X.; Cheng, K.T. Automated detection of clinically significant prostate cancer in mp-MRI images based on an end-to-end deep neural network. IEEE Trans. Med. Imaging 2018, 37, 1127–1139. [Google Scholar] [CrossRef]
- Cao, R.; Mohammadian Bajgiran, A.; Afshari Mirak, S.; Shakeri, S.; Zhong, X.; Enzmann, D.; Raman, S.; Sung, K. Joint Prostate Cancer Detection and Gleason Score Prediction in mp-MRI via FocalNet. IEEE Trans. Med. Imaging 2019, 38, 2496–2506. [Google Scholar] [CrossRef]
- Schelb, P.; Kohl, S.; Radtke, J.; Wiesenfarth, M.; Kickingereder, P.; Bickelhaupt, S.; Kuder, T.A.; Stenzinger, A.; Hohenfellner, M.; Schlemmer, H.; et al. Classification of cancer at prostate MRI: Deep Learning versus Clinical PI-RADS Assessment. Radiology 2019, 293, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Seah, J.C.Y.; Tang, J.S.N.; Kitchen, A. Detection of prostate cancer on multiparametric MRI. Med. Imaging 2017 Comput. Diagn. 2017, 10134, 1013429. [Google Scholar] [CrossRef]
- Parra, N.A.; Lu, H.; Li, Q.; Stoyanova, R.; Pollack, A.; Punnen, S.; Choi, J.; Abdalah, M.; Lopez, C.; Gage, K.; et al. Predicting clinically significant prostate cancer using DCE-MRI habitat descriptors. Oncotarget 2018, 9, 37125–37136. [Google Scholar] [CrossRef]
- Jensen, C.; Carl, J.; Boesen, L.; Langkilde, N.C.; Østergaard, L.R. Assessment of prostate cancer prognostic Gleason grade group using zonal-specific features extracted from biparametric MRI using a KNN classifier. J. Appl. Clin. Med. Phys. 2019, 20, 146–153. [Google Scholar] [CrossRef]
- Chen, Q.; Hu, S.; Long, P.; Lu, F.; Shi, Y.; Li, Y. A Transfer Learning Approach for Malignant Prostate Lesion Detection on Multiparametric MRI. Technol. Cancer Res. Treat. 2019, 18, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Cheng, J.; Guo, D.J.; He, X.J.; Luo, Y.D.; Zeng, Y.; Li, C.M. Prediction of prostate cancer aggressiveness with a combination of radiomics and machine learning-based analysis of dynamic contrast-enhanced MRI. Clin. Radiol. 2019, 74, 896.e1–896.e8. [Google Scholar] [CrossRef]
- Zhong, X.; Cao, R.; Shakeri, S.; Scalzo, F.; Lee, Y.; Enzmann, D.R.; Wu, H.H.; Raman, S.S.; Sung, K. Deep transfer learning-based prostate cancer classification using 3 Tesla multi-parametric MRI. Abdom. Radiol. 2019, 44, 2030–2039. [Google Scholar] [CrossRef]
- Toivonen, J.; Montoya Perez, I.; Movahedi, P.; Merisaari, H.; Pesola, M.; Taimen, P.; Boström, P.J.; Pohjankukka, J.; Kiviniemi, A.; Pahikkala, T. Radiomics and machine learning of multisequence multiparametric prostate MRI: Towards improved non-invasive prostate cancer characterization. PLoS ONE 2019, 14, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Qin, W.; Buyyounouski, M.; Ibragimov, B.; Hancock, S.; Han, B.; Xing, L. Prostate cancer classification with multiparametric MRI transfer learning model. Med. Phys. 2019, 46, 756–765. [Google Scholar] [CrossRef]
- Kwak, J.T.; Xu, S.; Wood, B.J.; Turkbey, B.; Choyke, P.L.; Pinto, P.A.; Wang, S.; Summers, R.M. Automated prostate cancer detection using T2-weighted and high-b-value diffusion-weighted magnetic resonance imaging. Med. Phys. 2015, 42, 2368–2378. [Google Scholar] [CrossRef] [PubMed]
- Cuocolo, R.; Cipullo, M.B.; Stanzione, A.; Romeo, V.; Green, R.; Cantoni, V.; Ponsiglione, A.; Ugga, L.; Imbriaco, M. Machine learning for the identification of clinically significant prostate cancer on MRI: A meta-analysis. Eur. Radiol. 2020, 30, 6877–6887. [Google Scholar] [CrossRef]
- Gao, J.; Jiang, Q.; Zhou, B.; Chen, D. Convolutional neural networks for computer-aided detection or diagnosis in medical image analysis: An overview. Math. Biosci. Eng. 2019, 16, 6536–6561. [Google Scholar] [CrossRef]
- Kohli, M.; Prevedello, L.M.; Filice, R.W.; Geis, J.R. Implementing machine learning in radiology practice and research. Am. J. Roentgenol. 2017, 208, 754–760. [Google Scholar] [CrossRef]
- Gawlitza, J.; Reiss-Zimmermann, M.; Thörmer, G.; Schaudinn, A.; Linder, N.; Garnov, N.; Horn, L.C.; Minh, D.H.; Ganzer, R.; Stolzenburg, J.U.; et al. Impact of the use of an endorectal coil for 3 T prostate MRI on image quality and cancer detection rate. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef]
- Barth, B.K.; Rupp, N.J.; Cornelius, A.; Nanz, D.; Grobholz, R.; Schmidtpeter, M.; Wild, P.J.; Eberli, D.; Donati, O.F. Diagnostic Accuracy of a MR Protocol Acquired with and without Endorectal Coil for Detection of Prostate Cancer: A Multicenter Study. Curr. Urol. 2019, 12, 88–96. [Google Scholar] [CrossRef]
- Dhatt, R.; Choy, S.; Co, S.J.; Ischia, J.; Kozlowski, P.; Harris, A.C.; Jones, E.C.; Black, P.C.; Goldenberg, S.L.; Chang, S.D. MRI of the Prostate With and Without Endorectal Coil at 3 T: Correlation With Whole-Mount Histopathologic Gleason Score. Am. J. Roentgenol. 2020, 215, 133–141. [Google Scholar] [CrossRef]
- Berman, R.M.; Brown, A.M.; Chang, S.D.; Sankineni, S.; Kadakia, M.; Wood, B.J.; Pinto, P.A.; Choyke, P.L.; Turkbey, B. DCE MRI of prostate cancer. Abdom. Radiol. 2016, 41, 844–853. [Google Scholar] [CrossRef]
- Castillo, J.M.T.; Arif, M.; Niessen, W.J.; Schoots, I.G.; Veenland, J.F. Automated classification of significant prostate cancer on MRI: A systematic review on the performance of machine learning applications. Cancers 2020, 12, 1606. [Google Scholar] [CrossRef]
- Palumbo, P.; Manetta, R.; Izzo, A.; Bruno, F.; Arrigoni, F.; De Filippo, M.; Splendiani, A.; Di Cesare, E.; Masciocchi, C.; Barile, A. Biparametric (bp) and multiparametric (mp) magnetic resonance imaging (MRI) approach to prostate cancer disease: A narrative review of current debate on dynamic contrast enhancement. Gland Surg. 2020, 9, 2235–2247. [Google Scholar] [CrossRef] [PubMed]
- Hulsen, T. An overview of publicly available patient-centered prostate cancer datasets. Transl. Androl. Urol. 2019, 8, S64–S77. [Google Scholar] [CrossRef]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The cancer imaging archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. Comment: The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 1–9. [Google Scholar] [CrossRef]
- Farahani, K.; Kalpathy-Cramer, J.; Chenevert, T.L.; Rubin, D.L.; Sunderland, J.J.; Nordstrom, R.J.; Buatti, J.; Hylton, N. Computational Challenges and Collaborative Projects in the NCI Quantitative Imaging Network. Tomography 2016, 2, 242–249. [Google Scholar] [CrossRef]
- Park, J.; Rho, M.J.; Park, Y.H.; Jung, C.K.; Chong, Y.; Kim, C.-S.; Go, H.; Jeon, S.S.; Kang, M.; Lee, H.J.; et al. PROMISE CLIP project: A retrospective, multicenter study for prostate cancer that integrates clinical, imaging and pathology data. Appl. Sci. 2019, 9, 2982. [Google Scholar] [CrossRef]
- Thompson, P.M.; Stein, J.L.; Medland, S.E.; Hibar, D.P.; Vasquez, A.A.; Renteria, M.E.; Toro, R.; Jahanshad, N.; Schumann, G.; Franke, B.; et al. The ENIGMA Consortium: Large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav. 2014, 8, 153–182. [Google Scholar] [CrossRef]
- Alfaro-Almagro, F.; Jenkinson, M.; Bangerter, N.K.; Andersson, J.; Griffanti, L.; Douaud, G.; Sotiropoulos, S.N.; Jbabdi, S.; Hernandez-Fernandez, M.; Vallee, E.; et al. Image processing and Quality Control for the first 10,000 brain imaging datasets from UK Biobank. Neuroimage 2018, 166, 400–424. [Google Scholar] [CrossRef]
- Hofman, M.S.; Lawrentschuk, N.; Francis, R.J.; Tang, C.; Vela, I.; Thomas, P.; Rutherford, N.; Martin, J.M.; Frydenberg, M.; Shakher, R.; et al. Prostate-specific membrane antigen PET-CT in patients with high-risk prostate cancer before curative-intent surgery or radiotherapy (proPSMA): A prospective, randomised, multicentre study. Lancet 2020, 395, 1208–1216. [Google Scholar] [CrossRef]
- Ceci, F.; Bianchi, L.; Borghesi, M.; Polverari, G.; Farolfi, A.; Briganti, A.; Schiavina, R.; Brunocilla, E.; Castellucci, P.; Fanti, S. Prediction nomogram for 68 Ga-PSMA-11 PET/CT in different clinical settings of PSA failure after radical treatment for prostate cancer. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 136–146. [Google Scholar] [CrossRef]
- Bianchi, L.; Borghesi, M.; Schiavina, R.; Castellucci, P.; Ercolino, A.; Bianchi, F.M.; Barbaresi, U.; Polverari, G.; Brunocilla, E.; Fanti, S.; et al. Predictive accuracy and clinical benefit of a nomogram aimed to predict 68 Ga-PSMA PET/CT positivity in patients with prostate cancer recurrence and PSA<1 ng/mL external validation on a single institution database. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 2100–2105. [Google Scholar] [CrossRef] [PubMed]
- Testa, C.; Pultrone, C.; Manners, D.N.; Schiavina, R.; Lodi, R. Metabolic imaging in prostate cancer: Where we are. Front. Oncol. 2016, 6, 225. [Google Scholar] [CrossRef][Green Version]
- Singh, S.P.; Wang, L.; Gupta, S.; Goli, H.; Padmanabhan, P.; Gulyás, B. 3d deep learning on medical images: A review. Sensors 2020, 20, 5097. [Google Scholar] [CrossRef]
| Paper | Dataset | MR Sequence | Hardware Setup | Image Processing |
|---|---|---|---|---|
| Liu, S. 2017 [17] | ProstateX (DICOM) | T2W, DWI, DCE | Siemens (Magnetom Trio and Skyra) at 3T without ERC | Registration |
| Mehrtash, A. 2017 [18] | ProstateX (DICOM) | DWI, DCE | Siemens (Magnetom Trio and Skyra) at 3T without ERC | Normalization |
| Seah, J. 2017 [27] | ProstateX (DICOM) | T2W, DWI, DCE | Siemens (Magnetom Trio and Skyra) at 3T without ERC | Normalization |
| Parra, A. 2018 [28] | 2 institutions (DICOM) | DWI, DCE | (I) Siemens and General Electric at 3T with external pelvic coil; (II) Siemens, Philips, General Electric at 3T and 1.5T with ERC | Registration, Normalization |
| Jensen, C. 2019 [29] | ProstateX (DICOM) | T2W, DWI, DCE | Siemens (Magnetom Trio and Skyra) at 3T without an ERC | Normalization |
| Chen, Q. 2019 [30] | ProstateX (DICOM) | T2W, DWI, DCE | Siemens (Magnetom Trio and Skyra) at 3T without an ERC | Normalization |
| Liu, B. 2019 [31] | 1 institution | DCE | General Electric (Signa Excite II) at 1.5T | N/A |
| Zhong, X. 2019 [32] | 1 institution | T2W, DWI, DCE | Siemens (Trio, Verio, Prisma or Skyra) at 3T with pelvic phased-array coil with or without ERC | Normalization |
| Toivonen, J. 2019 [33] | 1 institution | T2W, DWI | Philips (Ingenuity) at 3 T with 32 channel cardiac coils | Normalization |
| Yuan, Y. 2019 [35] | (I)1 institution (II) ProstateX (DICOM) | T2W, DWI | (I) N/A; (II) Siemens (Magnetom Trio and Skyra) at 3T without ERC | Normalization |
| Chen, T. 2019 [19] | 1 institution | T2W, DWI | Philips (Intera Achieva) at 3T with 32-channel body phased-array coil | N/A |
| Aldoj, N. 2020 [20] | ProstateX (DICOM) | T2W, DWI, DCE | Siemens (Magnetom Trio and Skyra) at 3T without ERC | Registration Normalization |
| Bernatz, S. 2020 [21] | 1 institution (DICOM) | T2W, DWI, DCE | Siemens (Magnetom Prisma FIT) at 3T with 32-channel body coil and spine phased-array coil | Limited image manipulation |
| Litjens, G. 2015 [22] | 1 institution | T2W, DWI, DCE | Siemens (Trio or Skyra) at 3T without ERC | N/A |
| Woznicki, P. 2020 [23] | 1 institution | T2W, DWI | Siemens (Magnetom Skyra and Trio) at 3T with pelvic phased-array coils | N/A |
| Wang, Z. 2018 [24] | (I)1 institution (II) ProstateX (DICOM) | T2W, DWI | (I) Siemens (Magnetom Skyra) at 3T (II) Siemens (Magnetom Trio and Skyra) at 3T without ERC | Registration |
| Cao, R. 2019 [25] | 1 institution | T2W, DWI | Siemens (Trio, Skyra, Prisma, Verio) at 3T with and without ERC | Registration Normalization |
| Schelb, P. 2019 [26] | 1 institution | T2W, DWI | Siemens (Prisma) at 3T with standard multichannel body coil and integrated spine phased-array coil | Registration |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corradini, D.; Brizi, L.; Gaudiano, C.; Bianchi, L.; Marcelli, E.; Golfieri, R.; Schiavina, R.; Testa, C.; Remondini, D. Challenges in the Use of Artificial Intelligence for Prostate Cancer Diagnosis from Multiparametric Imaging Data. Cancers 2021, 13, 3944. https://doi.org/10.3390/cancers13163944
Corradini D, Brizi L, Gaudiano C, Bianchi L, Marcelli E, Golfieri R, Schiavina R, Testa C, Remondini D. Challenges in the Use of Artificial Intelligence for Prostate Cancer Diagnosis from Multiparametric Imaging Data. Cancers. 2021; 13(16):3944. https://doi.org/10.3390/cancers13163944
Chicago/Turabian StyleCorradini, Daniele, Leonardo Brizi, Caterina Gaudiano, Lorenzo Bianchi, Emanuela Marcelli, Rita Golfieri, Riccardo Schiavina, Claudia Testa, and Daniel Remondini. 2021. "Challenges in the Use of Artificial Intelligence for Prostate Cancer Diagnosis from Multiparametric Imaging Data" Cancers 13, no. 16: 3944. https://doi.org/10.3390/cancers13163944
APA StyleCorradini, D., Brizi, L., Gaudiano, C., Bianchi, L., Marcelli, E., Golfieri, R., Schiavina, R., Testa, C., & Remondini, D. (2021). Challenges in the Use of Artificial Intelligence for Prostate Cancer Diagnosis from Multiparametric Imaging Data. Cancers, 13(16), 3944. https://doi.org/10.3390/cancers13163944








