Ethanol-Induced Cell Damage Can Result in the Development of Oral Tumors
Abstract
Simple Summary
Abstract
1. Alcohol Consumption and Its Adverse Effects Know a Long History
2. Oral Cancers Are Still Frequently Diagnosed despite Avoidable Risk Factors
3. Alcohol Is an Independent Risk Factor for Oral Carcinogenesis
3.1. Epidemiological Data Indicate a Strong Correlation between Alcohol and Oral Malignancies
3.2. In Vivo Data Support a Causal Effect of Alcohol on Oral Tumor Incidence
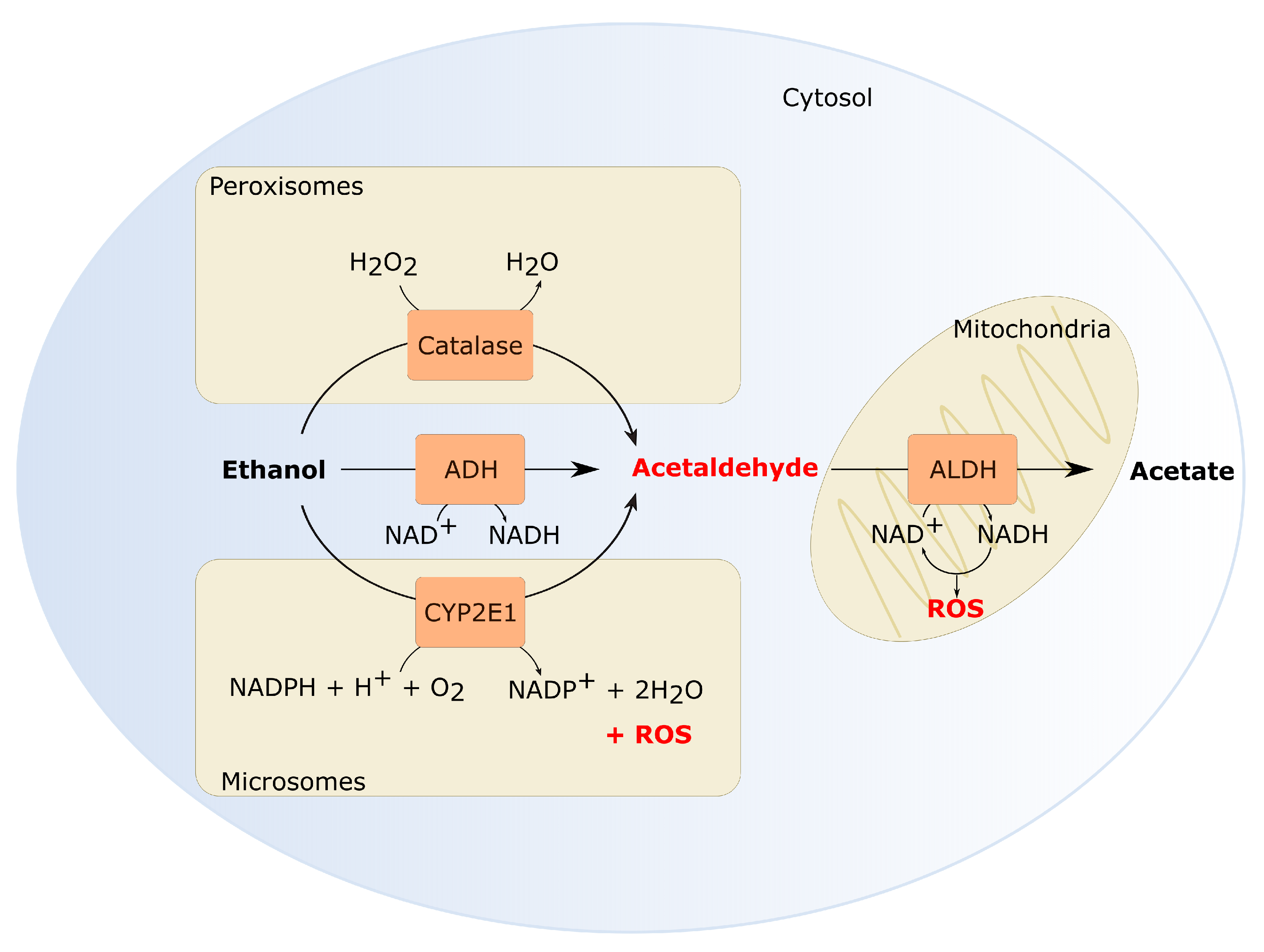
4. Various Molecular Alterations Have Been Attributed to Ethanol
4.1. Acetaldehyde Can Accumulate in the Oral Cavity
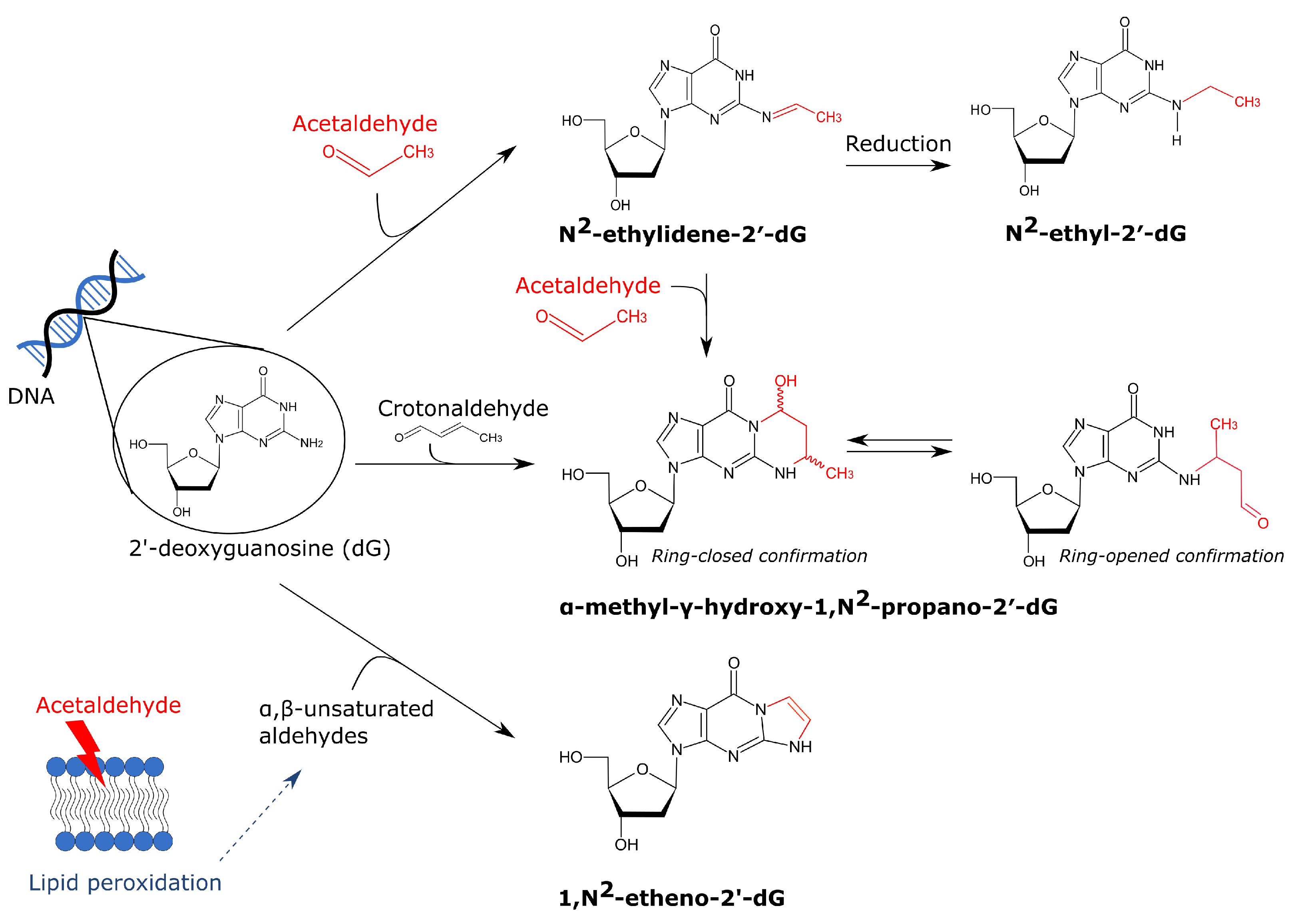
4.2. Metabolites of Ethanol Can Directly Affect the DNA by Formation of Adducts and Crosslinks
4.2.1. Acetaldehyde-Derived DNA Adducts and Crosslinks
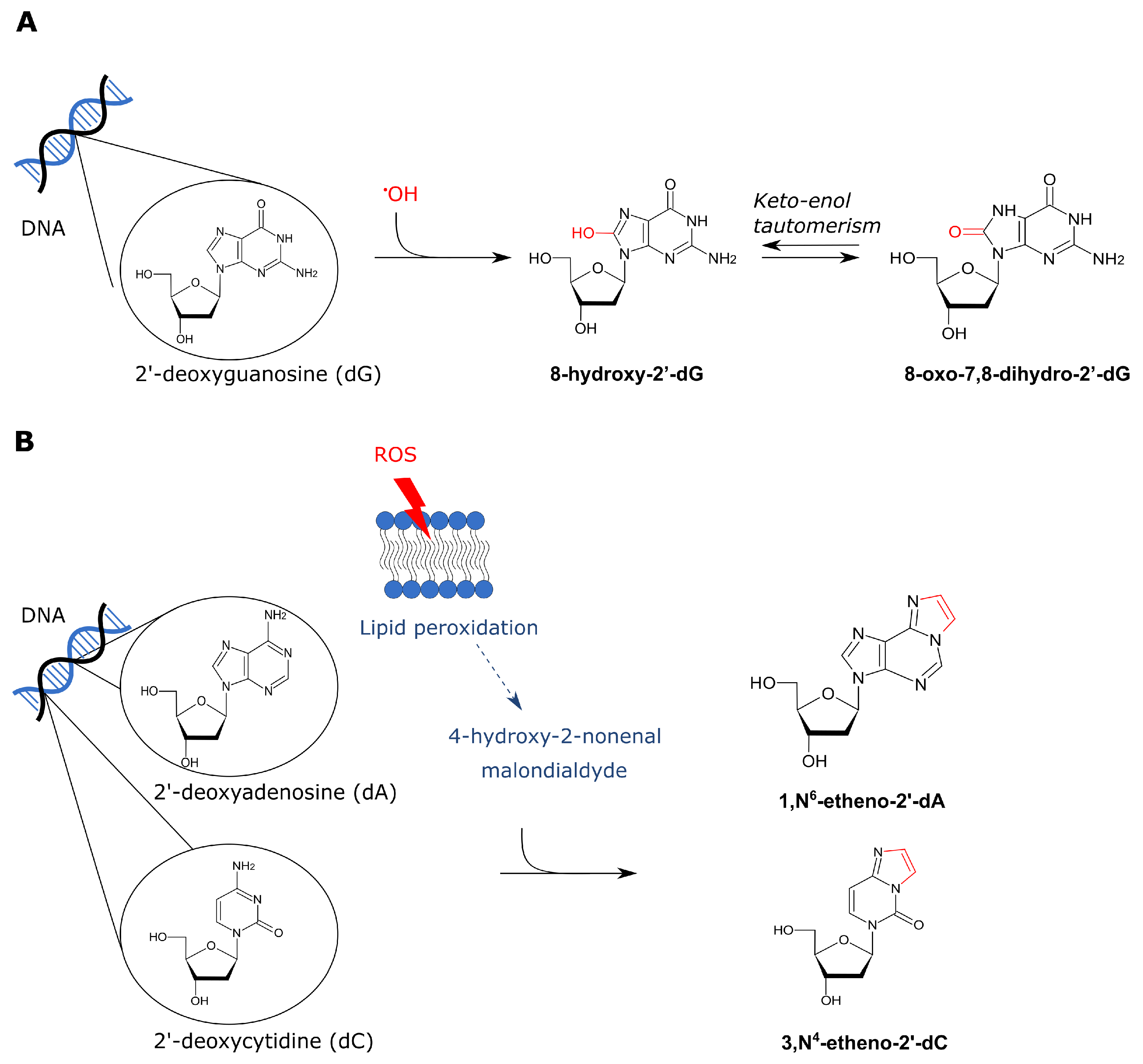
4.2.2. ROS-Derived DNA Adducts
4.3. Ethanol Exposure Alters the Epigenome
4.3.1. Ethanol Leads to DNA Hypomethylation
4.3.2. Patterns of Histone Modifications Can Change in the Presence of Ethanol
5. Mutational Signatures Give More Insight into Carcinogenesis
Several Ethanol-Related Mutational Signatures Have Been Identified
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- McGovern, P.E.; Zhang, J.; Tang, J.; Zhang, Z.; Hall, G.R.; Moreau, R.A.; Nuñez, A.; Butrym, E.D.; Richards, M.P.; Wang, C.S.; et al. Fermented beverages of pre- and proto-historic China. Proc. Natl. Acad. Sci. USA 2004, 101, 17593–17598. [Google Scholar] [CrossRef]
- Hanson, D.J. Historical evolution of alcohol consumption in society. In Alcohol: Science, Policy and Public Health; Boyle, P., Boffetta, P., Lowenfels, A.B., Burns, H., Brawley, O., Eds.; Oxford University Press: Oxford, UK, 2013; pp. 3–13. [Google Scholar] [CrossRef]
- Sournia, J.C. A History of Alcoholism, translated ed.; Basil Blackwell: Oxford, UK, 1990; p. 232. [Google Scholar]
- Le Daré, B.; Lagente, V.; Gicquel, T. Ethanol and its metabolites: Update on toxicity, benefits, and focus on immunomodulatory effects. Drug Metab. Rev. 2019, 51, 545–561. [Google Scholar] [CrossRef]
- Pearl, R. Alcohol and Longevity; Alfred A. Knopf.: New York, NY, USA, 1926; p. 273. [Google Scholar]
- Room, R.; Babor, T.; Rehm, J. Alcohol and public health. Lancet 2005, 365, 519–530. [Google Scholar] [CrossRef]
- Griswold, M.G.; Fullman, N.; Hawley, C.; Arian, N.; M Zimsen, S.R.; Tymeson, H.D.; Venkateswaran, V.; Douglas Tapp, A.; Forouzanfar, M.; Salama, J.; et al. Alcohol use and burden for 195 countries and territories, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2018, 392, 1015–1035. [Google Scholar] [CrossRef]
- World Health Organization. Global Status Report on Alcohol and Health 2018; Technical Report, Licence: CC BY-NC-SA 3.0 IGO; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Rehm, J.; Shield, K.D. Global burden of alcohol use disorders and alcohol liver disease. Biomedicines 2019, 7, 99. [Google Scholar] [CrossRef]
- Shield, K.; Manthey, J.; Rylett, M.; Probst, C.; Wettlaufer, A.; Parry, C.D.; Rehm, J. National, regional, and global burdens of disease from 2000 to 2016 attributable to alcohol use: A comparative risk assessment study. Lancet Public Health 2020, 5, e51–e61. [Google Scholar] [CrossRef]
- Praud, D.; Rota, M.; Rehm, J.; Shield, K.; Zatoński, W.; Hashibe, M.; La Vecchia, C.; Boffetta, P. Cancer incidence and mortality attributable to alcohol consumption. Int. J. Cancer 2016, 138, 1380–1387. [Google Scholar] [CrossRef]
- Rumgay, H.; Shield, K.; Charvat, H.; Ferrari, P.; Sornpaisarn, B.; Obot, I.; Islami, F.; Lemmens, V.E.P.P.; Rehm, J.; Soerjomataram, I. Global burden of cancer in 2020 attributable to alcohol consumption: A population-based study. Lancet Oncol. 2021, 21, S1470–S2045. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer. Personal Habits and Indoor Combustions; Technical Report; International Agency for Research on Cancer: Lyon, France, 2012. [Google Scholar]
- Shrestha, A.D.; Vedsted, P.; Kallestrup, P.; Neupane, D. Prevalence and incidence of oral cancer in low- and middle-income countries: A scoping review. Eur. J. Cancer Care 2020, 29, e13207. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.W.; Jayasekara, P.; Amarasinghe, A.A.; Hemantha, K. Squamous cell carcinoma and precursor lesions of the oral cavity: Epidemiology and aetiology. Periodontology 2000 2011, 57, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Rivera, C. Essentials of oral cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 11884–11894. [Google Scholar] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Neville, B.W.; Day, T.A. Oral Cancer and Precancerous Lesions. CA A Cancer J. Clin. 2002, 52, 195–215. [Google Scholar] [CrossRef]
- Tanaka, T.; Ishigamori, R. Understanding carcinogenesis for fighting oral cancer. J. Oncol. 2011, 2011, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Schwimmer, E. Die idiopathischen Schleim- hautplaques der Mundhöhle (Leukoplakia buc- calis). Arch. Dermatol. Res. 1877, 9, 570–611. [Google Scholar]
- Speight, P.M.; Khurram, S.A.; Kujan, O. Oral potentially malignant disorders: Risk of progression to malignancy. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2018, 125, 612–627. [Google Scholar] [CrossRef]
- Pinto, A.C.; Caramês, J.; Francisco, H.; Chen, A.; Azul, A.M.; Marques, D. Malignant transformation rate of oral leukoplakia—Systematic review. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2020, 129, 611. [Google Scholar] [CrossRef]
- Ganesh, D.; Sreenivasan, P.; Ohman, J.; Wallström, M.; Braz-Silva, P.H.; Giglio, D.; Kjeller, G.; Hasséus, B. Potentially malignant oral disorders and cancer transformation. Anticancer Res. 2018, 38, 3223–3229. [Google Scholar] [CrossRef]
- IARC. Tobacco smoking. IARC Monogr. Eval. Carcinog. Risks Chem. Hum. 1986, 38, 35–394. [Google Scholar]
- Vineis, P.; Alavanja, M.; Buffler, P.; Fontham, E.; Franceschi, S.; Gao, Y.T.; Gupta, P.C.; Hackshaw, A.; Matos, E.; Samet, J.; et al. Tobacco and Cancer: Recent Epidemiological Evidence. JNCI J. Natl. Cancer Inst. 2004, 96, 99–106. [Google Scholar] [CrossRef]
- Boffetta, P.; Hecht, S.; Gray, N.; Gupta, P.; Straif, K. Smokeless tobacco and cancer. Lancet Oncol. 2008, 9, 667–675. [Google Scholar] [CrossRef]
- Asthana, S.; Labani, S.; Kailash, U.; Sinha, D.N.; Mehrotra, R. Association of Smokeless Tobacco Use and Oral Cancer: A Systematic Global Review and Meta-Analysis. Nicotine Tob. Res. 2019, 21, 1162–1171. [Google Scholar] [CrossRef]
- Siddiqi, K.; Husain, S.; Vidyasagaran, A.; Readshaw, A.; Mishu, M.P.; Sheikh, A. Global burden of disease due to smokeless tobacco consumption in adults: An updated analysis of data from 127 countries. BMC Med. 2020, 18, 1–22. [Google Scholar] [CrossRef]
- Ko, Y.; Huang, Y.; Lee, C.; Chen, M.; Lin, L.; Tsai, C. Betel quid chewing, cigarette smoking and alcohol consumption related to oral cancer in Taiwan. J. Oral Pathol. Med. 1995, 24, 450–453. [Google Scholar] [CrossRef]
- Hashibe, M.; Brennan, P.; Chuang, S.; Boccia, S.; Castellsague, X.; Chen, C.; Curado, M.; Dal Maso, L.; Daudt, A.; Fabianova, E.; et al. Interaction between tobacco and alcohol use and the risk of head and neck cancer: Pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. Cancer Epidemiol. Biomarkers Prev. 2009, 18, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Dal Maso, L.; Torelli, N.; Biancotto, E.; Di Maso, M.; Gini, A.; Franchin, G.; Levi, F.; La Vecchia, C.; Serraino, D.; Polesel, J. Combined effect of tobacco smoking and alcohol drinking in the risk of head and neck cancers: A re-analysis of case–control studies using bi-dimensional spline models. Eur. J. Epidemiol. 2016, 31, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Mello, F.; Melo, G.; Pasetto, J.; Silva, C.; Warnakulasuriya, S.; Rivero, E. The synergistic effect of tobacco and alcohol consumption on oral squamous cell carcinoma: A systematic review and meta-analysis. Clin. Oral Investig. 2019, 23, 2849–2859. [Google Scholar] [CrossRef]
- Petti, S.; Scully, C. Oral cancer: The association between nation-based alcohol-drinking profiles and oral cancer mortality. Oral Oncol. 2005, 41, 828–834. [Google Scholar] [CrossRef] [PubMed]
- Boffetta, P.; Hashibe, M. Alcohol and cancer. Lancet Oncol. 2006, 7, 149–156. [Google Scholar] [CrossRef]
- Kanny, D.; Naimi, T.S.; Liu, Y.; Lu, H.; Brewer, R.D. Annual Total Binge Drinks Consumed by U.S. Adults, 2015. Am. J. Prev. Med. 2018, 54, 496. [Google Scholar] [CrossRef] [PubMed]
- Melikian, A.; Djordjevic, M.; Hosey, J.; Zhang, J.; Chen, S.; Zang, E.; Muscat, J.; Stellman, S. Gender differences relative to smoking behavior and emissions of toxins from mainstream cigarette smoke. Nicotine Tob. Res. 2007, 9, 377–387. [Google Scholar] [CrossRef]
- Marur, S.; D’Souza, G.; Westra, W.H.; Forastiere, A.A. HPV-associated head and neck cancer: A virus-related cancer epidemic. Lancet Oncol. 2010, 11, 781–789. [Google Scholar] [CrossRef]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Yete, S.; D’Souza, W.; Saranath, D. High-Risk Human Papillomavirus in Oral Cancer: Clinical Implications. Oncology 2018, 94, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Nauta, I.H.; Heideman, D.A.M.; Brink, A.; van der Steen, B.; Bloemena, E.; Koljenović, S.; de Jong, R.J.B.; Leemans, C.R.; Brakenhoff, R.H. The unveiled reality of human papillomavirus as risk factor for oral cavity squamous cell carcinoma. Int. J. Cancer 2021, 149, 430. [Google Scholar] [CrossRef]
- Khanna, S.; Palackdharry, S.; Roof, L.; Wicker, C.A.; Mark, J.; Zhu, Z.; Jandorav, R.; Molinolo, A.; Takiar, V.; Wise-Draper, T.M. Determining the molecular landscape and impact on prognosis in HPV-associated head and neck cancer. Cancers Head Neck 2020, 5, 11. [Google Scholar] [CrossRef]
- Kalinowski, A.; Humphreys, K. Governmental standard drink definitions and low-risk alcohol consumption guidelines in 37 countries. Addiction 2016, 111, 1293–1298. [Google Scholar] [CrossRef] [PubMed]
- Hoge Gezondheidsraad. Risico’s van Alcoholgebruik HGR NR 9438; Technical Report; Hoge Gezondheidsraad: Brussel, Belgium, 2018. [Google Scholar]
- Boffetta, P.; Hashibe, M.; La Vecchia, C.; Zatonski, W.; Rehm, J. The burden of cancer attributable to alcohol drinking. Int. J. Cancer 2006, 119, 884–887. [Google Scholar] [CrossRef] [PubMed]
- Baan, R.; Straif, K.; Grosse, Y.; Secretan, B.; El Ghissassi, F.; Bouvard, V.; Altieri, A.; Cogliano, V. Carcinogenicity of alcoholic beverages. Lancet Oncol. 2007, 8, 292–293. [Google Scholar] [CrossRef]
- Seitz, H.K.; Stickel, F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat. Rev. Cancer 2007, 7, 599–612. [Google Scholar] [CrossRef]
- Blot, W.J.; McLaughlin, J.K.; Winn, D.M.; Austin, D.F.; Greenberg, R.S.; Susan, S.; Preston, M.; Bernstein, L.; Schoenberg, J.B.; Stemhagen, A.; et al. Smoking and Drinking in Relation to Oral and Pharyngeal Cancer. Cancer Res. 1988, 48, 3282–3287. [Google Scholar]
- Yokoyama, A.; Kakiuchi, N.; Yoshizato, T.; Nannya, Y.; Suzuki, H.; Takeuchi, Y.; Shiozawa, Y.; Sato, Y.; Aoki, K.; Kim, S.K.; et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature 2019, 565, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Hashibe, M.; Brennan, P.; Benhamou, S.; Castellsague, X.; Chen, C.; Curado, M.P.; Dal Maso, L.; Daudt, A.W.; Fabianova, E.; Fernandez, L.; et al. Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers, and the risk of head and neck cancer: Pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. J. Natl. Cancer Inst. 2007, 99, 789. [Google Scholar] [CrossRef]
- Polesel, J.; Dal Maso, L.; Bagnardi, V.; Zucchetto, A.; Zambon, A.; Levi, F.; La Vecchia, C.; Franceschi, S. Estimating dose-response relationship between ethanol and risk of cancer using regression spline models. Int. J. Cancer 2005, 114, 836–841. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, B.Y.; Chang, S.C.; Hashibe, M.; La Vecchia, C.; Zhang, Z.F. Alcohol consumption and cancers of the oral cavity and pharynx from 1988 to 2009: An update. Eur. J. Cancer Prev. 2010, 19, 431–465. [Google Scholar] [CrossRef] [PubMed]
- Turati, F.; Garavello, W.; Tramacere, I.; Bagnardi, V.; Rota, M.; Scotti, L.; Islami, F.; Corrao, G.; Boffetta, P.; La Vecchia, C.; et al. A meta-analysis of alcohol drinking and oral and pharyngeal cancers. Part 2: Results by subsites. Oral Oncol. 2010, 46, 720–726. [Google Scholar] [CrossRef] [PubMed]
- Bagnardi, V.; Rota, M.; Botteri, E.; Tramacere, I.; Islami, F.; Fedirko, V.; Scotti, L.; Jenab, M.; Turati, F.; Pasquali, E.; et al. Light alcohol drinking and cancer: A meta-analysis. Ann. Oncol. 2013, 24, 301–308. [Google Scholar] [CrossRef]
- Bagnardi, V.; Rota, M.; Botteri, E.; Tramacere, I.; Islami, F.; Fedirko, V.; Scotti, L.; Jenab, M.; Turati, F.; Pasquali, E.; et al. Alcohol consumption and site-specific cancer risk: A comprehensive dose-response meta-analysis. Br. J. Cancer 2015, 112, 580–593. [Google Scholar] [CrossRef]
- Di Credico, G.; Polesel, J.; Dal Maso, L.; Pauli, F.; Torelli, N.; Luce, D.; Radoï, L.; Matsuo, K.; Serraino, D.; Brennan, P.; et al. Alcohol drinking and head and neck cancer risk: The joint effect of intensity and duration. Br. J. Cancer 2020, 123, 1456–1463. [Google Scholar] [CrossRef]
- Franceschi, S.; Talamini, R.; Barra, S.; Baron, A.E.; Negri, E.; Bidoli, E.; Serraino, D.; La Vecchia, C. Smoking and drinking in relation to cancers of the oral cavity, pharynx, larynx, and esophagus in northern Italy. Cancer Res. 1990, 50, 6502–6507. [Google Scholar] [CrossRef]
- Mashberg, A.; Boffetta, P.; Winkelman, R.; Garfinkel, L. Tobacco smoking, alcohol drinking, and cancer of the oral cavity and oropharynx among U.S. veterans. Cancer 1993, 72, 1369–1375. [Google Scholar] [CrossRef]
- Balaram, P.; Sridhar, H.; Rajkumar, T.; Vaccarella, S.; Herrero, R.; Nandakumar, A.; Ravichandran, K.; Ramdas, K.; Sankaranarayanan, R.; Gajalakshmi, V.; et al. Oral cancer in Southern India: The influence of smoking, drinking, paan-chewing and oral hygiene. Int. J. Cancer 2002, 98, 440–445. [Google Scholar] [CrossRef] [PubMed]
- Altieri, A.; Bosetti, C.; Gallus, S.; Franceschi, S.; Dal Maso, L.; Talamini, R.; Levi, F.; Negri, E.; Rodriguez, T.; La Vecchia, C. Wine, beer and spirits and risk of oral and pharyngeal cancer: A case-control study from Italy and Switzerland. Oral Oncol. 2004, 40, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Maasland, D.H.; van den Brandt, P.A.; Kremer, B.; Goldbohm, R.A.; Schouten, L.J. Alcohol consumption, cigarette smoking and the risk of subtypes of head-neck cancer: Results from the Netherlands Cohort Study. BMC Cancer 2014, 14, 187. [Google Scholar] [CrossRef] [PubMed]
- Tenore, G.; Nuvoli, A.; Mohsen, A.; Cassoni, A.; Battisti, A.; Terenzi, V.; Monaca, M.D.; Raponi, I.; Brauner, E.; De Felice, F.; et al. Tobacco, alcohol and family history of cancer as risk factors of Oral Squamous Cell Carcinoma: Case-control retrospective study. Appl. Sci. (Switz.) 2020, 10, 3896. [Google Scholar] [CrossRef]
- Sarich, P.; Canfell, K.; Egger, S.; Banks, E.; Joshy, G.; Grogan, P.; Weber, M.F. Alcohol consumption, drinking patterns and cancer incidence in an Australian cohort of 226,162 participants aged 45 years and over. Br. J. Cancer 2021, 124, 513–523. [Google Scholar] [CrossRef]
- Znaori, A.; Brennan, P.; Gajalakshmi, V.; Mathew, A.; Shanta, V.; Varghese, C.; Boffetta, P. Independent and combined effects of tobacco smoking, chewing and alcohol drinking on the risk of oral, pharyngeal and esophageal cancers in Indian men. Int. J. Cancer 2003, 105, 681–686. [Google Scholar] [CrossRef]
- Moreno-López, L.A.; Esparza-Gómez, G.C.; González-Navarro, A.; Cerero-Lapiedra, R.; González-Hernández, M.J.; Domínguez-Rojas, V. Risk of oral cancer associated with tobacco smoking, alcohol consumption and oral hygiene: A case-control study in Madrid, Spain. Oral Oncol. 2000, 36, 170–174. [Google Scholar] [CrossRef]
- Moore, S.R.; Johnson, N.W.; Pierce, A.M.; Wilson, D.F. The epidemiology of lip cancer: A review of global incidence and aetiology. Oral Dis. 1999, 5, 185–195. [Google Scholar] [CrossRef]
- López, E.P.M.; Moral, R.M.M.d.; Martínez-García, C.; Zanetti, R.; Rosso, S.; Serrano, S.; Aneiros, J.F.; Jimenez-Puente, A.; Redondo, M. Lifestyles, environmental and phenotypic factors associated with lip cancer: A case–control study in southern Spain. Br. J. Cancer 2003, 88, 1702–1707. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.J.; Jiang, R.S.; Wu, S.H.; Chen, F.J.; Liu, S.A. Smoking, alcohol, and betel quid and oral cancer: A prospective cohort study. J. Oncol. 2011, 2011, 525976. [Google Scholar] [CrossRef]
- Anantharaman, D.; Marron, M.; Lagiou, P.; Samoli, E.; Ahrens, W.; Pohlabeln, H.; Slamova, A.; Schejbalova, M.; Merletti, F.; Richiardi, L.; et al. Population attributable risk of tobacco and alcohol for upper aerodigestive tract cancer. Oral Oncol. 2011, 47, 725–731. [Google Scholar] [CrossRef]
- Ferreira Antunes, J.; Toporcov, T.; Biazevic, M.; Boing, A.; Scully, C.; Petti, S. Joint and independent effects of alcohol drinking and tobacco smoking on oral cancer: A large case-control study. PLoS ONE 2013, 8, e68132. [Google Scholar] [CrossRef]
- Wight, A.J.; Ogden, G.R. Possible mechanisms by which alcohol may influence the development of oral cancer—A review. Oral Oncol. 1998, 34, 441–447. [Google Scholar] [CrossRef]
- Maserejian, N.N.; Joshipura, K.J.; Rosner, B.A.; Giovannucci, E.; Zavras, A.I. Prospective study of alcohol consumption and risk of oral premalignant lesions in men. Cancer Epidemiol. Biomarkers Prev. 2006, 15, 774–781. [Google Scholar] [CrossRef]
- Li, L.; Psoter, W.J.; Buxó, C.J.; Elias, A.; Cuadrado, L.; Morse, D.E. Smoking and drinking in relation to oral potentially malignant disorders in Puerto Rico: A case-control study. BMC Cancer 2011, 11, 324. [Google Scholar] [CrossRef]
- Kumar, S.; Debnath, N.; Ismail, M.B.; Kumar, A.; Kumar, A.; Badiyani, B.K.; Dubey, P.K.; Sukhtankar, L.V. Prevalence and Risk Factors for Oral Potentially Malignant Disorders in Indian Population. Adv. Prev. Med. 2015, 2015, 1–7. [Google Scholar] [CrossRef]
- Chher, T.; Hak, S.; Kallarakkal, T.G.; Durward, C.; Ramanathan, A.; Ghani, W.M.N.; Razak, I.A.; Harun, M.H.; Ashar, N.A.M.; Rajandram, R.K.; et al. Prevalence of oral cancer, oral potentially malignant disorders and other oral mucosal lesions in Cambodia. Ethn. Health 2018, 23, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Aghbari, S.M.H.; Abushouk, A.I.; Attia, A.; Elmaraezy, A.; Menshawy, A.; Ahmed, M.S.; Elsaadany, B.A.; Ahmed, E.M. Malignant transformation of oral lichen planus and oral lichenoid lesions: A meta-analysis of 20095 patient data. Oral Oncol. 2017, 68, 92–102. [Google Scholar] [CrossRef] [PubMed]
- González-Moles, M.Á.; Ruiz-Ávila, I.; González-Ruiz, L.; Ayén, Á.; Gil-Montoya, J.A.; Ramos-García, P. Malignant transformation risk of oral lichen planus: A systematic review and comprehensive meta-analysis. Oral Oncol. 2019, 96, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Idrees, M.; Kujan, O.; Shearston, K.; Farah, C.S. Oral lichen planus has a very low malignant transformation rate: A systematic review and meta-analysis using strict diagnostic and inclusion criteria. J. Oral Pathol. Med. 2020, 1–12. [Google Scholar] [CrossRef]
- Smith, G.D.; Ebrahim, S. ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 2003, 32, 1–22. [Google Scholar] [CrossRef]
- Luczak, S.E.; Glatt, S.J.; Wall, T.J. Meta-analyses of ALDH2 and ADH1B with alcohol dependence in asians. Psychol. Bull. 2006, 132, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Hasin, D.; Aharonovich, E.; Liu, X.; Mamman, Z.; Matseoane, K.; Carr, L.; Li, T.K. Alcohol and ADH2 in Israel: Ashkenazis, Sephardics, and recent Russian immigrants. Am. J. Psychiatry 2002, 159, 1432–1434. [Google Scholar] [CrossRef]
- Liu, M.; Jiang, Y.; Wedow, R.; Li, Y.; Brazel, D.M.; Chen, F.; Datta, G.; Davila-Velderrain, J.; McGuire, D.; Tian, C.; et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 2019, 51, 237–244. [Google Scholar] [CrossRef]
- Gormley, M.; Dudding, T.; Sanderson, E.; Martin, R.M.; Thomas, S.; Tyrrell, J.; Ness, A.R.; Brennan, P.; Munafò, M.; Pring, M.; et al. A multivariable Mendelian randomization analysis investigating smoking and alcohol consumption in oral and oropharyngeal cancer. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Tang, X.H.; Knudsen, B.; Bemis, D.; Tickoo, S.; Gudas, L.J. Oral Cavity and Esophageal Carcinogenesis Modeled in Carcinogen-Treated Mice. Clin. Cancer Res. 2004, 10, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wang, X.; Zhang, X.; Sun, Z.; Chen, X. Ethanol Promotes Chemically Induced Oral Cancer in Mice through Activation of the 5-Lipoxygenase Pathway of Arachidonic Acid Metabolism. Cancer Prev. Res. 2011, 4, 1863–1872. [Google Scholar] [CrossRef]
- Nachiappan, V.; Mufti, S.I.; Chakravarti, A.; Eskelson, C.D.; Rajasekharan, R. Lipid peroxidation and ethanol-related tumor promotion in Fischer-344 rats treated with tobacco-specific nitrosamines. Alcohol Alcohol.) 1994, 29, 565–574. [Google Scholar]
- Soffritti, M.; Belpoggi, F.; Cevolani, D.; Guarino, M.; Padovani, M.; Maltoni, C. Results of Long-Term Experimental Studies on the Carcinogenicity of Methyl Alcohol and Ethyl Alcohol in Rats. Ann. N. Y. Acad. Sci. 2002, 982, 46–69. [Google Scholar] [CrossRef] [PubMed]
- Müller, P.; Hepke, B.; Meldau, U.; Raabe, G. Tissue damage in the rabbit oral mucosa by acute and chronic direct toxic action of different alcohol concentrations. Exp. Pathol. 1983, 24, 171–181. [Google Scholar] [CrossRef]
- Reidy, J.; McHugh, E.; Stassen, L.F. A review of the relationship between alcohol and oral cancer. Surgeon 2011, 9, 278–283. [Google Scholar] [CrossRef]
- Garcia Martins, R.H.; Marques Madeira, S.L.; Fabro, A.T.; Rocha, N.D.S.; De Oliveira Semenzati, G.; Alves, K.F. Effects to exposure of tobacco smoke and alcohol on the tongue and pharynx of rats. Inhal. Toxicol. 2012, 24, 153–160. [Google Scholar] [CrossRef]
- Wu, D.; Cederbaum, A.I. Alcohol, oxidative stress, and free radical damage. Alcohol Res. Health 2003, 27, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Bailey, S.M.; Cunningham, C.C. Contribution of mitochondria to oxidative stress associated with alcoholic liver disease. Free Radic. Biol. Med. 2002, 32, 11–16. [Google Scholar] [CrossRef]
- Crabb, D.W.; Matsumoto, M.; Chang, D.; You, M. Overview of the role of alcohol dehydrogenase and aldehyde dehydrogenase and their variants in the genesis of alcohol-related pathology. Proc. Nutr. Soc. 2004, 63, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Edenberg, H.J. The genetics of alcohol metabolism: Role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res. Health 2007, 30, 5–13. [Google Scholar]
- Vondracek, M.; Xi, Z.; Larsson, P.; Baker, V.; Mace, K.; Pfeifer, A.; Tjälve, H.; Donato, M.T.; Gomez-Lechon, M.J.; Graftström, R.C. Cytochrome P450 expression and related metabolism in human buccal mucosa. Carcinogenesis 2001, 22, 481–488. [Google Scholar] [CrossRef]
- Muto, M.; Hitomi, Y.; Ohtsu, A.; Shimada, H.; Kashiwase, Y.; Sasaki, H.; Yoshida, S.; Esumi, H. Acetaldehyde production by non-pathogenic Neisseria in human oral microflora: Implications for carcinogenesis in upper aerodigestive tract. Int. J. Cancer 2000, 88, 342–350. [Google Scholar] [CrossRef]
- Kurkivuori, J.; Salaspuro, V.; Kaihovaara, P.; Kari, K.; Rautemaa, R.; Grönroos, L.; Meurman, J.H.; Salaspuro, M. Acetaldehyde production from ethanol by oral streptococci. Oral Oncol. 2007, 43, 181–186. [Google Scholar] [CrossRef]
- Moritani, K.; Takeshita, T.; Shibata, Y.; Ninomiya, T.; Kiyohara, Y.; Yamashita, Y. Acetaldehyde production by major oral microbes. Oral Dis. 2015, 21, 748–754. [Google Scholar] [CrossRef]
- Homann, N.; Jousimies-Somer, H.; Jokelainen, K.; Heine, R.; Salaspuro, M. High acetaldehyde levels in saliva after ethanol consumption: Methodological aspects and pathogenetic implications. Carcinogenesis 1997, 18, 1743. [Google Scholar] [CrossRef] [PubMed]
- Salaspuro, V.; Salaspuro, M. Synergistic effect of alcohol drinking and smoking on in vivo acetaldehyde concentration in saliva. Int. J. Cancer 2004, 111, 480–483. [Google Scholar] [CrossRef] [PubMed]
- Lachenmeier, D.W.; Sohnius, E.M. The role of acetaldehyde outside ethanol metabolism in the carcinogenicity of alcoholic beverages: Evidence from a large chemical survey. Food Chem. Toxicol. 2008, 46, 2903–2911. [Google Scholar] [CrossRef]
- Balbo, S.; Brooks, P.J. Implications of Acetaldehyde-Derived DNA Adducts for Understanding Alcohol-Related Carcinogenesis. In Biological Basis of Alcohol-Induced Cancer. Advances in Experimental Medicine and Biology; Vasiliou, V., Zakhari, S., Seitz, H., Hoek, J., Eds.; Springer New York LLC: New York, NY, USA, 2015; Volume 815, pp. 71–88. [Google Scholar] [CrossRef]
- Homann, N.; Tillonen, J.; Meurman, J.H.; Rintamäki, H.; Lindqvist, C.; Rautio, M.; Jousimies-Somer, H.; Salaspuro, M. Increased salivary acetaldehyde levels in heavy drinkers and smokers: A microbiological approach to oral cavity cancer. Carcinogenesis 2000, 21, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Peters, B.A.; Jacobs, E.J.; Gapstur, S.M.; Purdue, M.P.; Freedman, N.D.; Alekseyenko, A.V.; Wu, J.; Yang, L.; Pei, Z.; et al. Drinking alcohol is associated with variation in the human oral microbiome in a large study of American adults. Microbiome 2018, 6. [Google Scholar] [CrossRef]
- Zakhari, S. Overview: How is alcohol metabolized by the body? Alcohol Res. Health 2006, 29, 245–254. [Google Scholar]
- Lieber, C.S.; DeCarli, L.M. Hepatic microsomal ethanol-oxidizing system. In vitro characteristics and adaptive properties in vivo. J. Biol. Chem. 1970, 245, 2512. [Google Scholar] [CrossRef]
- Salmela, K.S.; Kessova, I.G.; Tsyrlov, I.B.; Lieber, C.S. Respective Roles of Human Cytochrome P-4502E1, 1A2, and 3A4 in the Hepatic Microsomal Ethanol Oxidizing System. Alcohol. Clin. Exp. Res. 1998, 22, 2125–2132. [Google Scholar] [CrossRef]
- Lieber, C.S. The discovery of the microsomal ethanol oxidizing system and its physiologic and pathologic role. Drug Metab. Rev. 2004, 36, 511–529. [Google Scholar] [CrossRef]
- Lieber, C.S.; Decarli, L.M. The role of the hepatic microsomal ethanol oxidizing system (MEOS) for ethanol metabolism in vivo. J. Pharmacol. Exp. Ther. 1972, 181, 279–2887. [Google Scholar]
- Oneta, C.M.; Lieber, C.S.; Li, J.J.; Rüttimann, S.; Schmid, B.; Lattmann, J.; Rosman, A.S.; Seitz, H.K. Dynamics of cytochrome P4502E1 activity in man: Induction by ethanol and disappearance during withdrawal phase. J. Hepatol. 2002, 36, 47–52. [Google Scholar] [CrossRef]
- Kubiak-Tomaszewska, G.; Tomaszewski, P.; Pachecka, J.; Struga, M.; Olejarz, W.; Mielczarek-Puta, M.; Nowicka, G. Molecular mechanisms of ethanol biotransformation: Enzymes of oxidative and nonoxidative metabolic pathways in human. Xenobiotica 2020, 50, 1180–1201. [Google Scholar] [CrossRef] [PubMed]
- Goyal, M.M.; Basak, A. Hydroxyl radical generation theory: A possible explanation of unexplained actions of mammalian catalase. Int. J. Biochem. Mol. Biol. 2012, 3, 289. [Google Scholar] [CrossRef][Green Version]
- Aragon, C.M.; Rogan, F.; Amit, Z. Ethanol metabolism in rat brain homogenates by a catalase-H2O2 system. Biochem. Pharmacol. 1992, 44, 93–98. [Google Scholar] [CrossRef]
- Zimatkin, S.M.; Pronko, S.P.; Vasiliou, V.; Gonzalez, F.J.; Deitrich, R.A. Enzymatic mechanisms of ethanol oxidation in the brain. Alcohol. Clin. Exp. Res. 2006, 30, 1500–1505. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.L.; Vaca, C.E. Detection of DNA adducts of acetaldehyde in peripheral white blood cells of alcohol abusers. Carcinogenesis 1997, 18, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; McIntee, E.J.; Cheng, G.; Shi, Y.; Villalta, P.W.; Hecht, S.S. Identification of DNA adducts of acetaldehyde. Chem. Res. Toxicol. 2000, 13, 1149–1157. [Google Scholar] [CrossRef]
- Perrino, F.W.; Blans, P.; Harvey, S.; Gelhaus, S.L.; McGrath, C.; Akman, S.A.; Jenkins, G.S.; LaCourse, W.R.; Fishbein, J.C. The N2-Ethylguanine and the O6-Ethyl- and O6-Methylguanine Lesions in DNA: Contrasting Responses from the “Bypass” DNA Polymerase η and the Replicative DNA Polymerase α. Chem. Res. Toxicol. 2003, 16, 1616–1623. [Google Scholar] [CrossRef]
- Choi, J.Y.; Guengerich, F.P. Adduct size limits efficient and error-free bypass across bulky N2-guanine DNA lesions by human DNA polymerase η. J. Mol. Biol. 2005, 352, 72–90. [Google Scholar] [CrossRef] [PubMed]
- Upton, D.C.; Wang, X.; Blans, P.; Perrino, F.W.; Fishbein, J.C.; Akman, S.A. Replication of N2-ethyldeoxyguanosine DNA adducts in the human embryonic kidney cell line 293. Chem. Res. Toxicol. 2006, 19, 960–967. [Google Scholar] [CrossRef]
- Theruvathu, J.A.; Jaruga, P.; Nath, R.G.; Dizdaroglu, M.; Brooks, P.J. Polyamines stimulate the formation of mutagenic 1,N2-propanodeoxyguanosine adducts from acetaldehyde. Nucleic Acids Res. 2005, 33, 3520. [Google Scholar] [CrossRef]
- Garcia, C.C.M.; Angeli, J.P.F.; Freitas, F.P.; Gomes, O.F.; De Oliveira, T.F.; Loureiro, A.P.M.; Di Mascio, P.; Medeiros, M.H. [13C2]-acetaldehyde promotes unequivocal formation of 1, N2-propano-2′-deoxyguanosine in human cells. J. Am. Chem. Soc. 2011, 133, 9140–9143. [Google Scholar] [CrossRef] [PubMed]
- Brooks, P.J.; Zakhari, S. Acetaldehyde and the genome: Beyond nuclear DNA adducts and carcinogenesis. Environ. Mol. Mutagen. 2014, 55, 77–91. [Google Scholar] [CrossRef]
- Yang, I.Y.; Hossain, M.; Miller, H.; Khullar, S.; Johnson, F.; Grollman, A.; Moriya, M. Responses to the Major Acrolein-derived Deoxyguanosine Adduct in Escherichia coli. J. Biol. Chem. 2001, 276, 9071–9076. [Google Scholar] [CrossRef] [PubMed]
- Minko, I.G.; Washington, M.T.; Kanuri, M.; Prakash, L.; Prakash, S.; Lloyd, R.S. Translesion synthesis past acrolein-derived DNA adduct, γ-hydroxypropanodeoxyguanosine, by yeast and human DNA polymerase η. J. Biol. Chem. 2003, 278, 784–790. [Google Scholar] [CrossRef] [PubMed]
- Langevin, F.; Crossan, G.P.; Rosado, I.V.; Arends, M.J.; Patel, K.J. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 2011, 475, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Garaycoechea, J.I.; Crossan, G.P.; Langevin, F.; Mulderrig, L.; Louzada, S.; Yang, F.; Guilbaud, G.; Park, N.; Roerink, S.; Nik-Zainal, S.; et al. Alcohol and endogenous aldehydes damage chromosomes and mutate stem cells. Nature 2018, 553, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Tacconi, E.M.; Lai, X.; Folio, C.; Porru, M.; Zonderland, G.; Badie, S.; Michl, J.; Sechi, I.; Rogier, M.; Matía García, V.; et al. BRCA 1 and BRCA 2 tumor suppressors protect against endogenous acetaldehyde toxicity. EMBO Mol. Med. 2017, 9, 1398–1414. [Google Scholar] [CrossRef]
- Kim, H.; D’Andrea, A.D. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012, 26, 1393–1408. [Google Scholar] [CrossRef]
- Hodskinson, M.R.; Bolner, A.; Sato, K.; Kamimae-Lanning, A.N.; Rooijers, K.; Witte, M.; Mahesh, M.; Silhan, J.; Petek, M.; Williams, D.M.; et al. Alcohol-derived DNA crosslinks are repaired by two distinct mechanisms. Nature 2020, 579, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Loureiro, A.P.M.; Di Mascio, P.; Gomes, O.F.; Medeiros, M.H. trans,trans-2,4-Decadienal-induced 1,N2-etheno-2′-deoxyguanosine adduct formation. Chem. Res. Toxicol. 2000, 13, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.Y.; Zang, H.; Angel, K.C.; Kozekov, I.D.; Goodenough, A.K.; Rizzo, C.J.; Guengerich, F.P. Translesion synthesis across 1,N2-ethenoguanine by human DNA polymerases. Chem. Res. Toxicol. 2006, 19, 879–886. [Google Scholar] [CrossRef]
- Thelen, A.Z.; O’Brien, P.J. Recognition of 1,N2-ethenoguanine by alkyladenine DNA glycosylase is restricted by a conserved active-site residue. J. Biol. Chem. 2020, 295, 1685–1693. [Google Scholar] [CrossRef]
- Balbo, S.; Juanes, R.C.; Khariwala, S.; Baker, E.J.; Daunais, J.B.; Grant, K.A. Increased levels of the acetaldehyde-derived DNA adduct N2-ethyldeoxyguanosine in oral mucosa DNA from Rhesus monkeys exposed to alcohol. Mutagenesis 2016, 31, 553–558. [Google Scholar] [CrossRef] [PubMed]
- Yukawa, Y.; Ohashi, S.; Amanuma, Y.; Nakai, Y.; Tsurumaki, M.; Kikuchi, O.; Miyamoto, S.; Oyama, T.; Kawamoto, T.; Chiba, T.; et al. Impairment of aldehyde dehydrogenase 2 increases accumulation of acetaldehyde-derived DNA damage in the esophagus after ethanol ingestion. Am. J. Cancer Res. 2014, 4, 279–284. [Google Scholar]
- Balbo, S.; Meng, L.; Bliss, R.L.; Jensen, J.A.; Hatsukami, D.K.; Hecht, S.S. Kinetics of DNA adduct formation in the oral cavity after drinking alcohol. Cancer Epidemiol. Biomarkers Prev. 2012, 21, 601–608. [Google Scholar] [CrossRef]
- Matsuda, T.; Yabushita, H.; Kanaly, R.A.; Shibutani, S.; Yokoyama, A. Increased DNA damage in ALDH2-deficient alcoholics. Chem. Res. Toxicol. 2006, 19, 1374–1378. [Google Scholar] [CrossRef]
- Wu, D.; Cederbaum, A.I. Oxidative stress and alcoholic liver disease. Semin. Liver Dis. 2009, 29, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Kesarwala, A.; Krishna, M.; Mitchell, J. Oxidative stress in oral diseases. Oral Dis. 2016, 22, 9–18. [Google Scholar] [CrossRef]
- Enwonwu, C.O.; Meeks, V.I. Bionutrition and oral cancer in humans. Crit. Rev. Oral Biol. Med. 1995, 6, 5–17. [Google Scholar] [CrossRef]
- Irie, K.; Tomofuji, T.; Tamaki, N.; Sanbe, T.; Ekuni, D.; Azuma, T.; Maruyama, T.; Yamamoto, T. Effects of ethanol consumption on periodontal inflammation in rats. J. Dent. Res. 2008, 87, 456–460. [Google Scholar] [CrossRef]
- Cadet, J.; Richard Wagner, J. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb. Perspect. Biol. 2013, 5, a012559. [Google Scholar] [CrossRef]
- Valavanidis, A.; Vlachogianni, T.; Fiotakis, C. 8-Hydroxy-2′-deoxyguanosine (8-OHdG): A critical biomarker of oxidative stress and carcinogenesis. J. Environ. Sci. Health Part C Environ. Carcinog. Ecotoxicol. Rev. 2009, 27, 120–139. [Google Scholar] [CrossRef]
- Cheng, K.C.; Cahill, D.S.; Kasai, H.; Nishimura, S.; Loeb, L.A. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G → T and A → C substitutions. J. Biol. Chem. 1992, 267, 166–172. [Google Scholar] [CrossRef]
- Jaiswal, M.; LaRusso, N.F.; Nishioka, N.; Nakabeppu, Y.; Gores, G.J. Human Ogg1, a Protein Involved in the Repair of 8-Oxoguanine, Is Inhibited by Nitric Oxide. Cancer Res. 2001, 61, 6388–6393. [Google Scholar] [PubMed]
- Deng, X.S.; Deitrich, R. Ethanol Metabolism and Effects: Nitric Oxide and its Interaction. Curr. Clin. Pharmacol. 2007, 2, 145–153. [Google Scholar] [CrossRef]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.; Mazur, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef] [PubMed]
- Niemela, O. Distribution of ethanol-induced protein adducts in vivo: Relationship to tissue injury. Free Radic. Biol. Med. 2001, 31, 1533–1538. [Google Scholar] [CrossRef]
- Ghissassi, F.E.; Barbin, A.; Nair, J.; Bartsch, H. Formation of 1,N6-Ethenoadenine and 3,N4-Ethenocytosine by Lipid Peroxidation Products and Nucleic Acid Bases. Chem. Res. Toxicol. 1995, 8, 278–283. [Google Scholar] [CrossRef]
- O’Brien, P.J.; Ellenberger, T. Dissecting the Broad Substrate Specificity of Human 3-Methyladenine-DNA Glycosylase. J. Biol. Chem. 2004, 279, 9750–9757. [Google Scholar] [CrossRef]
- Kavli, B.; Sundheim, O.; Akbari, M.; Otterlei, M.; Nilsen, H.; Skorpen, F.; Aas, P.A.; Hagen, L.; Krokan, H.E.; Slupphaug, G. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem. 2002, 277, 39926–39936. [Google Scholar] [CrossRef]
- Goto, M.; Shinmura, K.; Matsushima, Y.; Ishino, K.; Yamada, H.; Totsuka, Y.; Matsuda, T.; Nakagama, H.; Sugimura, H. Human DNA glycosylase enzyme TDG repairs thymine mispaired with exocyclic etheno-DNA adducts. Free Radic. Biol. Med. 2014, 76, 136–146. [Google Scholar] [CrossRef]
- Linhart, K.; Bartsch, H.; Seitz, H.K. The role of reactive oxygen species (ROS) and cytochrome P-450 2E1 in the generation of carcinogenic etheno-DNA adducts. Redox Biol. 2014, 3, 56–62. [Google Scholar] [CrossRef]
- Hu, W.; Feng, Z.; Eveleigh, J.; Iyer, G.; Pan, J.; Amin, S.; Chung, F.L.; Tang, M.S. The major lipid peroxidation product, trans-4-hydroxy-2-nonenal, preferentially forms DNA adducts at codon 249 of human p53 gene, a unique mutational hotspot in hepatocellular carcinoma. Carcinogenesis 2002, 23, 1789. [Google Scholar] [CrossRef]
- Cederbaum, A.I.; Lu, Y.; Wu, D. Role of oxidative stress in alcohol-induced liver injury. Arch. Toxicol. 2009, 83, 519–548. [Google Scholar] [CrossRef]
- Tuma, D.J.; Casey, C.A. Dangerous byproducts of alcohol breakdown—Focus on adducts. Alcohol Res. Health 2003, 27, 285–290. [Google Scholar] [PubMed]
- Feller, L.; Altini, M.; Lemmer, J. Inflammation in the context of oral cancer. Oral Oncol. 2013, 49, 887–892. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T. Alcohol consumption and oxidative DNA damage. Int. J. Environ. Res. Public Health 2011, 8, 2895–2906. [Google Scholar] [CrossRef] [PubMed]
- Fagundes, N.C.F.; Fernandes, L.M.P.; Paraense, R.S.D.O.; De Farias-Junior, P.M.A.; Teixeira, F.B.; Alves, S.M.; Pinheiro, J.D.J.V.; Crespo-López, M.E.; Maia, C.S.F.; Lima, R.R. Binge Drinking of Ethanol during Adolescence Induces Oxidative Damage and Morphological Changes in Salivary Glands of Female Rats. Oxid. Med. Cell. Longev. 2016, 2016, 7323627. [Google Scholar] [CrossRef]
- Urvalek, A.M.; Osei-Sarfo, K.; Tang, X.H.; Zhang, T.; Scognamiglio, T.; Gudas, L.J. Identification of Ethanol and 4-Nitroquinoline-1-Oxide Induced Epigenetic and Oxidative Stress Markers During Oral Cavity Carcinogenesis. Alcohol. Clin. Exp. Res. 2015, 39, 1360–1372. [Google Scholar] [CrossRef]
- Kaur, J.; Politis, C.; Jacobs, R. Salivary 8-hydroxy-2-deoxyguanosine, malondialdehyde, vitamin C, and vitamin E in oral pre-cancer and cancer: Diagnostic value and free radical mechanism of action. Clin. Oral Investig. 2016, 20, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Warnakulasuriya, S.; Parkkila, S.; Nagao, T.; Preedy, V.R.; Pasanen, M.; Koivisto, H.; Niemelä, O. Demonstration of ethanol-induced protein adducts in oral leukoplakia (pre-cancer) and cancer. J. Oral Pathol. Med. 2008, 37, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Frank, A.; Seitz, H.K.; Bartsch, H.; Frank, N.; Nair, J. Immunohistochemical detection of 1, N6-ethenodeoxyadenosine in nuclei of human liver affected by disease predisposing to hepato-carcinogenesis. Carcinogenesis 2004, 25, 1027–1031. [Google Scholar] [CrossRef]
- Wang, Y.; Millonig, G.; Nair, J.; Patsenker, E.; Stickel, F.; Mueller, S.; Bartsch, H.; Seitz, H.K. Ethanol-induced cytochrome P4502E1 causes carcinogenic etheno-DNA lesions in alcoholic liver disease. Hepatology 2009, 50, 453–461. [Google Scholar] [CrossRef]
- Millonig, G.; Wang, Y.; Homann, N.; Bernhardt, F.; Qin, H.; Mueller, S.; Bartsch, H.; Seitz, H.K. Ethanol-mediated carcinogenesis in the human esophagus implicates CYP2E1 induction and the generation of carcinogenic DNA-lesions. Int. J. Cancer 2011, 128, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef]
- Shukla, S.D.; Velazquez, J.; French, S.W.; Lu, S.C.; Ticku, M.K.; Zakhari, S. Emerging role of epigenetics in the actions of alcohol. Alcohol. Clin. Exp. Res. 2008, 32, 1525–1534. [Google Scholar] [CrossRef]
- Varela-Rey, M.; Woodhoo, A.; Martinez-Chantar, M.L.; Mato, J.M.; Lu, S.C. Alcohol, DNA methylation, and cancer. Alcohol Res. Curr. Rev. 2012, 35, 25–35. [Google Scholar]
- Barak, A.J.; Beckenhauer, H.C.; Tuma, D.J. Betaine effects on hepatic methionine metabolism elicited by short-term ethanol feeding. Alcohol 1996, 13, 483–486. [Google Scholar] [CrossRef]
- Sánchez-Góngora, E.; Ruiz, F.; Mingorance, J.; An, W.; Corrales, F.J.; Mato, J.M. Interaction of liver methionine adenosyltransferase with hydroxyl radical. FASEB J. 1997, 11, 1013–1019. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, F.; Corrales, F.J.; Miqueo, C.; Mato, J.M. Nitric oxide inactivates rat hepatic methionine adenosyltransferase in vivo by S-nitrosylation. Hepatology 1998, 28, 1051–1057. [Google Scholar] [CrossRef]
- Lu, S.C.; Huang, Z.Z.; Yang, H.; Mato, J.M.; Avila, M.A.; Tsukamoto, H. Changes in methionine adenosyltransferase and S-adenosylmethionine homeostasis in alcoholic rat liver. Am. J. Physiol. Gastrointest. Liver Physiol. 2000, 279, G178–G185. [Google Scholar] [CrossRef] [PubMed]
- Garro, A.J.; McBeth, D.L.; Lima, V.; Lieber, C.S. Ethanol Consumption Inhibits Fetal DNA Methylation in Mice: Implications for the Fetal Alcohol Syndrome. Alcohol. Clin. Exp. Res. 1991, 15, 395–398. [Google Scholar] [CrossRef]
- Bönsch, D.; Lenz, B.; Fiszer, R.; Frieling, H.; Kornhuber, J.; Bleich, S. Lowered DNA methyltransferase (DNMT-3b) mRNA expression is associated with genomic DNA hypermethylation in patients with chronic alcoholism. J. Neural Transm. 2006, 113, 1299–1304. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y. Nutritional epigenetics: Impact of folate deficiency on DNA methylation and colon cancer susceptibility. J. Nutr. 2005, 135, 2703–2709. [Google Scholar] [CrossRef] [PubMed]
- Sharma, J.; Krupenko, S.A. Folate pathways mediating the effects of ethanol in tumorigenesis. Chem.-Biol. Interact. 2020, 324. [Google Scholar] [CrossRef]
- Smith, I.M.; Mydlarz, W.K.; Mithani, S.K.; Califano, J.A. DNA global hypomethylation in squamous cell head and neck cancer associated with smoking, alcohol consumption and stage. Int. J. Cancer 2007, 121, 1724–1728. [Google Scholar] [CrossRef]
- Chen, H.C.; Yang, C.M.; Cheng, J.T.; Tsai, K.W.; Fu, T.Y.; Liou, H.H.; Tseng, H.H.; Lee, J.H.; Li, G.C.; Wang, J.S.; et al. Global DNA hypomethylation is associated with the development and poor prognosis of tongue squamous cell carcinoma. J. Oral Pathol. Med. 2016, 45, 409–417. [Google Scholar] [CrossRef]
- Tanaka, C.; Uzawa, K.; Shibahara, T.; Yokoe, H.; Noma, H.; Tanzawa, H. Expression of an inhibitor of apoptosis, survivin, in oral carcinogenesis. J. Dent. Res. 2003, 82, 607–611. [Google Scholar] [CrossRef]
- Muzio, L.L.; Pannone, G.; Staibano, S.; Mignogna, M.D.; Rubini, C.; Mariggiò, M.A.; Procaccini, M.; Ferrari, F.; Rosa, G.D.; Altieri, D.C. Survivin expression in oral squamous cell carcinoma. Br. J. Cancer 2003, 89, 2244–2248. [Google Scholar] [CrossRef]
- Gaździcka, J.; Goła̧bek, K.; Strzelczyk, J.K.; Ostrowska, Z. Epigenetic Modifications in Head and Neck Cancer. Biochem. Genet. 2020, 58, 213–244. [Google Scholar] [CrossRef] [PubMed]
- Subbalekha, K.; Pimkhaokham, A.; Pavasant, P.; Chindavijak, S.; Phokaew, C.; Shuangshoti, S.; Matangkasombut, O.; Mutirangura, A. Detection of LINE-1s hypomethylation in oral rinses of oral squamous cell carcinoma patients. Oral Oncol. 2009, 45, 184–191. [Google Scholar] [CrossRef]
- Foy, J.P.; Pickering, C.R.; Papadimitrakopoulou, V.A.; Jelinek, J.; Lin, S.H.; William, W.N.; Frederick, M.J.; Wang, J.; Lang, W.; Feng, L.; et al. New DNA methylation markers and global DNA hypomethylation are associated with oral cancer development. Cancer Prev. Res. 2015, 8, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Ha, P.K.; Califano, J.A. Promoter methylation and inactivation of tumour-suppressor genes in oral squamous-cell carcinoma. Lancet Oncol. 2006, 7, 77–82. [Google Scholar] [CrossRef]
- Zakhari, S. Alcohol metabolism and epigenetics changes. Alcohol Res. Curr. Rev. 2013, 35, 6–16. [Google Scholar]
- Mews, P.; Egervari, G.; Nativio, R.; Sidoli, S.; Donahue, G.; Lombroso, S.I.; Alexander, D.C.; Riesche, S.L.; Heller, E.A.; Nestler, E.J.; et al. Alcohol metabolism contributes to brain histone acetylation. Nature 2019, 574, 717–721. [Google Scholar] [CrossRef] [PubMed]
- Park, P.H.; Lim, R.W.; Shukla, S.D. Involvement of histone acetyltransferase (HAT) in ethanol-induced acetylation of histone H3 in hepatocytes: Potential mechanism for gene expression. Am. J. Physiol. Gastrointest. Liver Physiol. 2005, 289, G1124–G1136. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, M.; Matassa, D.S.; Conte, M.; Colella, G.; Rana, G.; Fucci, L.; Piscopo, M. H3K4 histone methylation in oral squamous cell carcinoma. Acta Biochim. Pol. 2009, 56, 405–410. [Google Scholar] [CrossRef]
- Arif, M.; Vedamurthy, B.M.; Choudhari, R.; Ostwal, Y.B.; Mantelingu, K.; Kodaganur, G.S.; Kundu, T.K. Nitric oxide-mediated histone hyperacetylation in oral cancer: Target for a water-soluble HAT inhibitor, CTK7A. Chem. Biol. 2010, 17, 903–913. [Google Scholar] [CrossRef]
- Park, P.H.; Miller, R.; Shukla, S.D. Acetylation of histone H3 at lysine 9 by ethanol in rat hepatocytes. Biochem. Biophys. Res. Commun. 2003, 306, 501–504. [Google Scholar] [CrossRef]
- Kim, J.S.; Shukla, S.D. Acute in vivo effect of ethanol (binge drinking) on histone H3 modifications in rat tissues. Alcohol Alcohol. 2006, 41, 126–132. [Google Scholar] [CrossRef]
- Kurdistani, S.; Tavazoie, S.; Grunstein, M. Mapping global histone acetylation patterns to gene expression. Cell 2004, 117, 721–733. [Google Scholar] [CrossRef]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.; Schones, D.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-resolution profiling of histone methylations in the human genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Campbell, P.J.; Stratton, M.R. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013, 3, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, L.B.; Stratton, M.R. Mutational signatures: The patterns of somatic mutations hidden in cancer genomes. Curr. Opin. Genet. Dev. 2014, 24, 52. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Setton, J.; Lee, N.Y.; Riaz, N.; Powell, S.N. The therapeutic significance of mutational signatures from DNA repair deficiency in cancer. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Mutation Signatures v3. Available online: https://cancer.sanger.ac.uk/signatures/ (accessed on 14 June 2021).
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef]
- Roberts, S.A.; Lawrence, M.S.; Klimczak, L.J.; Grimm, S.A.; Fargo, D.; Stojanov, P.; Kiezun, A.; Kryukov, G.V.; Carter, S.L.; Saksena, G.; et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013, 45, 970–976. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Kim, J.; Haradhvala, N.J.; Huang, M.N.; Tian Ng, A.W.; Wu, Y.; Boot, A.; Covington, K.R.; Gordenin, D.A.; Bergstrom, E.N.; et al. The repertoire of mutational signatures in human cancer. Nature 2020, 578, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Kucab, J.E.; Zou, X.Q.; Morganella, S.; Joel, M.; Nanda, A.S.; Nagy, E.; Gomez, C.; Degasperi, A.; Harris, R.; Jackson, S.P.; et al. A Compendium of Mutational Signatures of Environmental Agents. Cell 2019, 117, 821–836. [Google Scholar] [CrossRef]
- Matsuda, T.; Kawanishi, M.; Yagi, T.; Matsui, S.; Takebe, H. Specific tandem GG to TT base substitutions induced by acetaldehyde are due to intra-strand crosslinks between adjacent guanine bases. PubMed—NCBI. Nucleic Acids Res. 1998, 26, 1769–1774. [Google Scholar] [CrossRef]
- Chang, J.; Tan, W.; Ling, Z.; Xi, R.; Shao, M.; Chen, M.; Luo, Y.; Zhao, Y.; Liu, Y.; Huang, X.; et al. Genomic analysis of oesophageal squamous-cell carcinoma identifies alcohol drinking-related mutation signature and genomic alterations. Nat. Commun. 2017, 8, 15290. [Google Scholar] [CrossRef]
- Li, X.C.; Wang, M.Y.; Yang, M.; Dai, H.J.; Zhang, B.F.; Wang, W.; Chu, X.L.; Wang, X.; Zheng, H.; Niu, R.F.; et al. A mutational signature associated with alcohol consumption and prognostically significantly mutated driver genes in esophageal squamous cell carcinoma. Ann. Oncol. 2018, 29, 938–944. [Google Scholar] [CrossRef]
- Letouzé, E.; Shinde, J.; Renault, V.; Couchy, G.; Blanc, J.F.; Tubacher, E.; Bayard, Q.; Bacq, D.; Meyer, V.; Semhoun, J.; et al. Mutational signatures reveal the dynamic interplay of risk factors and cellular processes during liver tumorigenesis. Nat. Commun. 2017, 8, 1315. [Google Scholar] [CrossRef]
- Supek, F.; Lehner, B. Clustered Mutation Signatures Reveal that Error-Prone DNA Repair Targets Mutations to Active Genes. Cell 2017, 170, 534–547.e23. [Google Scholar] [CrossRef] [PubMed]
- Sonohara, Y.; Yamamoto, J.; Tohashi, K.; Takatsuka, R.; Matsuda, T.; Iwai, S.; Kuraoka, I. Acetaldehyde forms covalent GG intrastrand crosslinks in DNA. Sci. Rep. 2019, 9, 660. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.M.; Férec, C.; Cooper, D.N. Patterns and mutational signatures of tandem base substitutions causing human inherited disease. Hum. Mutat. 2013, 34, 1119–1130. [Google Scholar] [CrossRef] [PubMed]
- Imielinski, M.; Berger, A.H.; Hammerman, P.S.; Hernandez, B.; Pugh, T.J.; Hodis, E.; Cho, J.; Suh, J.; Capelletti, M.; Sivachenko, A.; et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012, 150, 1107–1120. [Google Scholar] [CrossRef]
- Stransky, N.; Egloff, A.M.; Tward, A.D.; Kostic, A.D.; Cibulskis, K.; Sivachenko, A.; Kryukov, G.V.; Lawrence, M.S.; Sougnez, C.; McKenna, A.; et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011, 333, 1157–1160. [Google Scholar] [CrossRef]
- Plath, M.; Gass, J.; Hlevnjak, M.; Li, Q.; Feng, B.; Hostench, X.P.; Bieg, M.; Schroeder, L.; Holzinger, D.; Zapatka, M.; et al. Unraveling most abundant mutational signatures in head and neck cancer. Int. J. Cancer 2021, 148, 115–127. [Google Scholar] [CrossRef] [PubMed]
| Signature | Cancer Subsite | Proposed Etiology | References |
|---|---|---|---|
| DBS2 | Lung, Head and Neck | Acetaldehyde exposure | [199,200] |
| SBS16 | Esophagus, Liver | Acetaldehyde exposure | [201,202,203] |
| C4 | Liver, Head and Neck, Esophagus, Pancreas | Translesion polymerase η | [204] |
| E6 | Esophagus | Unknown | [201] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoes, L.; Dok, R.; Verstrepen, K.J.; Nuyts, S. Ethanol-Induced Cell Damage Can Result in the Development of Oral Tumors. Cancers 2021, 13, 3846. https://doi.org/10.3390/cancers13153846
Hoes L, Dok R, Verstrepen KJ, Nuyts S. Ethanol-Induced Cell Damage Can Result in the Development of Oral Tumors. Cancers. 2021; 13(15):3846. https://doi.org/10.3390/cancers13153846
Chicago/Turabian StyleHoes, Lore, Rüveyda Dok, Kevin J. Verstrepen, and Sandra Nuyts. 2021. "Ethanol-Induced Cell Damage Can Result in the Development of Oral Tumors" Cancers 13, no. 15: 3846. https://doi.org/10.3390/cancers13153846
APA StyleHoes, L., Dok, R., Verstrepen, K. J., & Nuyts, S. (2021). Ethanol-Induced Cell Damage Can Result in the Development of Oral Tumors. Cancers, 13(15), 3846. https://doi.org/10.3390/cancers13153846







