Transcriptomics-Based Phenotypic Screening Supports Drug Discovery in Human Glioblastoma Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells Used in This Study
2.2. Compounds Used in This Study
2.3. Proliferation Assay
2.4. Drug Combination Assays and Their Analysis
2.5. Microarray Analysis
3. Results
3.1. Established Cell Lines Used in These Studies
3.2. Compounds Used to Probe Drug-Induced Gene Expression
3.3. Growth Inhibition Characteristics of the Compounds
3.4. DIGEX Profiles for the Four Treatments
3.5. Upregulated Genes Accompanying Drug Treatments in U87MG Cells
3.6. Interpretation of the U87MG DIGEX Signatures
- The DIGEX signature for each cell/treatment combination is reproducible and specific, confirmed by the PCA analysis (Supplementary Figure S1).
- A GBM cell line such as U87MG, can radically change its DIGEX response when encountering different drug treatments, exhibiting considerable transcriptional plasticity.
3.7. Two Genes Upregulated by All Four Drug Treatments in U87MG
3.8. Which U87MG Genes Are Upregulated in Mardepodect and Regorafenib Treatments?
3.9. Pathway Enrichment Analysis for the Upregulated U87MG Gene Sets
3.10. Downregulated Genes Revealed by DIGEX
3.11. Pathway Analysis for the U87MG Downregulated Gene Sets
3.12. Summary of the U87MG Results
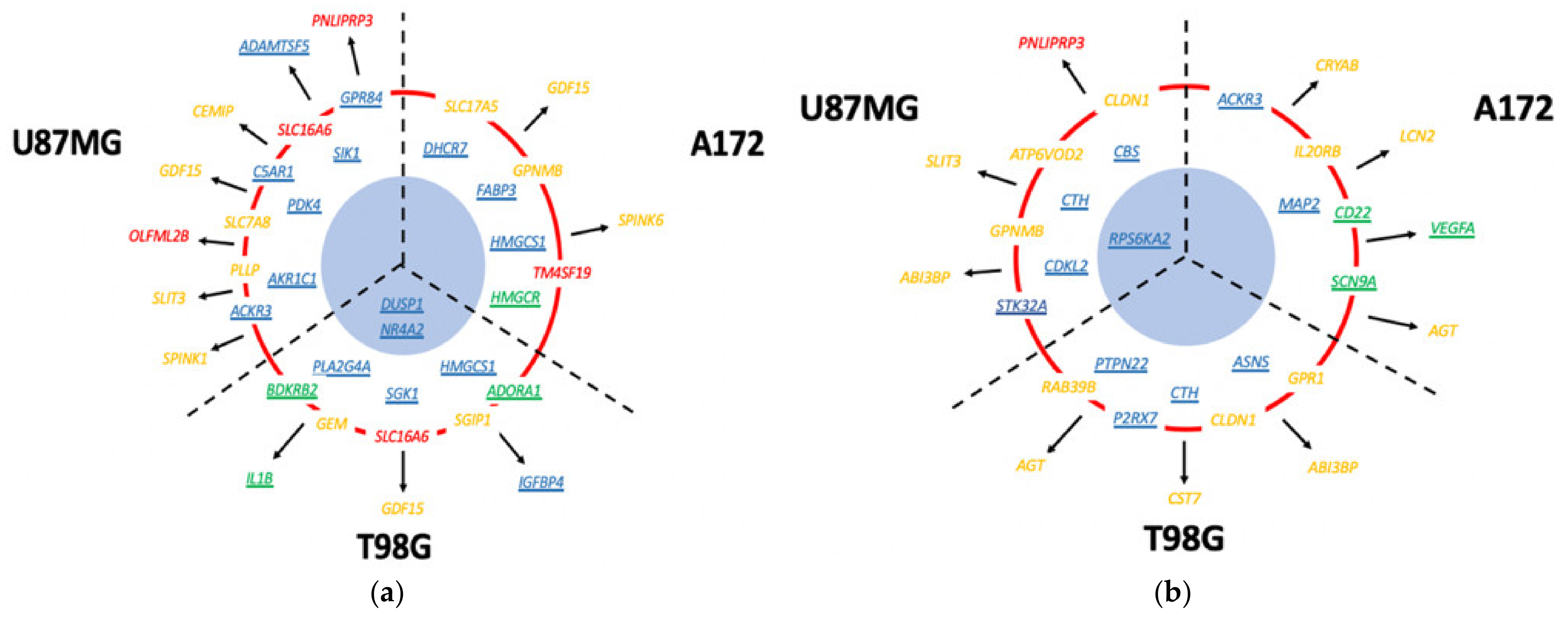
3.13. Which DIGEX Genes Are Shared between Mardepodect Treated U87MG, T98G, and A172 Cells?
3.13.1. Upregulated Genes
- GDF15 is a secreted growth factor, reportedly overexpressed in the cerebrospinal fluid (CSF) of GBM patients with poor treatment outcomes [42]. GDF15 binds to the GFRAL/RET receptor complex, stimulating cell growth through the ERK and AKT signaling pathways [68]. GDF15 has been suggested as a tumor-associated clinical biomarker suitable for liquid biopsy detection [69].
- DUS1/DUSP1 is a dual specificity phosphatase which dephosphorylates and inactivates the MAP kinase MAPK1/ERK2, leading amongst other effects to aberrant regulation of the cell cycle. DUS1 plays important roles in the initiation, progression, and recurrence of GBM [70].
- SIK1 is a serine/threonine protein kinase that regulates transcription by phosphorylating transcriptional coactivators such as the CRTCs and HDACs. When cAMP levels increase, SIKs are phosphorylated by activated PKA and sequestrated by phosphorylated 14-3-3 proteins as inactive complexes in the cytoplasm [71]. Increased SIK1 transcription in Mardepodect-treated GBM cells may reflect changes in these cAMP-driven processes.
- KS6A2/RPS6KA2, also known as RSK/RSK3, is a member of the RSK serine/threonine-protein kinase family that acts as a downstream effector of ERK in the MAPK1/ERK2 and MAPK3/ERK1 signaling pathway, mediating cellular proliferation and survival in prostate cancer [75]. The related RSK kinase, KS6A3/RSK2, encoded by RPS6KA3, has been reported to regulate growth and invasion in GBM [76].
- S1PR1 is the GPCR for the bioactive lyso-sphingolipid sphingosine 1-phosphate (S1P) which is coupled to the Gi subclass of heteromeric G proteins. In cancer cells, signaling through S1PR1 leads to the activation of RAC1, SRC, PTK2/FAK1, as well as MAP kinases, and influences cell proliferation and survival in GBM [77].
3.13.2. Downregulated Genes Shared between Cells
3.14. Differential Gene Expression Is Recapitulated in the Corresponding Pathway Analyses
3.15. How Do the DIGEX Signatures of Mardepodect-Treated Cells Compare to Those Seen in Regorafenib-Treated Cells?
3.16. What Can Be Inferred from the Cell-Specific DIGEX Signatures?
3.17. Which Genes Encode Proteins That Could Be Viable Drug Targets in GBM?
3.17.1. U87MG Cells Treated with Mardepodect
3.17.2. T98G and A172 Cells Treated with Mardepodect
3.18. Downregulated Genes May Indicate Cell Cycle Control Imposed by Drug Treatment
3.19. Summary of Drug-Induced Gene Expression (DIGEX) Analysis Results
3.20. Validating Individual Drug-Inducible Genes as Pharmacological Targets in GBM as Monotherapies and Drug Combinations
- Two FDA approved inhibitors for HMGCoA reductase, Atorvastatin and Simvastatin. The gene encoding HMGCoA reductase (HMGCR) is in the Top 200 genes upregulated by Mardepodect in A172 and T98G cells but not U87MG. HMGCR is absent from the Regorafenib Top 200.
- Two exploratory inhibitors of the salt-inducible kinase SIK1, HG-9-91-01, and WH-4-023. The gene encoding SIK1 is present in the Top 200 genes upregulated by Mardepodect in all three GBM cells but absent from the Top 200 in Regorafenib-treated cells.
- Two inhibitors of the Janus Kinase JAK2, the FDA approved drug Ruxolitinib and the exploratory compound AZD1480. The gene encoding JAK2 is induced in U87MG by LY-294002 [20] but absent from the Top 200 upregulated genes in both Mardepodect and Regorafenib treated cells.
- Two inhibitors of the bradykinin B2 receptor, Icatibant and WIN 64338. The BDKRB2 gene is in the Top 200 upregulated genes in Mardepodect treated T98G cells.
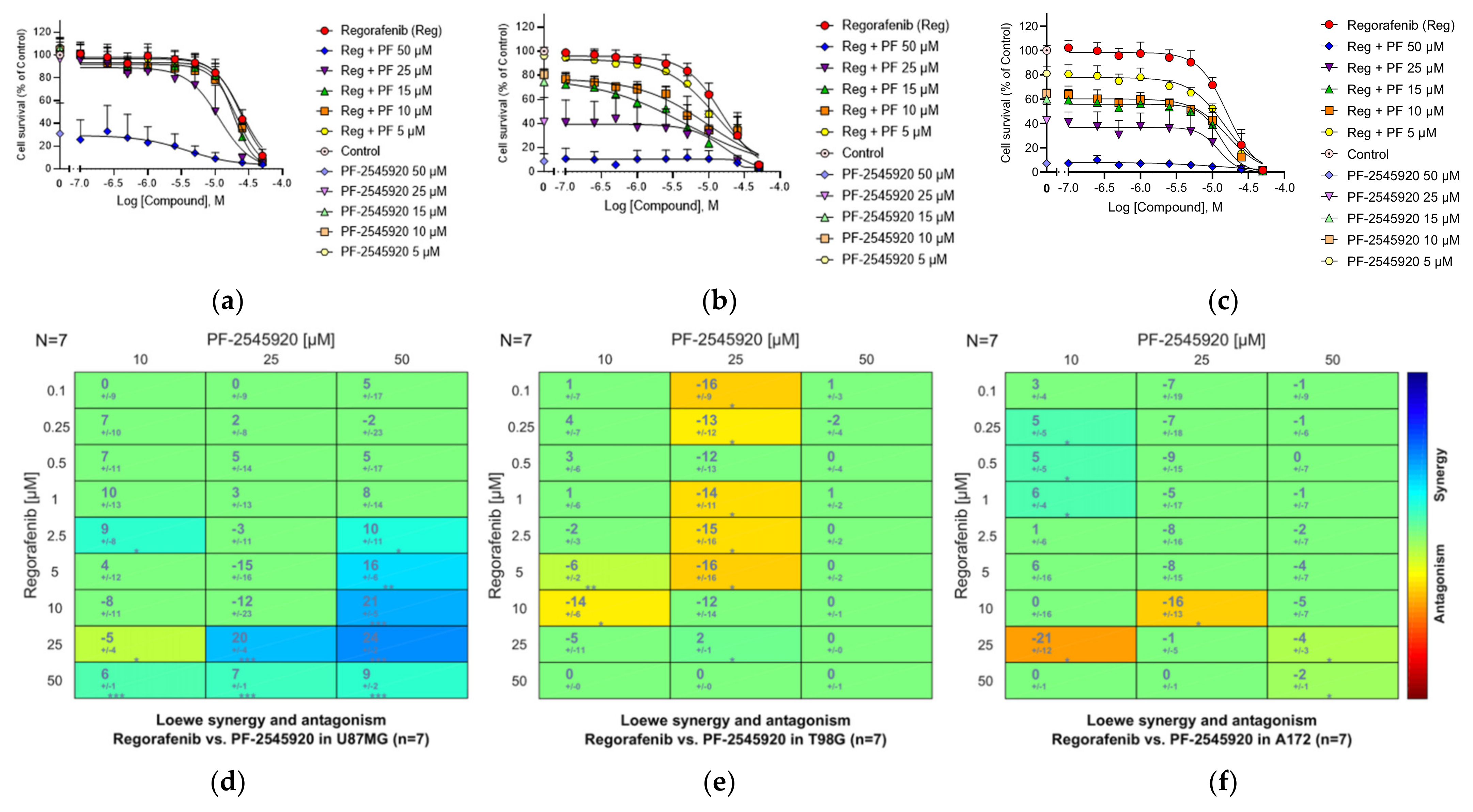
3.21. Combinations of the PDE10A Inhibitor Mardepodect and Regorafenib
3.22. HMGCoA Reductase
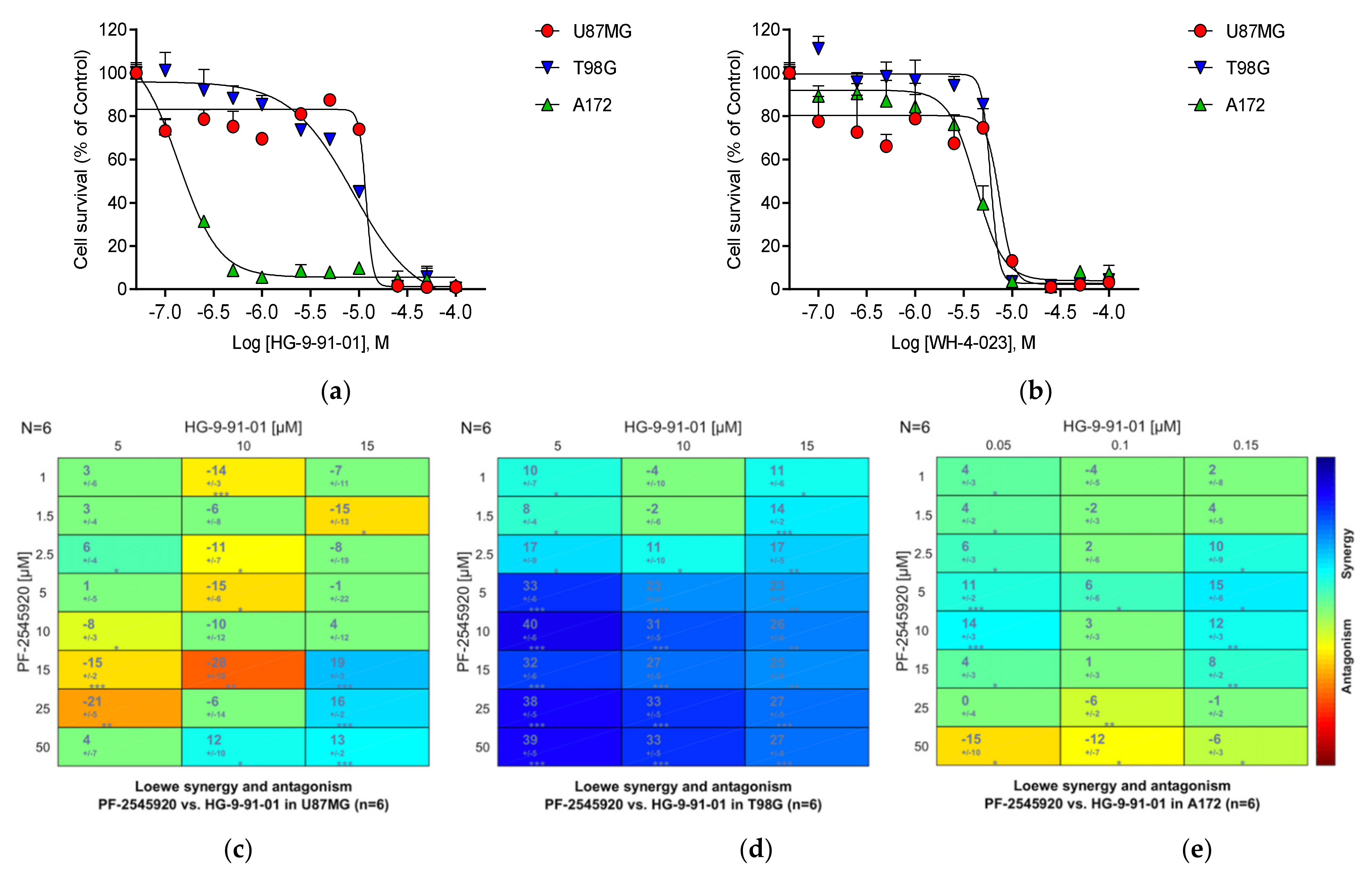
3.23. Salt-Inducible Kinase SIK1
3.24. Janus Kinase JAK2
3.25. GPCRs
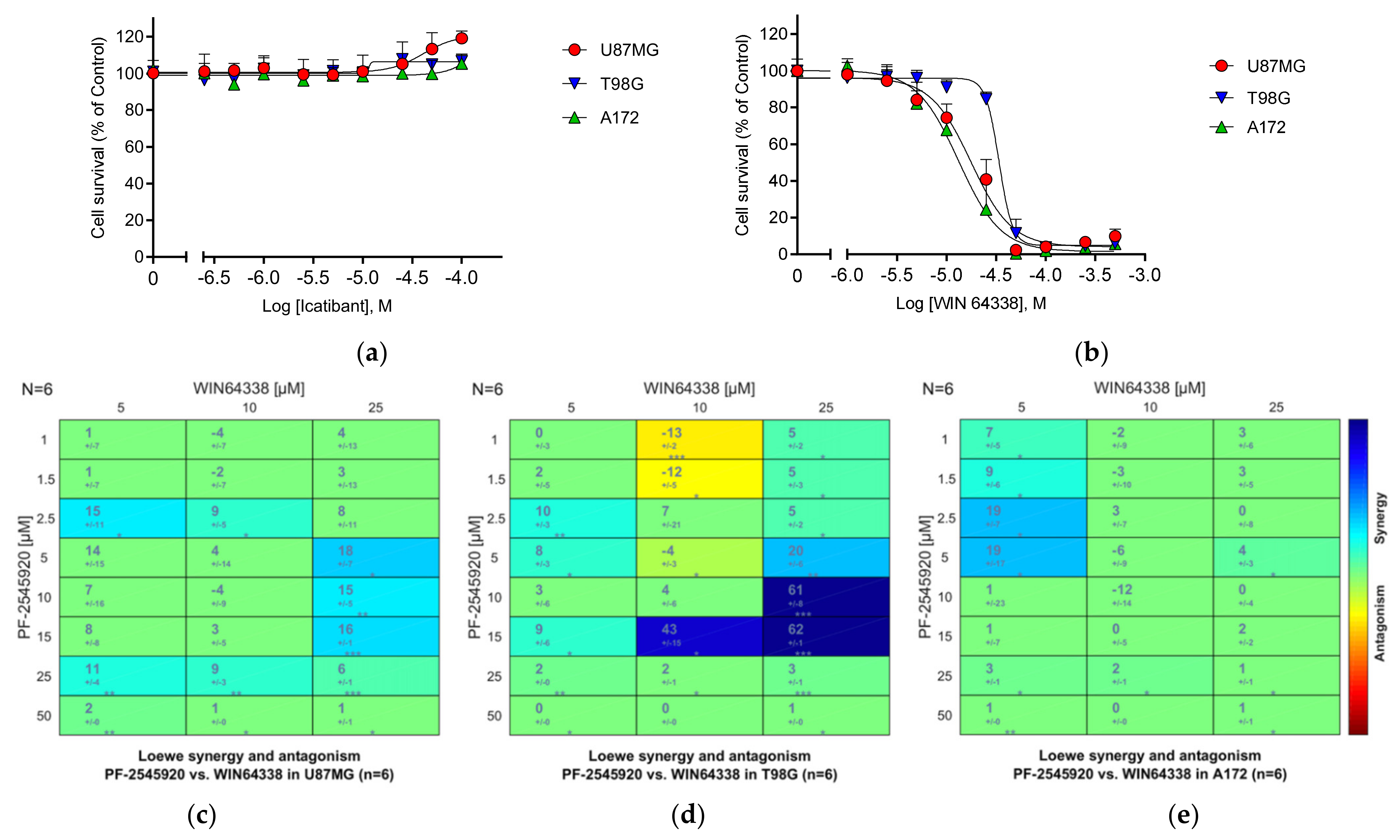
3.25.1. Bradykinin B2 Receptor (B2R)
3.25.2. Other GPCRs
3.26. Summary of the Compound Synergy Studies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sottoriva, A.; Spiteri, I.; Piccirillo, S.G.M.; Touloumis, A.; Collins, V.P.; Marioni, J.C.; Curtis, C.; Watts, C.; Tavare, S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. USA 2013, 110, 4009–4014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carén, H.; Stricker, S.H.; Bulstrode, H.; Gagrica, S.; Johnstone, E.; Bartlett, T.E.; Feber, A.; Wilson, G.; Teschendorff, A.E.; Bertone, P.; et al. Glioblastoma stem cells respond to differentiation cues but fail to undergo commitment and terminal cell-cycle arrest. Stem Cell Rep. 2015, 5, 829–842. [Google Scholar] [CrossRef] [Green Version]
- Engström, P.G.; Tommei, D.; Stricker, S.H.; Ender, C.; Pollard, S.M.; Bertone, P. Digital transcriptome profiling of normal and glioblastoma-derived neural stem cells identifies genes associated with patient survival. Genome Med. 2012, 4, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Babikir, H.; Müller, S.; Yagnik, G.; Shamardani, K.; Catalan, F.; Kohanbash, G.; Alvarado, B.; Di Lullo, E.; Kriegstein, A.; et al. The phenotypes of proliferating glioblastoma cells reside on a single axis of variation. Cancer Discov. 2019, 9, 1708–1719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neftel, C.; Laffy, J.; Filbin, M.G.; Hara, T.; Shore, M.E.; Rahme, G.J.; Richman, A.R.; Silverbush, D.; Shaw, M.L.; Hebert, C.M.; et al. An integrative model of cellular states, plasticity and genetics for glioblastoma. Cell 2019, 178, 835–849.e21. [Google Scholar] [CrossRef] [PubMed]
- The ICGC/TCGA pan-cancer analysis of whole genomes consortium. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef] [Green Version]
- Zanders, E.D.; Svensson, F.; Bailey, D.S. Therapy for glioblastoma: Is it working? Drug Discov. Today 2019, 24, 1193–1201. [Google Scholar] [CrossRef] [PubMed]
- Saleem, H.; Abdul, K.U.; Küçükosmanoglu, A.; Houweling, M.; Cornelissen, F.M.G.; Heiland, D.H.; Hegi, M.E.; Kouwenhoven, M.C.M.; Bailey, D.; Würdinger, T.; et al. The ticking clock of EGFR therapy resistance in glioblastoma: Target independence or target compensation. Drug Resist. Updates 2019, 43, 29–37. [Google Scholar] [CrossRef]
- Kast, R.E.; Boockvar, J.A.; Brüning, A.; Cappello, F.; Chang, W.-W.; Cvek, B.; Dou, Q.P.; Duenas-Gonzalez, A.; Efferth, T.; Focosi, D.; et al. A Conceptually New Treatment Approach for Relapsed Glioblastoma: Coordinated Undermining of Survival Paths with Nine Repurposed Drugs (CUSP9) by the International Initiative for Accelerated Improvement of Glioblastoma Care. Oncotarget 2013, 4, 502–530. [Google Scholar] [CrossRef] [Green Version]
- Gromeier, M.; Brown, M.C.; Zhang, G.; Lin, X.; Chen, Y.; Wei, Z.; Beaubier, N.; Yan, H.; He, Y.; Desjardins, A.; et al. Very low mutation burden is a feature of inflamed recurrent glioblastomas responsive to cancer immunotherapy. Nat. Commun. 2021, 12, 352. [Google Scholar] [CrossRef] [PubMed]
- Agliardi, G.; Liuzzi, A.R.; Hotblack, A.; De Feo, D.; Núñez, N.; Stowe, C.L.; Friebel, E.; Nannini, F.; Rindlisbacher, L.; Roberts, T.A.; et al. Intratumoral IL-12 delivery empowers CAR-t cell immunotherapy in a pre-clinical model of glioblastoma. Nat. Commun. 2021, 12, 444. [Google Scholar] [CrossRef]
- Gupta, P.B.; Pastushenko, I.; Skibinski, A.; Blanpain, C.; Kuperwasser, C. Phenotypic plasticity: Driver of cancer initiation, progression and therapy resistance. Cell Stem Cell 2019, 24, 65–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Yu, H.; Yoo, S.; Lee, E.; Laganà, A.; Parekh, S.; Schadt, E.E.; Wang, L.; Zhu, J. A network analysis of multiple myeloma related gene signatures. Cancers 2019, 11, 1452. [Google Scholar] [CrossRef] [Green Version]
- Boumahdi, S.; Sauvage, D.F.J. The great escape: Tumour cell plasticity in resistance to targeted therapy. Nat. Rev. Drug Discov. 2020, 19, 39–56. [Google Scholar] [CrossRef]
- Swinney, D.C.; Anthony, J. How were new medicines discovered? Nat. Rev. Drug Discov. 2011, 10, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Moffat, J.G.; Rudolph, J.; Bailey, D. Phenotypic screening in cancer drug discovery-past, present and future. Nat. Rev. Drug Discov. 2014, 13, 588–602. [Google Scholar] [CrossRef] [PubMed]
- Blagg, J.; Workman, P. Choose and use your chemical probe wisely to explore cancer biology. Cancer Cell 2017, 32, 9–25. [Google Scholar] [CrossRef] [Green Version]
- Jörg, M.; Madden, K.S. The right tools for the job: The central role for next generation chemical probes and chemistry-based target deconvolution methods in phenotypic drug discovery. RSC Med. Chem. 2021, 12, 646–665. [Google Scholar] [CrossRef]
- Lyne, S.B.; Yamini, B. An alternative pipeline for glioblastoma therapeutics: A systematic review of drug repurposing in glioblastoma. Cancers 2021, 13, 1953. [Google Scholar] [CrossRef] [PubMed]
- Pruteanu, L.-L.; Kopanitsa, L.; Módos, D.; Kletnieks, E.; Samarova, E.; Bender, A.; Gomez, L.D.; Bailey, D.S. Transcriptomics predicts compound synergy in drug and natural product treated glioblastoma cells. PLoS ONE 2020, 15, e0239551. [Google Scholar] [CrossRef] [PubMed]
- Menniti, F.S.; Chappie, T.A.; Humphrey, J.M.; Schmidt, C.J. Phosphodiesterase 10A inhibitors: A novel approach to the treatment of the symptoms of schizophrenia. Curr. Opin. Investig. Drugs 2007, 8, 54–59. [Google Scholar] [PubMed]
- Verhoest, P.R.; Chapin, D.S.; Corman, M.; Fonseca, K.; Harms, J.F.; Hou, X.; Marr, E.S.; Menniti, F.S.; Nelson, F.; O’Connor, R.; et al. Discovery of a novel class of phosphodiesterase 10a inhibitors and identification of clinical candidate 2-[4-(1-Methyl-4-Pyridin-4-Yl-1H-Pyrazol-3-Yl)-Phenoxymethyl]-Quinoline (PF-2545920) for the treatment of schizophrenia. J. Med. Chem. 2009, 52, 5188–5196. [Google Scholar] [CrossRef]
- Walling, D.P.; Banerjee, A.; Dawra, V.; Boyer, S.; Schmidt, C.J.; de Martinis, N. Phosphodiesterase 10A inhibitor monotherapy is not an effective treatment of acute schizophrenia. J. Clin. Psychopharmacol. 2019, 39, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, F.B.; Wild, E.J. Clinical trials corner: September 2017. J. Huntingt. Dis. 2017, 6, 255–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grothey, A.; Blay, J.-Y.; Pavlakis, N.; Yoshino, T.; Bruix, J. Evolving role of regorafenib for the treatment of advanced cancers. Cancer Treat. Rev. 2020, 86, 101993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilhelm, S.M.; Dumas, J.; Adnane, L.; Lynch, M.; Carter, C.A.; Schütz, G.; Thierauch, K.-H.; Zopf, D. Regorafenib (BAY 73-4506): A new oral multikinase inhibitor of angiogenic, stromal and oncogenic receptor tyrosine kinases with potent preclinical antitumor activity. Int. J. Cancer 2011, 129, 245–255. [Google Scholar] [CrossRef]
- Lombardi, G.; De Salvo, G.L.; Brandes, A.A.; Eoli, M.; Rudà, R.; Faedi, M.; Lolli, I.; Pace, A.; Daniele, B.; Pasqualetti, F.; et al. Regorafenib compared with lomustine in patients with relapsed glioblastoma (REGOMA): A multicentre, open-label, randomised, controlled, phase 2 trial. Lancet Oncol. 2019, 20, 110–119. [Google Scholar] [CrossRef]
- Alexander, B.M.; Ba, S.; Berger, M.S.; Berry, D.A.; Cavenee, W.K.; Chang, S.M.; Cloughesy, T.F.; Jiang, T.; Khasraw, M.; Li, W.; et al. Adaptive global innovative learning environment for glioblastoma: GBM AGILE. Clin. Cancer Res. 2018, 24, 737–743. [Google Scholar] [CrossRef] [Green Version]
- Safitri, D.; Harris, M.; Potter, H.; Yan Yeung, H.; Winfield, I.; Kopanitsa, L.; Svensson, F.; Rahman, T.; Harper, M.T.; Bailey, D.; et al. Elevated intracellular cAMP concentration mediates growth suppression in glioma cells. Biochem. Pharmacol. 2020, 174, 113823. [Google Scholar] [CrossRef]
- Veroli, D.G.Y.; Fornari, C.; Wang, D.; Mollard, S.; Bramhall, J.L.; Richards, F.M.; Jodrell, D.I. Combenefit: An interactive platform for the analysis and visualization of drug combinations. Bioinformatics 2016, 32, 2866–2868. [Google Scholar] [CrossRef] [PubMed]
- Irizarry, R.A. Exploration, normalization and summaries of high density oligonucleotide array probe level data. Biostatistics 2003, 4, 249–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, D.-T.; Mathias, S.; Bologa, C.; Brunak, S.; Fernandez, N.; Gaulton, A.; Hersey, A.; Holmes, J.; Jensen, L.J.; Karlsson, A.; et al. Pharos: Collating protein information to shed light on the druggable genome. Nucleic Acids Res. 2017, 45, D995–D1002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svensson, F.; Westerman, B.; Würdinger, T.; Bailey, D. GBM drug bank—A new resource for glioblastoma drug discovery and informatics research. Neuro Oncol. 2018, 20, 1680–1681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saelee, P.; Wongkham, S.; Puapairoj, A.; Khuntikeo, N.; Petmitr, S.; Chariyalertsak, S.; Sumethchotimaytha, W.; Karalak, A. Novel PNLIPRP3 and DOCK8 gene expression and prognostic implications of DNA loss on chromosome 10q25.3 in hepatocellular carcinoma. Asian Pac. J. Cancer Prev. 2009, 10, 501–506. [Google Scholar] [PubMed]
- Junes-Gill, K.S.; Lawrence, C.E.; Wheeler, C.J.; Cordner, R.; Gill, T.G.; Mar, V.; Shiri, L.; Basile, L.A. Human hematopoietic signal peptide-containing secreted 1 (HHSS1) modulates genes and pathways in glioma: Implications for the regulation of tumorigenicity and angiogenesis. BMC Cancer 2014, 14, 920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Human Protein Atlas: PNLIPRP3. Available online: https://www.proteinatlas.org/ENSG00000203837-PNLIPRP3 (accessed on 2 June 2021).
- Liu, T.-T.; Liu, X.-T.; Chen, Q.-X.; Shi, Y. Lipase inhibitors for obesity: A review. Biomed. Pharmacother. 2020, 128, 110314. [Google Scholar] [CrossRef]
- Whitelaw, J.A.; Lilla, S.; Paul, N.R.; Fort, L.; Zanivan, S.; Machesky, L.M. CYRI/Fam49 proteins represent a new class of rac1 interactors. Commun. Integr. Biol. 2019, 12, 112–118. [Google Scholar] [CrossRef]
- Fort, L.; Batista, J.M.; Thomason, P.A.; Spence, H.J.; Whitelaw, J.A.; Tweedy, L.; Greaves, J.; Martin, K.J.; Anderson, K.I.; Brown, P.; et al. Fam49/CYRI interacts with rac1 and locally suppresses protrusions. Nat. Cell Biol. 2018, 20, 1159–1171. [Google Scholar] [CrossRef] [Green Version]
- The Human Protein Atlas: FAM49A. Available online: https://www.proteinatlas.org/ENSG00000197872-FAM49A (accessed on 2 June 2021).
- Kaplan, E.; Stone, R.; Hume, P.J.; Greene, N.P.; Koronakis, V. Structure of CYRI-B (FAM49B), a key regulator of cellular actin assembly. Acta Crystallogr. Sect. D Struct. Biol. 2020, 76, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Shnaper, S.; Desbaillets, I.; Brown, D.A.; Murat, A.; Migliavacca, E.; Schluep, M.; Ostermann, S.; Hamou, M.-F.; Stupp, R.; Breit, S.N.; et al. Elevated levels of MIC-1/GDF15 in the cerebrospinal fluid of patients are associated with glioblastoma and worse outcome. Int. J. Cancer 2009, 125, 2624–2630. [Google Scholar] [CrossRef]
- Codó, P.; Weller, M.; Kaulich, K.; Schraivogel, D.; Silginer, M.; Reifenberger, G.; Meister, G.; Roth, P. Control of glioma cell migration and invasiveness by GDF-15. Oncotarget 2016, 7, 7732–7746. [Google Scholar] [CrossRef]
- Roth, P.; Junker, M.; Tritschler, I.; Mittelbronn, M.; Dombrowski, Y.; Breit, S.N.; Tabatabai, G.; Wick, W.; Weller, M.; Wischhusen, J. GDF-15 contributes to proliferation and immune escape of malignant gliomas. Clin. Cancer Res. 2010, 16, 3851–3859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wischhusen, J.; Melero, I.; Fridman, W.H. Growth/Differentiation Factor-15 (GDF-15): From biomarker to novel targetable immune checkpoint. Front. Immunol. 2020, 11, 951. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Chen, H.; Zhong, D.; Wei, W.; Liu, L.; Duan, Q.; Han, B.; Li, G. TRIB3 facilitates glioblastoma progression via restraining autophagy. Aging 2020, 12, 25020–25034. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, C.; Li, X.; Hu, Y.; Gou, R.; Guo, Q.; Nie, X.; Liu, J.; Zhu, L.; Lin, B. Down-Regulation of TRIB3 inhibits the progression of ovarian cancer via mek/erk signaling pathway. Cancer Cell Int. 2020, 20, 418. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Li, L.; Chen, L.; Gao, Y.; Chen, X.; Cao, Y. TRIB3 confers glioma cell stemness via interacting with Β-catenin. Environ. Toxicol. 2020, 35, 697–706. [Google Scholar] [CrossRef]
- Li, Z.; Chen, Y.; An, T.; Liu, P.; Zhu, J.; Yang, H.; Zhang, W.; Dong, T.; Jiang, J.; Zhang, Y.; et al. Nuciferine inhibits the progression of glioblastoma by suppressing the SOX2-AKT/STAT3-Slug signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 139. [Google Scholar] [CrossRef] [Green Version]
- Sui, A.; Xu, Y.; Yang, J.; Pan, B.; Wu, J.; Guo, T.; Shen, Y.; Guo, X. The histone H3 Lys 27 demethylase KDM6B promotes migration and invasion of glioma cells partly by regulating the expression of SNAI1. Neurochem. Int. 2019, 124, 123–129. [Google Scholar] [CrossRef]
- Min, D.-J.; Zhao, Y.; Monks, A.; Palmisano, A.; Hose, C.; Teicher, B.A.; Doroshow, J.H.; Simon, R.M. Identification of pharmacodynamic biomarkers and common molecular mechanisms of response to genotoxic agents in cancer cell lines. Cancer Chemother Pharm. 2019, 84, 771–780. [Google Scholar] [CrossRef]
- Tang, Q.; Ren, L.; Liu, J.; Li, W.; Zheng, X.; Wang, J.; Du, G. Withaferin A triggers G2/M arrest and intrinsic apoptosis in glioblastoma cells via ATF4-ATF3-CHOP axis. Cell Prolif. 2020, 53. [Google Scholar] [CrossRef] [Green Version]
- Van Noorden, C.J.F.; Hira, V.V.V.; van Dijck, A.J.; Novak, M.; Breznik, B.; Molenaar, R.J. Energy metabolism in IDH1 wild-type and IDH1-Mutated glioblastoma stem cells: A novel target for therapy? Cells 2021, 10, 705. [Google Scholar] [CrossRef] [PubMed]
- Perissi, V.; Aggarwal, A.; Glass, C.K.; Rose, D.W.; Rosenfeld, M.G. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 2004, 116, 511–526. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Ylivinkka, I.; Chen, P.; Li, L.; Hautaniemi, S.; Nyman, T.A.; Keski-Oja, J.; Hyytiäinen, M. Netrin-4 promotes glioblastoma cell proliferation through integrin Β4 signaling. Neoplasia 2012, 14, 219–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.; Jang, S.-W.; Okada, M.; Chan, C.-B.; Feng, Y.; Liu, Y.; Luo, S.-W.; Hong, Y.; Rama, N.; Xiong, W.-C.; et al. Netrin-1 mediates neuronal survival through pike-l interaction with the dependence receptor UNC5B. Nat. Cell Biol. 2008, 10, 698–706. [Google Scholar] [CrossRef] [Green Version]
- Pyrzynska, B.; Banach-Orlowska, M.; Teperek-Tkacz, M.; Miekus, K.; Drabik, G.; Majka, M.; Miaczynska, M. Multifunctional protein APPL2 contributes to survival of human glioma cells. Mol. Oncol. 2013, 7, 67–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, A.; Nakayama, H.; Wang, P.; König, C.; Akino, T.; Sandlund, J.; Coma, S.; Italiano, J.E.; Mammoto, A.; Bielenberg, D.R.; et al. Netrin-1 promotes glioblastoma cell invasiveness and angiogenesis by multiple pathways including activation of RhoA, Cathepsin B and CAMP-Response element-binding protein. J. Biol. Chem. 2013, 288, 2210–2222. [Google Scholar] [CrossRef] [Green Version]
- Ylivinkka, I.; Hu, Y.; Chen, P.; Rantanen, V.; Hautaniemi, S.; Nyman, T.A.; Keski-Oja, J.; Hyytiäinen, M. Netrin-1 induced activation of notch signaling mediates glioblastoma cell invasion. J. Cell Sci. 2013, 126, 2459–2469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emanuele, M.J.; Enrico, T.P.; Mouery, R.D.; Wasserman, D.; Nachum, S.; Tzur, A. Complex cartography: Regulation of E2F Transcription factors by Cyclin F and ubiquitin. Trends Cell Biol. 2020, 30, 640–652. [Google Scholar] [CrossRef]
- Lee, S.; Park, Y.R.; Kim, S.; Park, E.; Kang, M.J.; So, I.; Chun, J.N.; Jeon, J. Geraniol suppresses prostate cancer growth through down-regulation of E2F8. Cancer Med. 2016, 5, 2899–2908. [Google Scholar] [CrossRef]
- Teng, J.; Hejazi, S.; Hiddingh, L.; Carvalho, L.; de Gooijer, M.C.; Wakimoto, H.; Barazas, M.; Tannous, M.; Chi, A.S.; Noske, D.P.; et al. Recycling drug screen repurposes hydroxyurea as a sensitizer of glioblastomas to temozolomide targeting de novo DNA synthesis, irrespective of molecular subtype. Neuro Oncol. 2018, 20, 642–654. [Google Scholar] [CrossRef] [Green Version]
- Mondal, P.; Sen, S.; Klein, B.J.; Tiwary, N.; Gadad, S.S.; Kutateladze, T.G.; Roy, S.; Das, C. TCF19 promotes cell proliferation through binding to the histone H3K4me3 mark. Biochemistry 2020, 59, 389–399. [Google Scholar] [CrossRef]
- Hoffmann, S.; Pentakota, S.; Mund, A.; Haahr, P.; Coscia, F.; Gallo, M.; Mann, M.; Taylor, N.M.; Mailand, N. FAM111 protease activity undermines cellular fitness and is amplified by gain-of-function mutations in human disease. EMBO Rep. 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, K.; Nojima, S.; Hijiki, S.; Tahara, S.; Ohshima, K.; Matsui, T.; Hori, Y.; Kurashige, M.; Umeda, D.; Kiyokawa, H.; et al. FAM111B enhances proliferation of KRAS-driven lung adenocarcinoma by degrading P16. Cancer Sci. 2020, 111, 2635–2646. [Google Scholar] [CrossRef] [PubMed]
- Richter, J.D.; Bassell, G.J.; Klann, E. Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat. Rev. Neurosci. 2015, 16, 595–605. [Google Scholar] [CrossRef] [Green Version]
- Tejero, R.; Huang, Y.; Katsyv, I.; Kluge, M.; Lin, J.Y.; Tome-Garcia, J.; Daviaud, N.; Wang, Y.; Zhang, B.; Tsankova, N.M.; et al. Gene signatures of quiescent glioblastoma cells reveal mesenchymal shift and interactions with niche microenvironment. EBioMedicine 2019, 42, 252–269. [Google Scholar] [CrossRef] [Green Version]
- Emmerson, P.J.; Duffin, K.L.; Chintharlapalli, S.; Wu, X. GDF15 and growth control. Front. Physiol. 2018, 9, 1712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, K.; Shi, J.; Liu, Z.; Jia, Y.; Qin, Q.; Zhang, H.; Wan, S.; Niu, Z.; Lu, L.; Sun, J.; et al. A panel of five plasma proteins for the early diagnosis of hepatitis B virus-related hepatocellular carcinoma in individuals at risk. EBioMedicine 2020, 52, 102638. [Google Scholar] [CrossRef] [Green Version]
- Dedobbeleer, M.; Willems, E.; Freeman, S.; Lombard, A.; Goffart, N.; Rogister, B. Phosphatases and solid tumors: Focus on glioblastoma initiation, progression and recurrences. Biochem. J. 2017, 474, 2903–2924. [Google Scholar] [CrossRef]
- Sonntag, T.; Vaughan, J.M.; Montminy, M. 14-3-3 proteins mediate inhibitory effects of cAMP on Salt-inducible Kinases (SIK s). FEBS J. 2018, 285, 467–480. [Google Scholar] [CrossRef] [Green Version]
- Ahmadi, M.; Amiri, S.; Pecic, S.; Machaj, F.; Rosik, J.; Łos, M.J.; Alizadeh, J.; Mahdian, R.; da Silva Rosa, S.C.; Schaafsma, D.; et al. Pleiotropic effects of statins: A focus on cancer. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2020, 1866, 165968. [Google Scholar] [CrossRef]
- Sunde, M.; McGrath, K.C.Y.; Young, L.; Matthews, J.M.; Chua, E.L.; Mackay, J.P.; Death, A.K. TC-1 is a novel tumorigenic and natively disordered protein associated with thyroid cancer. Cancer Res. 2004, 64, 2766–2773. [Google Scholar] [CrossRef] [Green Version]
- Lei, J.; Li, W.; Yang, Y.; Lu, Q.; Zhang, N.; Bai, G.; Zhong, D.; Su, K.; Liu, B.; Li, X.; et al. TC-1 overexpression promotes cell proliferation in human non-small cell lung cancer that can be inhibited by PD173074. PLoS ONE 2014, 9, e100075. [Google Scholar] [CrossRef] [PubMed]
- Cronin, R.; Brooke, G.N.; Prischi, F. The role of the P90 ribosomal S6 kinase family in prostate cancer progression and therapy resistance. Oncogene 2021, 40, 3775–3785. [Google Scholar] [CrossRef] [PubMed]
- Sulzmaier, F.J.; Young-Robbins, S.; Jiang, P.; Geerts, D.; Prechtl, A.M.; Matter, M.L.; Kesari, S.; Ramos, J.W. RSK2 activity mediates glioblastoma invasiveness and is a potential target for new therapeutics. Oncotarget 2016, 7, 79869–79884. [Google Scholar] [CrossRef] [Green Version]
- Mahajan-Thakur, S.; Bien-Möller, S.; Marx, S.; Schroeder, H.; Rauch, B. Sphingosine 1-Phosphate (S1P) signaling in glioblastoma multiforme-A systematic review. Int. J. Mol. Sci. 2017, 18, 2448. [Google Scholar] [CrossRef] [Green Version]
- Loeffler, S.; Fayard, B.; Weis, J.; Weissenberger, J. Interleukin-6 induces transcriptional activation of Vascular Endothelial Growth Factor (VEGF) in astrocytesin vivo and regulatesVEGF promoter activity in glioblastoma cells via direct interaction between STAT3 and Sp1. Int. J. Cancer 2005, 115, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Sferrazzo, G.; Di Rosa, M.; Barone, E.; Li Volti, G.; Musso, N.; Tibullo, D.; Barbagallo, I. Heme Oxygenase-1 in central nervous system malignancies. J. Clin. Med. 2020, 9, 1562. [Google Scholar] [CrossRef]
- Shono, K.; Yamaguchi, I.; Mizobuchi, Y.; Kagusa, H.; Sumi, A.; Fujihara, T.; Nakajima, K.; Kitazato, K.T.; Matsuzaki, K.; Saya, H.; et al. Downregulation of the CCL2/CCR2 and CXCL10/CXCR3 axes contributes to antitumor effects in a mouse model of malignant glioma. Sci. Rep. 2020, 10, 15286. [Google Scholar] [CrossRef]
- Groblewska, M.; Litman-Zawadzka, A.; Mroczko, B. The role of selected chemokines and their receptors in the development of gliomas. Int. J. Mol. Sci. 2020, 21, 3704. [Google Scholar] [CrossRef]
- Ströbele, S.; Schneider, M.; Schneele, L.; Siegelin, M.D.; Nonnenmacher, L.; Zhou, S.; Karpel-Massle, G.; Westhoff, M.-A.; Halatsch, M.-E.; Debatin, K.-M. A potential role for the inhibition of PI3K signaling in glioblastoma therapy. PLoS ONE 2015, 10, e0131670. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, F.; Sun, Q.; Patel, D.; Stommel, J. Cholesterol metabolism: A potential therapeutic target in glioblastoma. Cancers 2019, 11, 146. [Google Scholar] [CrossRef] [Green Version]
- Nakakuki, M.; Kawano, H.; Notsu, T.; Imada, K.; Mizuguchi, K.; Shimano, H. A novel processing system of sterol regulatory element-binding protein-1c regulated by polyunsaturated fatty acid. J. Biochem. 2014, 155, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Pharos: PNLIPRP3 (Pancreatic Lipase-Related Protein 3). Available online: https://pharos.nih.gov/targets/Q17RR3 (accessed on 2 June 2021).
- Hattermann, K.; Held-Feindt, J.; Lucius, R.; Müerköster, S.S.; Penfold, M.E.T.; Schall, T.J.; Mentlein, R. The chemokine receptor CXCR7 is highly expressed in human glioma cells and mediates antiapoptotic effects. Cancer Res. 2010, 70, 3299–3308. [Google Scholar] [CrossRef] [Green Version]
- Karki, K.; Li, X.; Jin, U.-H.; Mohankumar, K.; Zarei, M.; Michelhaugh, S.K.; Mittal, S.; Tjalkens, R.; Safe, S. Nuclear Receptor 4A2 (NR4A2) is a druggable target for glioblastomas. J. Neuro Oncol. 2020, 146, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Löw, S.; Vougioukas, V.I.; Hielscher, T.; Schmidt, U.; Unterberg, A.; Halatsch, M.E. Pathogenetic pathways leading to glioblastoma multiforme: Association between gene expressions and resistance to erlotinib. Anticancer Res. 2008, 28, 3729–3732. [Google Scholar] [PubMed]
- Rahane, C.S.; Kutzner, A.; Heese, K. A cancer tissue-specific FAM72 expression profile defines a novel glioblastoma multiform (GBM) gene-mutation signature. J. Neurooncol. 2019, 141, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Banelli, B.; Carra, E.; Barbieri, F.; Würth, R.; Parodi, F.; Pattarozzi, A.; Carosio, R.; Forlani, A.; Allemanni, G.; Marubbi, D.; et al. The histone demethylase KDM5A is a key factor for the resistance to temozolomide in glioblastoma. Cell Cycle 2015, 14, 3418–3429. [Google Scholar] [CrossRef] [Green Version]
- McAllister, T.E.; England, K.S.; Hopkinson, R.J.; Brennan, P.E.; Kawamura, A.; Schofield, C.J. Recent progress in histone demethylase inhibitors. J. Med. Chem. 2016, 59, 1308–1329. [Google Scholar] [CrossRef]
- Banelli, B.; Daga, A.; Forlani, A.; Allemanni, G.; Marubbi, D.; Pistillo, M.P.; Profumo, A.; Romani, M. Small molecules targeting histone demethylase genes (KDMs) inhibit growth of temozolomide-resistant glioblastoma cells. Oncotarget 2017, 8, 34896–34910. [Google Scholar] [CrossRef] [Green Version]
- Mahdessian, D.; Cesnik, A.J.; Gnann, C.; Danielsson, F.; Stenström, L.; Arif, M.; Zhang, C.; Le, T.; Johansson, F.; Shutten, R.; et al. Spatiotemporal dissection of the cell cycle with single-cell proteogenomics. Nature 2021, 590, 649–654. [Google Scholar] [CrossRef]
- Fung, N.H.; Grima, C.A.; Widodo, S.S.; Kaye, A.H.; Whitehead, C.A.; Stylli, S.S.; Mantamadiotis, T. Understanding and exploiting cell signalling convergence nodes and pathway cross-talk in malignant brain cancer. Cell. Signal. 2019, 57, 2–9. [Google Scholar] [CrossRef]
- Qiu, Z.; Yuan, W.; Chen, T.; Zhou, C.; Liu, C.; Huang, Y.; Han, D.; Huang, Q. HMGCR positively regulated the growth and migration of glioblastoma cells. Gene 2016, 576, 22–27. [Google Scholar] [CrossRef]
- Cote, D.J.; Rosner, B.A.; Smith-Warner, S.A.; Egan, K.M.; Stampfer, M.J. Statin use, hyperlipidemia and risk of glioma. Eur. J. Epidemiol. 2019, 34, 997–1011. [Google Scholar] [CrossRef]
- Taub, M. Salt inducible kinase signaling networks: Implications for acute kidney injury and therapeutic potential. Int. J. Mol. Sci. 2019, 20, 3219. [Google Scholar] [CrossRef] [Green Version]
- Leone, P.E.; González, M.B.; Elosua, C.; Gómez-Moreta, J.A.; Lumbreras, E.; Robledo, C.; Santos-Briz, A.; Valero, J.M.; de la Guardia, R.D.; Gutiérrez, N.C.; et al. Integration of global spectral karyotyping, CGH arrays and expression arrays reveals important genes in the pathogenesis of glioblastoma multiforme. Ann. Surg. Oncol. 2012, 19, 2367–2379. [Google Scholar] [CrossRef] [PubMed]
- Sundberg, T.B.; Liang, Y.; Wu, H.; Choi, H.G.; Kim, N.D.; Sim, T.; Johannessen, L.; Petrone, A.; Khor, B.; Graham, D.B.; et al. Development of chemical probes for investigation of salt-inducible kinase function in vivo. ACS Chem. Biol. 2016, 11, 2105–2111. [Google Scholar] [CrossRef] [Green Version]
- Martin, M.W.; Newcomb, J.; Nunes, J.J.; McGowan, D.C.; Armistead, D.M.; Boucher, C.; Buchanan, J.L.; Buckner, W.; Chai, L.; Elbaum, D.; et al. Novel 2-Aminopyrimidine carbamates as potent and orally active inhibitors of Lck: Synthesis, SAR and in vivo antiinflammatory activity. J. Med. Chem. 2006, 49, 4981–4991. [Google Scholar] [CrossRef]
- Tan, S.K.; Jermakowicz, A.; Mookhtiar, A.K.; Nemeroff, C.B.; Schürer, S.C.; Ayad, N.G. Drug repositioning in glioblastoma: A pathway perspective. Front. Pharmacol. 2018, 9, 218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeter, J.M.; Bowles, T.L.; Curiel-Lewandrowski, C.; Swetter, S.M.; Filipp, F.V.; Abdel-Malek, Z.A.; Geskin, L.J.; Brewer, J.D.; Arbiser, J.L.; Gershenwald, J.E.; et al. Chemoprevention agents for melanoma: A path forward into phase 3 clinical trials. Cancer 2019, 125, 18–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishimori, S.; O’Meara, M.J.; Castro, C.D.; Noda, H.; Cetinbas, M.; da Silva Martins, J.; Ayturk, U.; Brooks, D.J.; Bruce, M.; Nagata, M.; et al. Salt-Inducible kinases dictate parathyroid hormone 1 receptor action in bone development and remodeling. J. Clin. Investig. 2019, 129, 5187–5203. [Google Scholar] [CrossRef]
- Wein, M.N.; Foretz, M.; Fisher, D.E.; Xavier, R.J.; Kronenberg, H.M. Salt-Inducible kinases: Physiology, regulation by CAMP, and therapeutic potential. Trends Endocrinol. Metab. 2018, 29, 723–735. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.; MacKenzie, K.F.; Petkevicius, K.; Kristariyanto, Y.; Zhang, J.; Choi, H.G.; Peggie, M.; Plater, L.; Pedrioli, P.G.A.; McIver, E.; et al. Phosphorylation of CRTC3 by the salt-inducible kinases controls the interconversion of classically activated and regulatory macrophages. Proc. Natl. Acad. Sci. USA 2012, 109, 16986–16991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sundberg, T.B.; Choi, H.G.; Song, J.-H.; Russell, C.N.; Hussain, M.M.; Graham, D.B.; Khor, B.; Gagnon, J.; O’Connell, D.J.; Narayan, K.; et al. Small-Molecule screening identifies inhibition of salt-inducible kinases as a therapeutic strategy to enhance immunoregulatory functions of dendritic cells. Proc. Natl. Acad. Sci. USA 2014, 111, 12468–12473. [Google Scholar] [CrossRef] [Green Version]
- Brantley, E.C.; Benveniste, E.N. Signal transducer and activator of transcription-3: A molecular hub for signaling pathways in gliomas. Mol. Cancer Res. 2008, 6, 675–684. [Google Scholar] [CrossRef] [Green Version]
- Mukthavaram, R.; Ouyang, X.; Saklecha, R.; Jiang, P.; Nomura, N.; Pingle, S.C.; Guo, F.; Makale, M.; Kesari, S. Effect of the JAK2/STAT3 inhibitor SAR317461 on human glioblastoma tumorspheres. J. Transl. Med. 2015, 13, 269. [Google Scholar] [CrossRef] [Green Version]
- Ioannidis, S.; Lamb, M.L.; Wang, T.; Almeida, L.; Block, M.H.; Davies, A.M.; Peng, B.; Su, M.; Zhang, H.-J.; Hoffmann, E.; et al. Discovery of 5-Chloro-N2-[(1 S)-1-(5-Fluoropyrimidin-2-Yl)Ethyl]-N4-(5-Methyl-1 H-Pyrazol-3-Yl)Pyrimidine-2,4-Diamine (AZD1480) as a novel inhibitor of the Jak/Stat pathway. J. Med. Chem. 2011, 54, 262–276. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.A.; Luo, X.; Lu, M.; An, Z.; Haas-Kogan, D.A.; Phillips, J.J.; Shokat, K.M.; Weiss, W.A.; Fan, Q.W. Cooperative blockade of PKCα and JAK2 drives apoptosis in glioblastoma. Cancer Res. 2020, 80, 709–718. [Google Scholar] [CrossRef] [PubMed]
- Quintás-Cardama, A.; Vaddi, K.; Liu, P.; Manshouri, T.; Li, J.; Scherle, P.A.; Caulder, E.; Wen, X.; Li, Y.; Waeltz, P.; et al. Preclinical characterization of the selective JAK1/2 inhibitor INCB018424: Therapeutic implications for the treatment of myeloproliferative neoplasms. Blood 2010, 115, 3109–3117. [Google Scholar] [CrossRef]
- Roskoski, R. Janus Kinase (JAK) inhibitors in the treatment of inflammatory and neoplastic diseases. Pharmacol. Res. 2016, 111, 784–803. [Google Scholar] [CrossRef]
- Darling, N.J.; Toth, R.; Arthur, J.S.C.; Clark, K. Inhibition of SIK2 and SIK3 during Differentiation enhances the anti-inflammatory phenotype of macrophages. Biochem. J. 2017, 474, 521–537. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Wang, J.; Shi, F.; Shan, A.; Xu, S.; Lv, W. BDKRB2 is a novel EMT-related biomarker and predicts poor survival in glioma. Aging 2021, 13, 7499–7516. [Google Scholar] [CrossRef] [PubMed]
- Seifert, S.; Sontheimer, H. Bradykinin enhances invasion of malignant glioma into the brain parenchyma by inducing cells to undergo amoeboid migration. J. Physiol. 2014, 592, 5109–5127. [Google Scholar] [CrossRef] [PubMed]
- Balmforth, A.J.; Parkinson, F.E.; Altiok, N.; Fredholm, B.B. Identification of a B2-Bradykinin receptor linked to phospholipase c and inhibition of dopamine stimulated cyclic AMP accumulation in the human astrocytoma cell line D384. Naunyn Schmiedeberg’s Arch. Pharmacol. 1992, 346. [Google Scholar] [CrossRef]
- Lesage, A.; Gibson, C.; Marceau, F.; Ambrosi, H.-D.; Saupe, J.; Katzer, W.; Loenders, B.; Charest-Morin, X.; Knolle, J. In Vitro pharmacological profile of a new small molecule bradykinin B2 receptor antagonist. Front. Pharmacol. 2020, 11, 916. [Google Scholar] [CrossRef] [PubMed]
- Hock, F.J.; Wirth, K.; Albus, U.; Linz, W.; Gerhards, H.J.; Wiemer, G.; Henke, S.; Breipohl, G.; König, W.; Knolle, J.; et al. Hoe 140 a new potent and long acting bradykinin-antagonist: In Vitro studies. Br. J. Pharmacol. 1991, 102, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Sawutz, D.G.; Salvino, J.M.; Dolle, R.E.; Casiano, F.; Ward, S.J.; Houck, W.T.; Faunce, D.M.; Douty, B.D.; Baizman, E.; Awad, M.M. The nonpeptide WIN 64338 is a bradykinin B2 receptor antagonist. Proc. Natl. Acad. Sci. USA 1994, 91, 4693–4697. [Google Scholar] [CrossRef] [Green Version]
- Hauser, A.S.; Chavali, S.; Masuho, I.; Jahn, L.J.; Martemyanov, K.A.; Gloriam, D.E.; Babu, M.M. Pharmacogenomics of GPCR drug targets. Cell 2018, 172, 41–54.e19. [Google Scholar] [CrossRef] [Green Version]
- Wu, V.; Yeerna, H.; Nohata, N.; Chiou, J.; Harismendy, O.; Raimondi, F.; Inoue, A.; Russell, R.B.; Tamayo, P.; Gutkind, J.S. Illuminating the Onco-GPCRome: Novel G protein–coupled receptor-driven oncocrine networks and targets for cancer immunotherapy. J. Biol. Chem. 2019, 294, 11062–11086. [Google Scholar] [CrossRef] [Green Version]
- Meyer, R.C.; Giddens, M.M.; Schaefer, S.A.; Hall, R.A. GPR37 and GPR37L1 are receptors for the neuroprotective and glioprotective factors prosaptide and prosaposin. Proc. Natl. Acad. Sci. USA 2013, 110, 9529–9534. [Google Scholar] [CrossRef] [Green Version]
- Yin, J.; Chen, K.-Y.M.; Clark, M.J.; Hijazi, M.; Kumari, P.; Bai, X.; Sunahara, R.K.; Barth, P.; Rosenbaum, D.M. Structure of a D2 dopamine receptor–G-protein complex in a lipid membrane. Nature 2020, 584, 125–129. [Google Scholar] [CrossRef]
- Lundius, E.G.; Vukojević, V.; Hertz, E.; Stroth, N.; Cederlund, A.; Hiraiwa, M.; Terenius, L.; Svenningsson, P. GPR37 protein trafficking to the plasma membrane regulated by prosaposin and GM1 gangliosides promotes cell viability. J. Biol. Chem. 2014, 289, 4660–4673. [Google Scholar] [CrossRef] [Green Version]
- Dunham, J.H.; Meyer, R.C.; Garcia, E.L.; Hall, R.A. GPR37 surface expression enhancement via N-terminal truncation or protein−protein interactions. Biochemistry 2009, 48, 10286–10297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hertz, E.; Terenius, L.; Vukojević, V.; Svenningsson, P. GPR37 and GPR37L1 differently interact with dopamine 2 receptors in live cells. Neuropharmacology 2019, 152, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.E.; Krigsfeld, G.; Mayes, P.A.; Patel, L.; Dicker, D.T.; Patel, A.S.; Dolloff, N.G.; Messaris, E.; Scata, K.A.; Wang, W.; et al. Dual inactivation of Akt and ERK by TIC10 signals Foxo3a nuclear translocation, trail gene induction and potent antitumor effects. Sci. Transl. Med. 2013, 5, 171ra17. [Google Scholar] [CrossRef] [Green Version]
- Prabhu, V.V.; Morrow, S.; Rahman Kawakibi, A.; Zhou, L.; Ralff, M.; Ray, J.; Jhaveri, A.; Ferrarini, I.; Lee, Y.; Parker, C.; et al. ONC201 and imipridones: Anti-Cancer compounds with clinical efficacy. Neoplasia 2020, 22, 725–744. [Google Scholar] [CrossRef]
- He, Y.; Li, J.; Koga, T.; Ma, J.; Dhawan, S.; Suzuki, Y.; Furnari, F.; Prabhu, V.V.; Allen, J.E.; Chen, C.C. Epidermal growth factor receptor as a molecular determinant of glioblastoma response to dopamine receptor D2 inhibitors. Neuro Oncol. 2021, 23, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, L.; Leduc, M.; Thibodeau, J.-F.; Zhang, M.-Z.; Grouix, B.; Sarra-Bournet, F.; Gagnon, W.; Hince, K.; Tremblay, M.; Geerts, L.; et al. A newly discovered antifibrotic pathway regulated by two fatty acid receptors. Am. J. Pathol. 2018, 188, 1132–1148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lynch, J.; Wang, J. G protein-coupled receptor signaling in stem cells and cancer. Int. J. Mol. Sci. 2016, 17, 707. [Google Scholar] [CrossRef] [Green Version]
- Marsango, S.; Barki, N.; Jenkins, L.; Tobin, A.B.; Milligan, G. Therapeutic validation of an orphan g protein-coupled receptor: The case of GPR84. Br. J. Pharm. 2020. [Google Scholar] [CrossRef]
- Hayakawa, M.; Kaizawa, H.; Moritomo, H.; Koizumi, T.; Ohishi, T.; Okada, M.; Ohta, M.; Tsukamoto, S.; Parker, P.; Workman, P.; et al. Synthesis and biological evaluation of 4-morpholino-2-phenylquinazolines and related derivatives as novel PI3 kinase P110α inhibitors. Bioorg. Med. Chem. 2006, 14, 6847–6858. [Google Scholar] [CrossRef]
- Gharbi, S.I.; Zvelebil, M.J.; Shuttleworth, S.J.; Hancox, T.; Saghir, N.; Timms, J.F.; Waterfield, M.D. Exploring the specificity of the PI3K family inhibitor LY294002. Biochem. J. 2007, 404, 15–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Méresse, S.; Fodil, M.; Fleury, F.; Chénais, B. Fucoxanthin, a marine-derived carotenoid from brown seaweeds and microalgae: A promising bioactive compound for cancer therapy. Int. J. Mol. Sci. 2020, 21, 9273. [Google Scholar] [CrossRef] [PubMed]
- Noorani, I.; Bradley, A.; de la Rosa, J. CRISPR and transposon in vivo screens for cancer drivers and therapeutic targets. Genome Biol. 2020, 21, 204. [Google Scholar] [CrossRef] [PubMed]
- Noorani, I.; de la Rosa, J.; Choi, Y.H.; Strong, A.; Ponstingl, H.; Vijayabaskar, M.S.; Lee, J.; Lee, E.; Richard-Londt, A.; Friedrich, M.; et al. PiggyBac mutagenesis and exome sequencing identify genetic driver landscapes and potential therapeutic targets of EGFR-mutant gliomas. Genome Biol. 2020, 21, 181. [Google Scholar] [CrossRef]
- Tejeda, G.S.; Whiteley, E.L.; Deeb, T.Z.; Bürli, R.W.; Moss, S.J.; Sheridan, E.; Brandon, N.J.; Baillie, G.S. Chorea-related mutations in PDE10A result in aberrant compartmentalization and functionality of the enzyme. Proc. Natl. Acad. Sci. USA 2020, 117, 677–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]

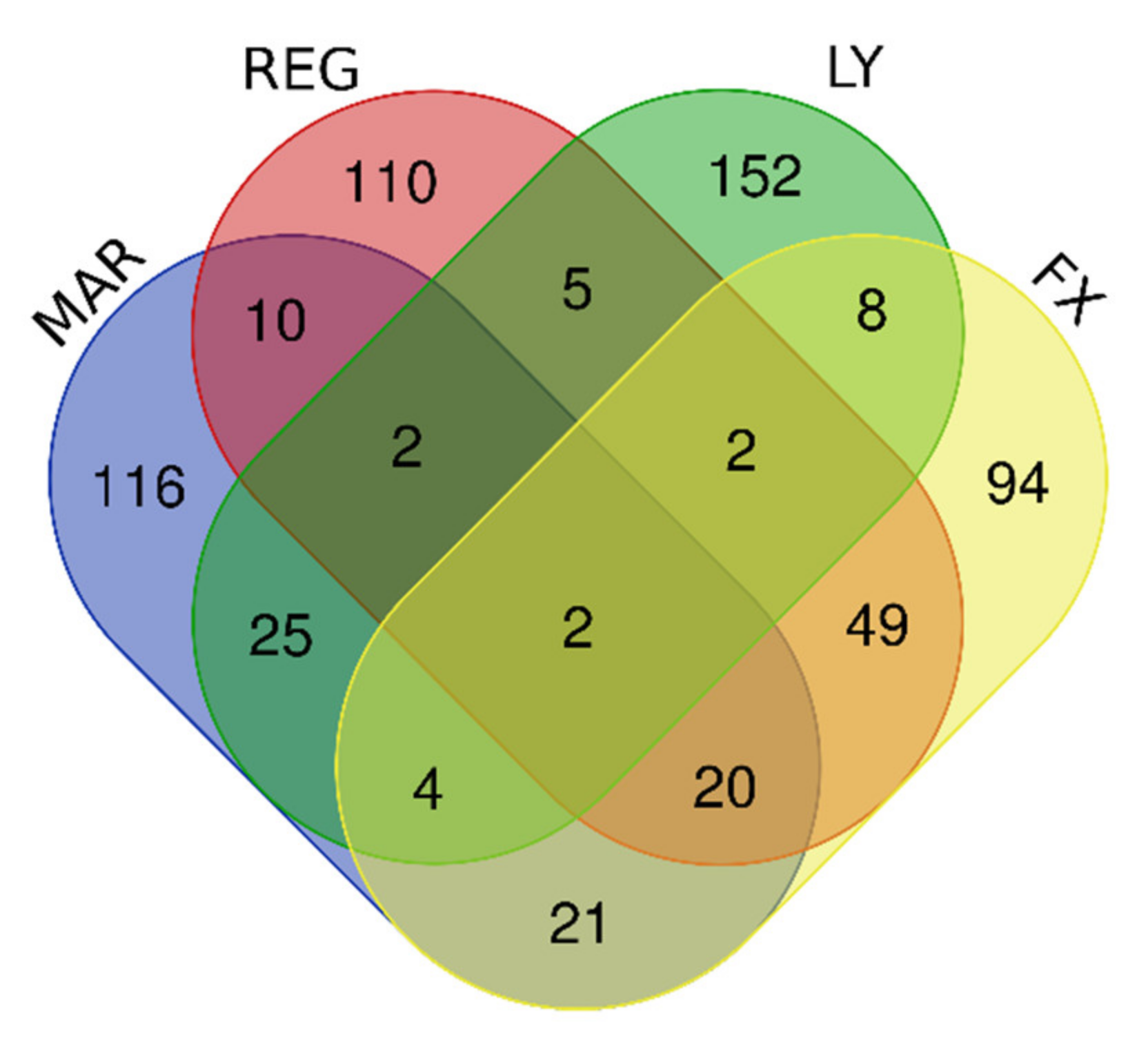
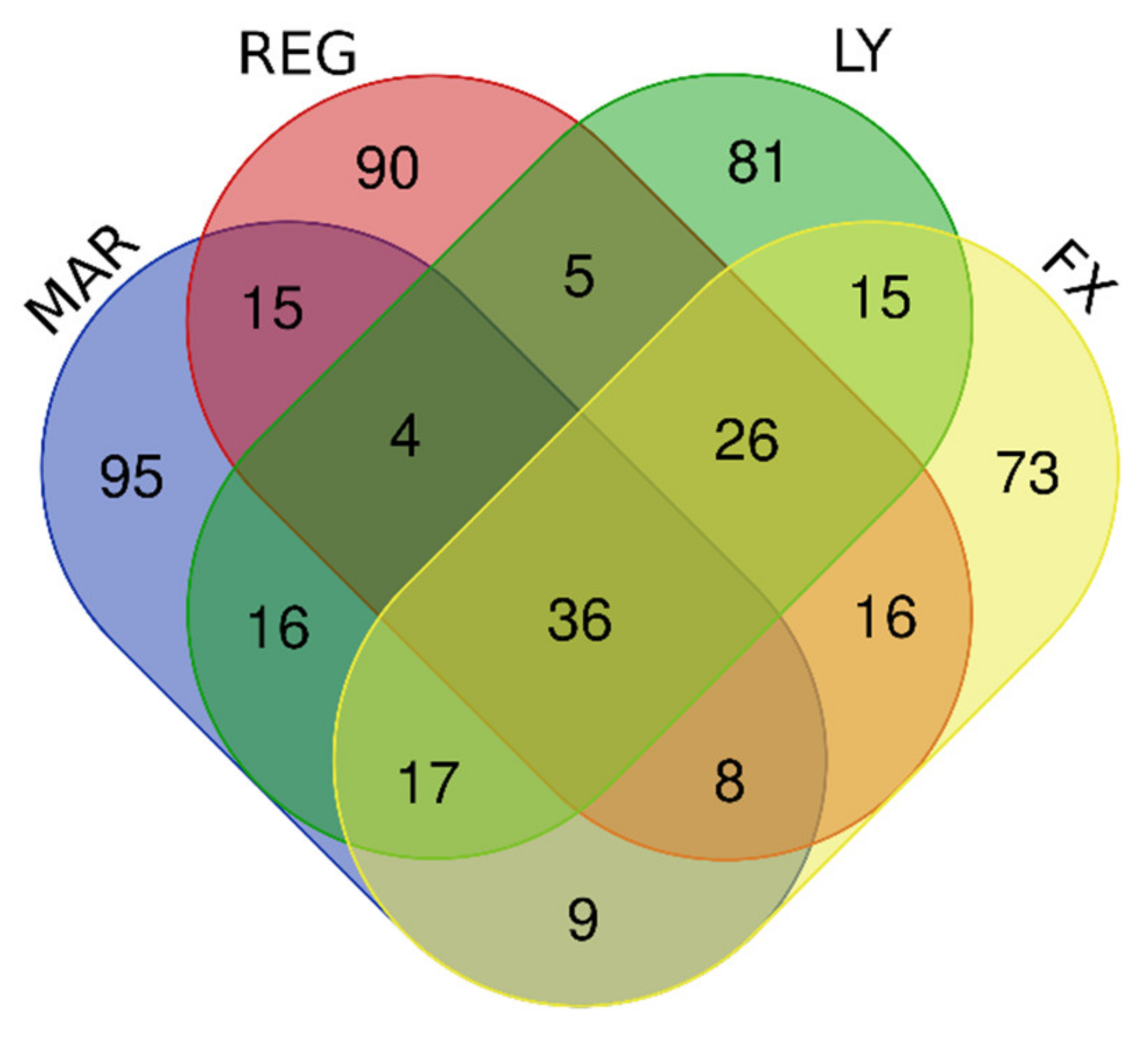
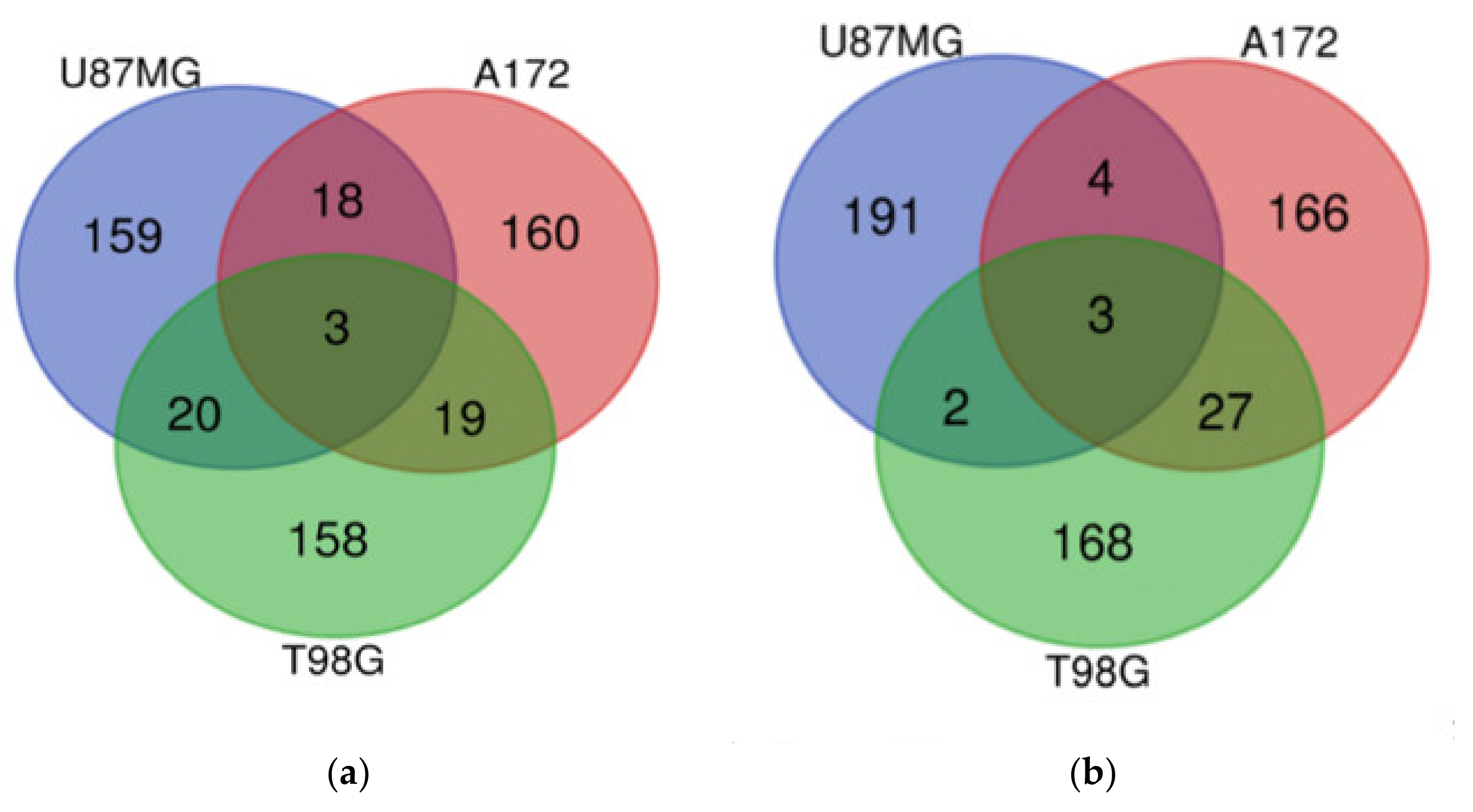
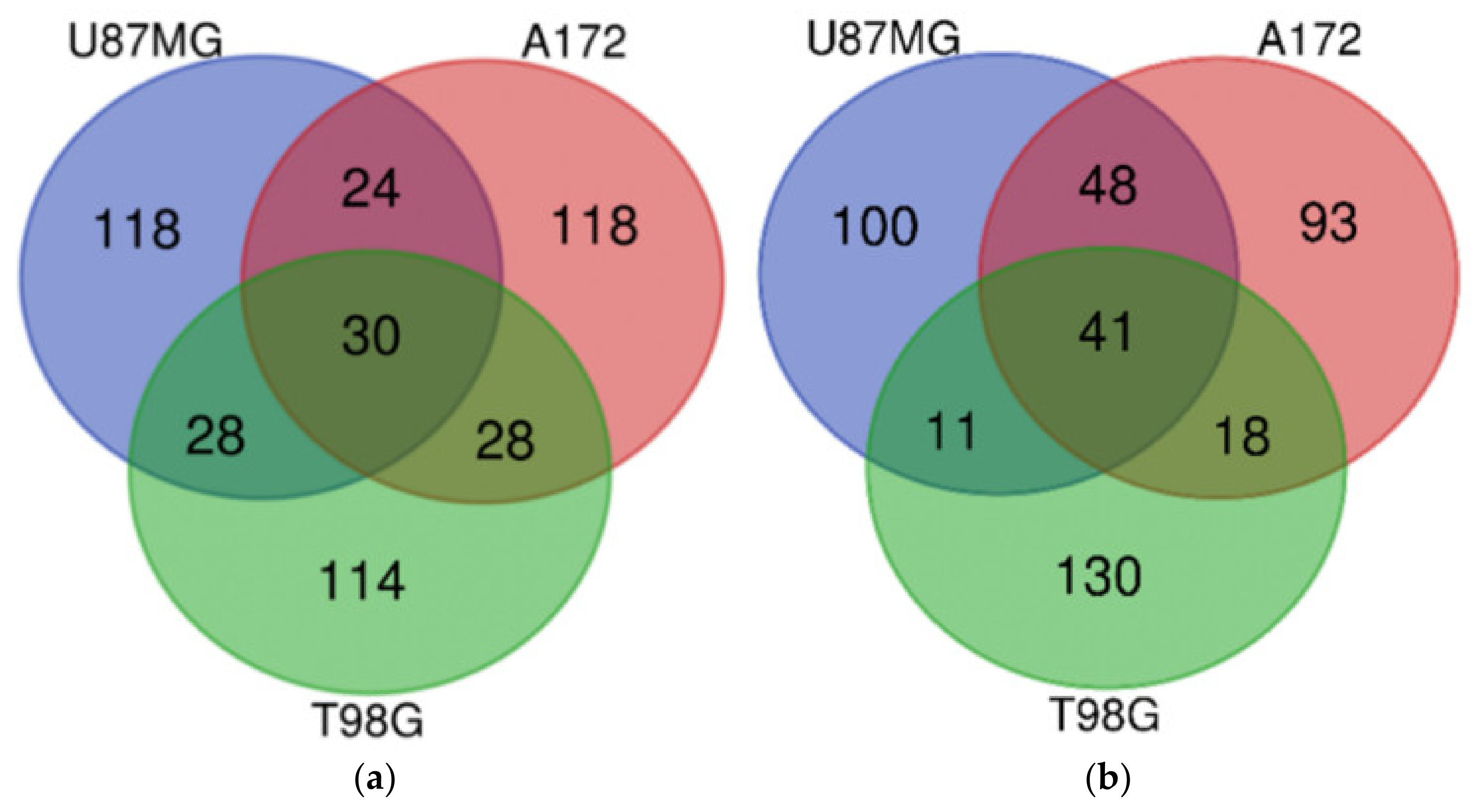


| Gene Group | Gene Number | Gene Names |
|---|---|---|
| Upregulated by Mardepodect, Regorafenib, LY-294002 and Fucoxanthin | 2 | PNLIPRP3, FAM49A |
| Upregulated by Mardepodect and Regorafenib | 34 | PNLIPRP3, FAM49A, PFKFB2, WDR78, GDF15, HMOX1, MSC, TRIB3, GPNMB, ERICH2, CRYM, SLC22A15, NUPR1, LURAP1L, ATP6V0D2, CLEC2D, GCNT3, SLIT3, IDH1, CTH, TM4SF19, RFTN2, KCP, RPS6KA2, KIF26B, UNC5B, PLK2, PLXDC2, FLYWCH1, THBS2, PPARGC1A, PLEKHF1, SLFN5, HECW1 |
| Upregulated by the two multi-kinase inhibitors Regorafenib and LY-294002 | 11 | ADARB1, SOD2, TTLL1, RSPO3, PPIL6, GPCPD1, H1F0, PFKFB2, WDR78, PNLIPRP3, FAM49A |
| Upregulated by Mardepodect only, not by Regorafenib, LY-294002 or Fucoxanthin | 116 | Gene names are found in Supplementary Table S1 |
| Upregulated by Regorafenib only, not by Mardepodect, LY-294002 or Fucoxanthin | 110 | Gene names are found in Supplementary Table S1 |
| Gene Group | Gene Number | Gene Names |
|---|---|---|
| Downregulated by Mardepodect, Regorafenib, LY-294002, and Fucoxanthin | 36 | KIAA1524, ESCO2, E2F8, HIST1H1B, LMNB1, HIST1H2BB, CDCA3, HIST2H3A, HIST1H2BM, TCF19, FBXO5, HIST1H3B, TYMS, DNA2, ORC1, HIST1H2BI, FAM111B, RRM2, ZWINT, HIST1H3A, ASF1B, HIST1H2BH, GPR19, HELLS, PLK4, HIST1H2AG, RAD54L, CDC45, HIST1H3F, HIST1H2AI, SPC25, KIFC1, KIF15, GINS2, UBE2T, HJURP |
| Downregulated by Mardepodect and Regorafenib | 63 | ARL14EPL, HIST1H4D, PBK, HIST1H2AB, RFC3, ATAD2, BARD1, KIF20A, MCM7, KIF11, HIST1H2BJ, TRMU, MKI67, CENPE, ASPM, SPAG5, KIF4A ANGPTL4, ANLN, TACC3, HIST2H4A, HIST2H4B, CPA4, PLEKHG4B, H2AFX, GTSE1, NCAPG, KIAA1524, ESCO2, E2F8, HIST1H1B, LMNB1, HIST1H2BB, CDCA3, HIST2H3A, HIST1H2BM, TCF19, FBXO5, HIST1H3B, TYMS, DNA2, ORC1, HIST1H2BI, FAM111B, RRM2, ZWINT, HIST1H3A, ASF1B, HIST1H2BH, GPR19, HELLS, PLK4, HIST1H2AG, RAD54L, CDC45, HIST1H3F, HIST1H2AI, SPC25, KIFC1, KIF15, GINS2, UBE2T, HJURP |
| Downregulated by Mardepodect only, not Regorafenib, LY-294002, or Fucoxanthin | 95 | Gene names are found in Supplementary Table S4 |
| Downregulated by Regorafenib only, not Mardepodect, LY-294002, or Fucoxanthin | 90 | Gene names are found in Supplementary Table S4 |
| Sharing Groups | Upregulated | Downregulated |
|---|---|---|
| U87MG, A172, and T98G | GDF15, DUSP1, SIK1 | CPA4, FAM111B, CCL2 |
| U87MG and A172 | HMOX1, SLC11A2, GPNMB, GPR183, UAP1L1, PLEKHO1, DUSP4, LIPG, NUPR1, PPARGC1A, LURAP1L, AK5, FAM49A, RRAGD, RRAGC, TM4SF19, FBXO32, RFTN2 | E2F8, TNFRSF11B, PI3, TXNIP |
| U87MG and T98G | NR4A2, CD55, HES1, SLC16A6, S1PR1, NR4A1, C8orf4, CEMIP, DNAJB9, TNFAIP6, SNAI1, SGK1, ITGB3, RND3, TRIB1, GRAMD4, NR4A3, ZCWPW2, IL6, RPS6KA2 | CD84, HIST1H2BM |
| A172 and T98G | HMGCR, DDIT4, FASN, CLCN5, SLC2A3, JAKMIP2, IDI1, AGT, CYP51A1, HLA-DMA, SCD, FDFT1, MVD, HMGCS1, ST3GAL5, DHCR7, ZBED8, RELL2, INSIG1 | EDN1, IRF1, PLXNA2, TRIM22, SERTAD4, TNFRSF9, TNFSF10, RARRES3, LGALS9, CCNE2, KRT18, IL7R, VCAM1, TNFAIP2, ENC1, RNF150, ANKRD1, ROR1, APOL3, CYR61, GBP4, CTGF, PRDM1, ALPK2, LYPD1, BIRC3, IL2RG |
| Cell Line Groups | Upregulated Genes | Downregulated Genes |
|---|---|---|
| U87MG, A172, and T98G | TUBE1, GDF15, TRIB3, PTPDC1, WARS, ERICH2, SLC22A18, SLC6A9, CD22, ATF3, FAM49A, CBS, SLFN5, TMEM159, DDIT3, PSAT1, IL20RB, SOHLH2, TTLL1, PCK2, P2RX7, ASNS, NUPR1, DFNA5, AARS, CCDC169, GTPBP2, PPIL6, RAB39B, KCNH1 | IL7R, ESCO2, E2F8, MCM3, CLSPN, DTL, HIST1H1B, LMNB1, PCNA, EXO1, GINS1, MCM6, ATAD2, BARD1, HIST1H2BM, SERTAD4, MCM10, FBXO5, POLE2, TYMS, DNA2, MCM5, F3, ORC1, UHRF1, FAM111B, RRM2, HIST1H3A, ATAD5, HELLS, E2F1, H2AFX, CCNE2, SPC25, MCM2, MCM4, FANCB, GINS2, WDR76, HIST1H2AB, CDC25A |
| U87MG and A172 | ESRP1, PKD1L2, HMOX1, TBL1X, KCNT2, MSC, LURAP1L, ANK2, UNC5B, GPNMB, STK32A, PHGDH, IDH1, PIP5KL1, THBS4, PLPPR4, SLC43A1, HKDC1, TPK1, TM4SF19, MOCOS, PTPN13, SCN9A, CLIP4 | TMPO, KIAA1524, MKI67, TGFBR2, KIF20B, ZGRF1, RAD51, ASPM, LDLR, SPAG5, RFC3, DUSP6, CDCA3, HIST2H3A, LRR1, CENPI, BRIP1, TACC3, TCF19, SGOL2, STIL, MCM7, CASC5, HIST1H3B, STARD13, KIF11, ZWINT, ASF1B, FEN1, HIST1H2BO, PLK4, RAD54L, ZNF367, CDC45, NCAPD2, POLQ, PBK, NCAPG, CDC6, HIST1H2BJ, POLA2, KIFC1, ARL6IP6, CDCA5, UBE2T, LIN9, HJURP, XRCC2 |
| U87MG and T98G | TSPAN1, PRELID3A, DUS4L, PPARGC1A, TSLP, UHRF1BP1, STAT2, CCDC113, TUFT1, RCAN1, GADD45A, SH3BGR, CLDN1, C6orf48, GARNL3, TNFRSF9, ABI3BP, CTH, DDR2, SLC22A15, CCPG1, GPR1, CCNB1IP1, DMGDH, GPCPD1, ERN1, CYP2R1, ACAD11 | STC1, MEST, CCNF, EGLN3, SPRY1, HIST1H4D, HIST1H2BI, FAM20C, HIST1H2BH, EGR1, CDK2 |
| A172 and T98G | GRB10, FYN, PCDH1, PPP1R3B, HOXB9, FAM129A, SYCP2L, SYT14, SEL1L3, S1PR1, SLC1A4, THBS3, VEGFA, OSBPL6, ULBP1, ARHGEF2, SESN2, AGT, DTNA, MAP2, CHAC1, C10orf107, LCA5L, CREB5, STEAP1, CYP4V2, ADGRG1, AFF3 | EDN1, MMP13, HIST1H3H, MYCBP, HIST1H4L, GMNN, CCND1, CENPW, TNFRSF11B, HIST1H4A, MIS18BP1, CCL2, TRIB2, CDC7, PRDM1, CSNK1G1, MYB, HIST1H2BF |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shapovalov, V.; Kopanitsa, L.; Pruteanu, L.-L.; Ladds, G.; Bailey, D.S. Transcriptomics-Based Phenotypic Screening Supports Drug Discovery in Human Glioblastoma Cells. Cancers 2021, 13, 3780. https://doi.org/10.3390/cancers13153780
Shapovalov V, Kopanitsa L, Pruteanu L-L, Ladds G, Bailey DS. Transcriptomics-Based Phenotypic Screening Supports Drug Discovery in Human Glioblastoma Cells. Cancers. 2021; 13(15):3780. https://doi.org/10.3390/cancers13153780
Chicago/Turabian StyleShapovalov, Vladimir, Liliya Kopanitsa, Lavinia-Lorena Pruteanu, Graham Ladds, and David S. Bailey. 2021. "Transcriptomics-Based Phenotypic Screening Supports Drug Discovery in Human Glioblastoma Cells" Cancers 13, no. 15: 3780. https://doi.org/10.3390/cancers13153780
APA StyleShapovalov, V., Kopanitsa, L., Pruteanu, L.-L., Ladds, G., & Bailey, D. S. (2021). Transcriptomics-Based Phenotypic Screening Supports Drug Discovery in Human Glioblastoma Cells. Cancers, 13(15), 3780. https://doi.org/10.3390/cancers13153780






