Characterization and Clinical Utility of BRAFV600 Mutation Detection Using Cell-Free DNA in Patients with Advanced Melanoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Patients
2.3. Samples and Mutation Analysis
2.4. Treatments
2.5. Outcomes and Assessments
2.6. Statistical Considerations
3. Results
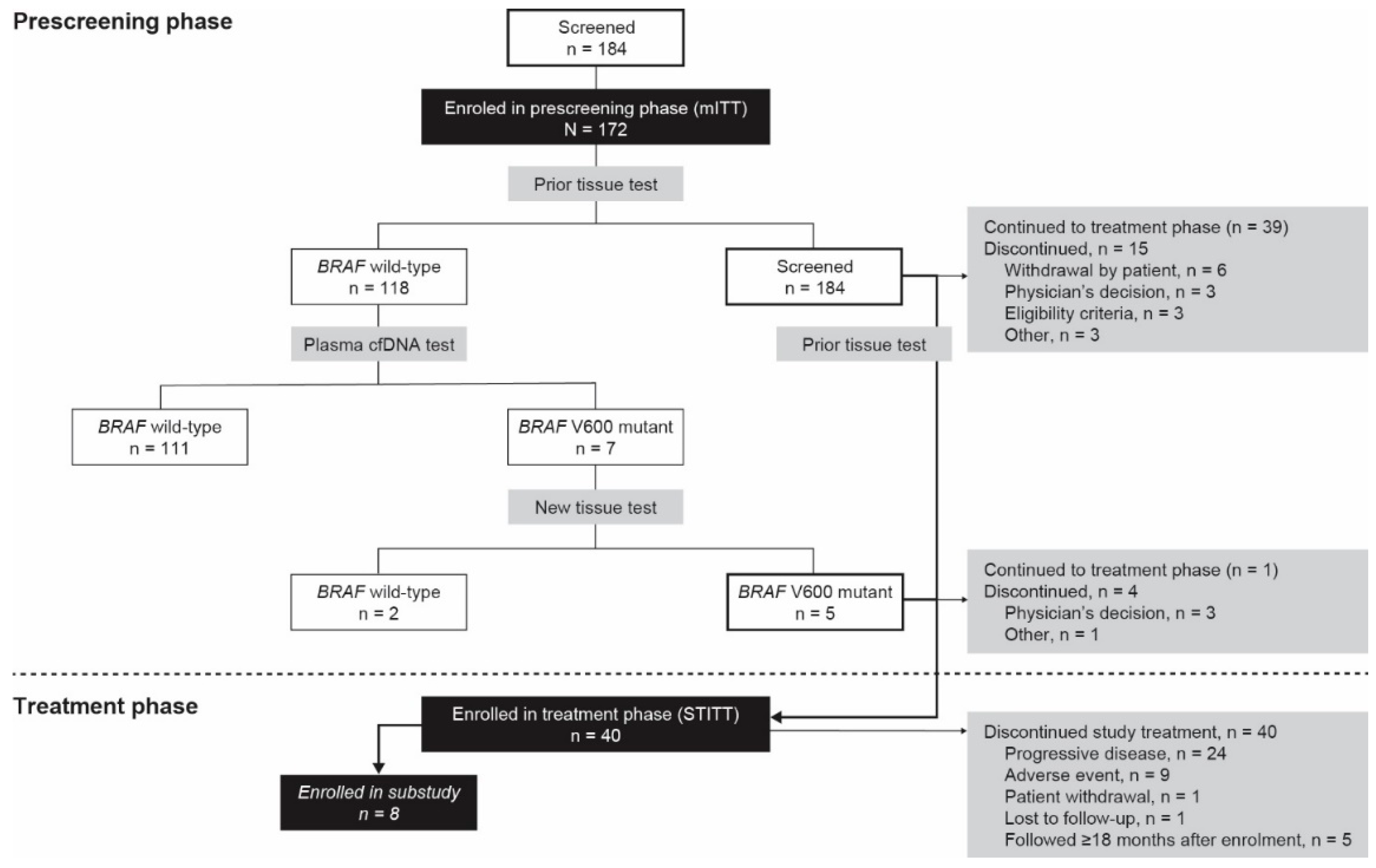
3.1. Patient Disposition
3.2. Prescreening Phase
3.2.1. Primary Endpoint
3.2.2. Secondary Endpoint
3.2.3. Exploratory Endpoint
3.3. Treatment Phase
3.3.1. Efficacy Outcomes
3.3.2. Correlation of Mutation Testing and Clinical Outcome
3.3.3. Substudy
3.3.4. Exposure and Safety
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bisschop, C.; Ter, E.A.; Bosman, L.J.; Platteel, I.; Jalving, M.; van den Berg, A.; Diepstra, A.; van Hemel, B.; Diercks, G.F.H.; Hospers, G.A.P.; et al. Rapid BRAF mutation tests in patients with advanced melanoma: Comparison of immunohistochemistry, Droplet Digital PCR, and the Idylla Mutation Platform. Melanoma Res. 2018, 28, 96–104. [Google Scholar] [CrossRef] [Green Version]
- Bruno, W.; Martinuzzi, C.; Andreotti, V.; Pastorino, L.; Spagnolo, F.; Dalmasso, B.; Cabiddu, F.; Gualco, M.; Ballestrero, A.; Bianchi-Scarrà, G.; et al. Heterogeneity and frequency of BRAF mutations in primary melanoma: Comparison between molecular methods and immunohistochemistry. Oncotarget 2017, 8, 8069–8082. [Google Scholar] [CrossRef] [Green Version]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef]
- Li, Y.; Umbach, D.M.; Li, L. Putative genomic characteristics of BRAF V600K versus V600E cutaneous melanoma. Melanoma Res. 2017, 27, 527–535. [Google Scholar] [CrossRef]
- Keilholz, U.; Ascierto, P.A.; Dummer, R.; Robert, C.; Lorigan, P.; van Akkooi, A.; Arance, A.; Blank, C.U.; Sileni, V.C.; Donia, M.; et al. ESMO consensus conference recommendations on the management of metastatic melanoma: Under the auspices of the ESMO Guidelines Committee. Ann. Oncol. 2020, 31, 1435–1448. [Google Scholar] [CrossRef] [PubMed]
- Swetter, S.M.; Thompson, J.A.; Albertini, M.R.; Barker, C.A.; Boland, G.; Carson, W.E. National Comprehensive Cancer Network® (NCCN®) Clinical Practice Guidelines in Oncology—Cutaneous Melanoma Version 4.2020. 2020. Available online: https://www.nccn.org/professionals/physician_gls/pdf/cutaneous_melanoma.pdf (accessed on 16 September 2020).
- Cohen, J.V.; Buchbinder, E.I. The evolution of adjuvant therapy for melanoma. Curr. Oncol. Rep. 2019, 21, 106. [Google Scholar] [CrossRef] [PubMed]
- Dimitriou, F.; Long, G.V.; Menzies, A.M. Novel adjuvant options for cutaneous melanoma. Ann. Oncol. 2021, 32, 854–865. [Google Scholar] [CrossRef] [PubMed]
- Blankenstein, S.A.; van Akkooi, A.C.J. Adjuvant systemic therapy in high-risk melanoma. Melanoma Res. 2019, 29, 358–364. [Google Scholar] [CrossRef]
- Noor, R.; Trinh, V.; Kim, K.; Hwu, W.J. BRAF-targeted therapy for metastatic melanoma: Rationale, clinical activity and safety. Clin. Investig. 2011, 1, 1127–1139. [Google Scholar] [CrossRef]
- Cheng, L.; Lopez-Beltran, A.; Massari, F.; MacLennan, G.T.; Montironi, R. Molecular testing for BRAF mutations to inform melanoma treatment decisions: A move toward precision medicine. Mod. Pathol. 2018, 31, 24–38. [Google Scholar] [CrossRef]
- Cheng, L.Y.; Haydu, L.E.; Song, P.; Nie, J.; Tetzlaff, M.T.; Kwong, L.N.; Gershenwald, J.E.; Davies, M.A.; Zhang, D.Y. High sensitivity sanger sequencing detection of BRAF mutations in metastatic melanoma FFPE tissue specimens. Sci. Rep. 2021, 11, 9043. [Google Scholar] [CrossRef]
- Harle, A.; Salleron, J.; Franczak, C.; Dubois, C.; Filhine-Tressarieu, P.; Leroux, A.; Merlin, J.-L. Detection of BRAF mutations using a fully automated platform and comparison with high resolution melting, real-time allele specific amplification, immunohistochemistry and next generation sequencing assays, for patients with metastatic melanoma. PLoS ONE 2016, 11, e0153576. [Google Scholar] [CrossRef]
- Huang, W.; Kuo, T.T.; Wu, C.E.; Cheng, H.-Y.; Hsieh, C.-H.; Hsieh, J.-J.; Shen, Y.-C.; Hou, M.-M.; Hsu, T.; Chang, J.W.-C. A comparison of immunohistochemical and molecular methods used for analyzing the BRAFV600E gene mutation in malignant melanoma in Taiwan. Asia Pac. J. Clin. Oncol. 2016, 12, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Janku, F.; Claes, B.; Huang, H.J.; Falchook, G.S.; Devogelaere, B.; Kockx, M.; Bempt, I.V.; Reijans, M.; Naing, A.; Fu, S.; et al. BRAF mutation testing with a rapid, fully integrated molecular diagnostics system. Oncotarget 2015, 6, 26886–26894. [Google Scholar] [CrossRef] [Green Version]
- Alrabadi, N.; Gibson, N.; Curless, K.; Cheng, L.; Kuhar, M.; Chen, S.; Warren, S.J.P.; Alomari, A.K. Detection of driver mutations in BRAF can aid in diagnosis and early treatment of dedifferentiated metastatic melanoma. Mod. Pathol. 2019, 32, 330–337. [Google Scholar] [CrossRef]
- Kong, B.Y.; Carlino, M.S.; Menzies, A.M. Biology and treatment of BRAF mutant metastatic melanoma. Melanoma Manag. 2016, 3, 33–45. [Google Scholar] [CrossRef]
- Vanni, I.; Tanda, E.T.; Spagnolo, F.; Andreotti, V.; Bruno, W.; Ghiorzo, P. The current state of molecular testing in the BRAF-mutated melanoma landscape. Front. Mol. Biosci. 2020, 7, 113. [Google Scholar] [CrossRef] [PubMed]
- Donadio, E.; Giusti, L.; Cetani, F.; Da Valle, Y.; Ciregia, F.; Giannaccini, G.; Pardi, E.; Saponaro, F.; Torregrossa, L.; Basolo, F.; et al. Evaluation of formalin-fixed paraffin-embedded tissues in the proteomic analysis of parathyroid glands. Proteome Sci. 2011, 9, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janku, F.; Huang, H.J.; Claes, B.; Falchook, G.S.; Fu, S.; Hong, D.; Ramzanali, N.M.; Nitti, G.; Cabrilo, G.; Tsimberidou, A.M.; et al. BRAF mutation testing in cell-free DNA from the plasma of patients with advanced cancers using a rapid, automated molecular diagnostics system. Mol. Cancer Ther. 2016, 15, 1397–1404. [Google Scholar] [CrossRef] [Green Version]
- Calapre, L.; Warburton, L.; Millward, M.; Ziman, M.; Gray, E.S. Circulating tumour DNA (ctDNA) as a liquid biopsy for melanoma. Cancer Lett. 2017, 404, 62–69. [Google Scholar] [CrossRef]
- Balch, C.M.; Gershenwald, J.E.; Soong, S.J.; Thompson, J.F.; Atkins, M.B.; Byrd, D.R.; Buzaid, A.C.; Cochran, A.J.; Coit, D.G.; Ding, S.; et al. Final version of 2009 AJCC melanoma staging and classification. J. Clin. Oncol. 2009, 27, 6199–6206. [Google Scholar] [CrossRef] [Green Version]
- National Institutes of Health. Common Terminology Criteria for Adverse Events (CTCAE) v4.03. 2009. Available online: https://www.eortc.be/services/doc/ctc/CTCAE_4.03_2010-06-14_QuickReference_5x7.pdf (accessed on 16 September 2020).
- Long-Mira, E.; Ilie, M.; Chamorey, E.; Leduff-Blanc, F.; Montaudié, H.; Tanga, V.; Allégra, M.; Lespinet-Fabre, V.; Bordone, O.; Bonnetaud, C.; et al. Monitoring BRAF and NRAS mutations with cell-free circulating tumor DNA from metastatic melanoma patients. Oncotarget 2018, 9, 36238–36249. [Google Scholar] [CrossRef] [Green Version]
- Molina-Vila, M.A.; de-Las-Casas, C.M.; Bertran-Alamillo, J.; Jordana-Ariza, N.; González-Cao, M.; Rosell, R. cfDNA analysis from blood in melanoma. Ann. Transl. Med. 2015, 3, 309. [Google Scholar] [CrossRef]
- Burjanivova, T.; Malicherova, B.; Grendar, M.; Minarikova, E.; Dusenka, R.; Vanova, B.; Bobrovska, M.; Pecova, T.; Homola, I.; Lasabova, Z.; et al. Detection of BRAFV600E mutation in melanoma patients by digital PCR of circulating DNA. Genet. Test. Mol. Biomark. 2019, 23, 241–245. [Google Scholar] [CrossRef]
- Knol, A.C.; Vallée, A.; Herbreteau, G.; Nguyen, J.M.; Varey, E.; Gaultier, A.; Théoleyre, S.; Saint-Jean, M.; Peuvrel, L.; Brocard, A.; et al. Clinical significance of BRAF mutation status in circulating tumor DNA of metastatic melanoma patients at baseline. Exp. Dermatol. 2016, 25, 783–788. [Google Scholar] [CrossRef]
- Kozak, K.; Kowalik, A.; Gos, A.; Wasag, B.; Lugowska, I.; Jurkowska, M.; Krawczynska, N.; Kosela-Paterczyk, H.; Switaj, T.; Teterycz, P.; et al. Cell-free DNA BRAF V600E measurements during BRAF inhibitor therapy of metastatic melanoma: Long-term analysis. Tumori 2020. [Google Scholar] [CrossRef]
- Larkin, J.; Ascierto, P.A.; Dreno, B.; Atkinson, V.; Liszkay, G.; Maio, M.; Mandalà, M.; Demidov, L.; Stroyakovskiy, D.; Thomas, L.; et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N. Engl. J. Med. 2014, 371, 1867–1876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schreuer, M.; Jansen, Y.; Planken, S.; Chevolet, I.; Seremet, T.; Kruse, V.; Neyns, B. Combination of dabrafenib plus trametinib for BRAF and MEK inhibitor pretreated patients with advanced BRAF(V600)-mutant melanoma: An open-label, single arm, dual-centre, phase 2 clinical trial. Lancet Oncol. 2017, 18, 464–472. [Google Scholar] [CrossRef]
- Schreuer, M.; Meersseman, G.; Van Den Herrewegen, S.; Jansen, Y.; Chevolet, I.; Bott, A.; Wilgenhof, S.; Seremet, T.; Jacobs, B.; Buyl, R.; et al. Quantitative assessment of BRAFV600 mutant circulating cell-free tumor DNA as a tool for therapeutic monitoring in metastatic melanoma patients treated with BRAF/MEK inhibitors. J. Transl. Med. 2016, 14, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ascierto, P.A.; McArthur, G.A.; Dréno, B.; Atkinson, V.; Liszkay, G.; Di Giacomo, A.M.; Mandalà, M.; Demidov, L.; Stroyakovskiy, D.; Thomas, L.; et al. Cobimetinib combined with vemurafenib in advanced BRAF(V600)-mutant melanoma (coBRIM): Updated efficacy results from a randomised, double-blind, phase 3 trial. Lancet Oncol. 2016, 17, 1248–1260. [Google Scholar] [CrossRef]
| mITT | STITT | |
|---|---|---|
| n = 172 | n = 40 | |
| Age, median years (range) | 62.5 (20–93) | 56.5 (26–82) |
| Sex, n (%) | ||
| Male | 84 (48.8) | 18 (45.0) |
| Female | 88 (51.2) | 22 (55.0) |
| ECOG score, n (%) | ||
| 0 | NE | 21 (52.5) |
| 1 | NE | 16 (40.0) |
| 2 | NE | 3 (7.5) |
| Time since diagnosis of metastases, median months (range) | 19.3 (0.1–260.7) | 12.4 (0.8–260.7) |
| Age at diagnosis of metastases, median years (range) | 59.1 (18–91) | 53.5 (26–83) |
| Disease stage at study entry, n (%) | ||
| Unresectable stage IIIC | 16 (9.3) | 1 (2.5) |
| Stage IV | 156 (90.7) | 29 (97.5) |
| Measurable disease at study entry, n (%) | 147 (85.5) | 37 (92.5) |
| Number of target lesions, n (%) | ||
| 0–3 | NE | 22 (55.0) |
| >3 | NE | 18 (45.0) |
| Type of tissue material, n (%) | ||
| Archival | 140 (81.4) | 30 (75.0) |
| Recent | 32 (18.6) | 10 (25.0) |
| Prior tissue BRAF mutation test result, n (%) | ||
| BRAF wild-type | 118 (68.6) | 1 (2.5) |
| BRAFV600 mutation | 54 (31.4) | 39 (97.5) |
| Prior therapy, n (%) | ||
| Immunotherapy | – | 9 (33) |
| Targeted therapy | – | 0 |
| Other systemic therapy | – | 7 (17.5) |
| Investigational treatment | – | 1 (2.5) |
| Radiotherapy | – | 8 (20.0) |
| A. BRAF | Plasma cfDNA Test Result | |||
|---|---|---|---|---|
| Wild-Type | Mutant | |||
| Tissue test result | Wild-type | 111 | 7 | Patients with wild-type tissue test = 118 |
| Mutant | 19 | 35 | Patients with mutant tissue test = 54 | |
| Patients with wild-type plasma test = 130 | Patients with mutant plasma test = 42 | |||
| B. Plasma | BRAF cfDNA test result | |||
| Wild-type | Mutant | |||
| NRAS cfDNA test result | Wild-type | 107 | 42 | Patients with wild-type NRAS = 149 |
| Mutant | 22 | 0 | Patients with mutant NRAS = 22 | |
| Patients with wild-type plasma test = 129 | Patients with mutant plasma test = 42 | |||
| STITT n = 40 | Archival Tissue * n = 29 | Recent Tissue † n = 10 | |
|---|---|---|---|
| Objective response rate, ‡ n (%) | |||
| CR | 3 (8.3) | 3 (11.5) | – |
| PR | 26 (72.2) | 20 (76.9) | 6 (60.0) |
| SD | 2 (5.6) | – | 2 (20.0) |
| PD | 1 (2.8) | – | 1 (10.0) |
| Other nonresponders | 4 (11.1) | 3 (11.5) | 1 (10.0) |
| Responders (CR + PR), n/n′ (%) | 29/36 (80.6) | 23/26 (88.5) | 6/10 (60.0) |
| 95% CI | 64.0–91.8 | 69.8–97.6 | 26.2–87.8 |
| Duration of Response, n′ ‡ | 29 | 23 | n′ = 6 |
| Median, months (95% CI) | 11.0 (9.2–NE) | 11.0 (9.2–NE) | 8.8 (3.6–NE) |
| Progression-free survival | |||
| Median months (95% CI) | 13.6 (9.5–16.5) | 14.5 (10.8–NE) | 6.2 (3.6–NE) |
| System Organ Class/Preferred Term | n = 40 |
|---|---|
| Any treatment-emergent adverse event, n (%) | 39 (97.5) |
| Skin and subcutaneous tissue disorders, n (%) | 33 (82.5) |
| Rash | 19 (47.5) |
| Photosensitivity reaction | 11 (27.5) |
| Maculopapular rash | 9 (22.5) |
| Alopecia | 4 (10.0) |
| Investigations, n (%) | 22 (55.0) |
| Blood creatinine phosphokinase increased | 13 (32.5) |
| Gamma-glutamyltransferase increased | 5 (12.5) |
| C-reactive protein increased | 4 (10.0) |
| General disorders and administration site conditions, n (%) | 21 (52.5) |
| Pyrexia | 11 (27.5) |
| Fatigue | 8 (20.0) |
| Oedema peripheral | 5 (12.5) |
| Infections and infestations, n (%) | 21 (52.5) |
| Upper respiratory tract infection | 8 (20.0) |
| Conjunctivitis | 6 (15.0) |
| Urinary tract infection | 4 (10.0) |
| Gastrointestinal disorders, n (%) | 20 (50.0) |
| Diarrhea | 13 (32.5) |
| Vomiting | 6 (15.0) |
| Nausea | 5 (12.5) |
| Musculoskeletal and connective tissue disorders, n (%) | 14 (35.0) |
| Arthralgia | 11 (27.5) |
| Musculoskeletal pain | 4 (10.0) |
| Myalgia | 4 (10.0) |
| Pain in extremity | 4 (10.0) |
| Eye disorders, n (%) | 12 (30.0) |
| Vision blurred | 6 (15.0) |
| Chorioretinopathy | 4 (10.0) |
| Nervous system disorders, n (%) | 12 (30.0) |
| Headache | 6 (15.0) |
| Metabolism and nutrition disorders, n (%) | 7 (17.5) |
| Decreased appetite | 4 (10.0) |
| Hypokalemia | 4 (10.0) |
| Hypertension | 5 (12.5) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rutkowski, P.; Pauwels, P.; Kerger, J.; Jacobs, B.; Maertens, G.; Gadeyne, V.; Thielemans, A.; de Backer, K.; Neyns, B. Characterization and Clinical Utility of BRAFV600 Mutation Detection Using Cell-Free DNA in Patients with Advanced Melanoma. Cancers 2021, 13, 3591. https://doi.org/10.3390/cancers13143591
Rutkowski P, Pauwels P, Kerger J, Jacobs B, Maertens G, Gadeyne V, Thielemans A, de Backer K, Neyns B. Characterization and Clinical Utility of BRAFV600 Mutation Detection Using Cell-Free DNA in Patients with Advanced Melanoma. Cancers. 2021; 13(14):3591. https://doi.org/10.3390/cancers13143591
Chicago/Turabian StyleRutkowski, Piotr, Patrick Pauwels, Joseph Kerger, Bart Jacobs, Geert Maertens, Valerie Gadeyne, Anne Thielemans, Katrien de Backer, and Bart Neyns. 2021. "Characterization and Clinical Utility of BRAFV600 Mutation Detection Using Cell-Free DNA in Patients with Advanced Melanoma" Cancers 13, no. 14: 3591. https://doi.org/10.3390/cancers13143591
APA StyleRutkowski, P., Pauwels, P., Kerger, J., Jacobs, B., Maertens, G., Gadeyne, V., Thielemans, A., de Backer, K., & Neyns, B. (2021). Characterization and Clinical Utility of BRAFV600 Mutation Detection Using Cell-Free DNA in Patients with Advanced Melanoma. Cancers, 13(14), 3591. https://doi.org/10.3390/cancers13143591








