Severe Phenotype in Patients with Large Deletions of NF1
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. Phenotypes
2.3. NF1 Molecular Analysis
2.4. Statistical Analyses
2.5. Characterization of the Deletions
3. Results
3.1. Description of the Cohort
3.1.1. Pigmentary Manifestations
3.1.2. Neurofibromas
3.1.3. OPGs and Neurological Findings
3.1.4. Malignancies
3.1.5. Cardiovascular Abnormalities
3.1.6. Other Findings
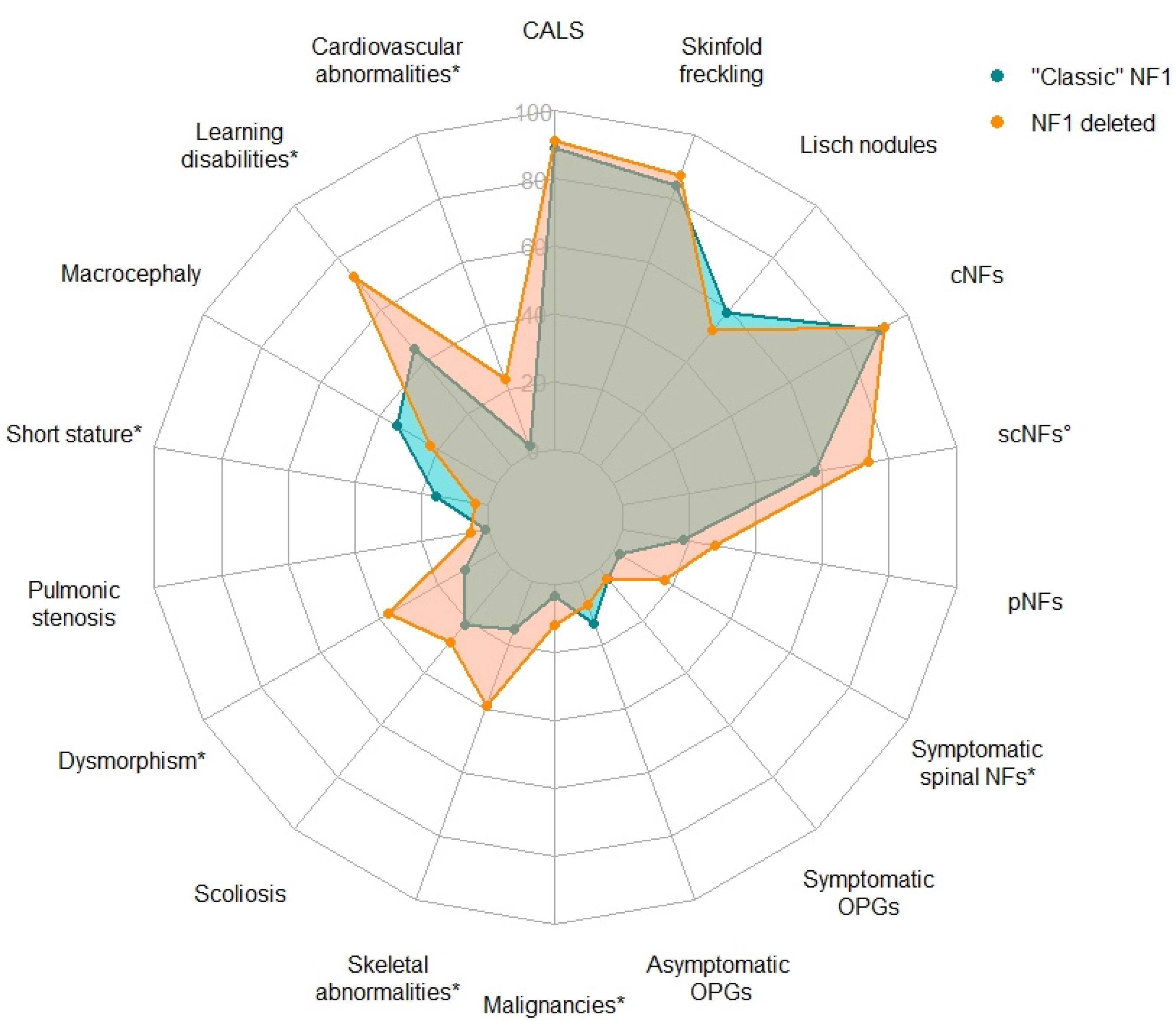
3.2. Comparison of the Clinical Findings in the French NF1-Deleted Cohort with “Classic NF1” Phenotype
3.3. Characterization of the Deletions of the NF1 Locus
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Williams, V.C.; Lucas, J.; Babcock, M.A.; Gutmann, D.H.; Korf, B.; Maria, B.L. Neurofibromatosis Type 1 Revisited. Pediatrics 2009, 123, 124–133. [Google Scholar] [CrossRef] [PubMed]
- Sabbagh, A.; Pasmant, E.; Imbard, A.; Luscan, A.; Soares, M.; Blanché, H.; Laurendeau, I.; Ferkal, S.; Vidaud, M.; Pinson, S.; et al. NF1 Molecular Characterization and Neurofibromatosis Type I Genotype-Phenotype Correlation: The French Experience. Hum. Mutat. 2013, 34, 1510–1518. [Google Scholar] [CrossRef] [PubMed]
- Messiaen, L.M.; Callens, T.; Mortier, G.; Beysen, D.; Vandenbroucke, I.; Van Roy, N.; Speleman, F.; Paepe, A.D. Exhaustive Mutation Analysis of the NF1 Gene Allows Identification of 95% of Mutations and Reveals a High Frequency of Unusual Splicing Defects. Hum. Mutat. 2000, 15, 541–555. [Google Scholar] [CrossRef]
- von Deimling, A.; Krone, W.; Menon, A.G. Neurofibromatosis Type 1: Pathology, Clinical Features and Molecular Genetics. Brain Pathol. 1995, 5, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.F.; O’Connell, P.; Viskochil, D.; Cawthon, R.; Robertson, M.; Culver, M.; Dunn, D.; Stevens, J.; Gesteland, R.; White, R. The Neurofibromatosis Type 1 Gene Encodes a Protein Related to GAP. Cell 1990, 62, 599–608. [Google Scholar] [CrossRef]
- National Institutes of Health Consensus Development Conference Statement: Neurofibromatosis. Bethesda, MD, USA, July 13–15, 1987. Neurofibromatosis 1988, 1, 172–178.
- Tucker, T.; Wolkenstein, P.; Revuz, J.; Zeller, J.; Friedman, J.M. Association between Benign and Malignant Peripheral Nerve Sheath Tumors in NF1. Neurology 2005, 65, 205–211. [Google Scholar] [CrossRef]
- Patel, N.B.; Stacy, G.S. Musculoskeletal Manifestations of Neurofibromatosis Type 1. AJR Am. J. Roentgenol. 2012, 199, W99–W106. [Google Scholar] [CrossRef]
- Bayat, M.; Bayat, A. Neurological Manifestations of Neurofibromatosis: A Review. Neurol. Sci. 2020, 41, 2685–2690. [Google Scholar] [CrossRef]
- Lin, A.E.; Birch, P.H.; Korf, B.R.; Tenconi, R.; Niimura, M.; Poyhonen, M.; Armfield Uhas, K.; Sigorini, M.; Virdis, R.; Romano, C.; et al. Cardiovascular Malformations and Other Cardiovascular Abnormalities in Neurofibromatosis 1. Am. J. Med. Genet. 2000, 95, 108–117. [Google Scholar] [CrossRef]
- Cutruzzolà, A.; Irace, C.; Frazzetto, M.; Sabatino, J.; Gullace, R.; De Rosa, S.; Spaccarotella, C.; Concolino, D.; Indolfi, C.; Gnasso, A. Functional and Morphological Cardiovascular Alterations Associated with Neurofibromatosis 1. Sci. Rep. 2020, 10, 12070. [Google Scholar] [CrossRef]
- Bizzarri, C.; Bottaro, G. Endocrine Implications of Neurofibromatosis 1 in Childhood. Horm. Res. Paediatr. 2015, 83, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Torres Nupan, M.M.; Velez Van Meerbeke, A.; López Cabra, C.A.; Herrera Gomez, P.M. Cognitive and Behavioral Disorders in Children with Neurofibromatosis Type 1. Front. Pediatrics 2017, 5, 227. [Google Scholar] [CrossRef]
- Carey, J.C. Neurofibromatosis-Noonan Syndrome. Am. J. Med. Genet. 1998, 75, 263–264. [Google Scholar] [CrossRef]
- Costa, A.D.A.; Gutmann, D.H. Brain Tumors in Neurofibromatosis Type 1. Neurooncol. Adv. 2019, 1, vdz040. [Google Scholar] [CrossRef]
- Seminog, O.O.; Goldacre, M.J. Risk of Benign Tumours of Nervous System, and of Malignant Neoplasms, in People with Neurofibromatosis: Population-Based Record-Linkage Study. Br. J. Cancer 2013, 108, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Easton, D.F.; Ponder, M.A.; Huson, S.M.; Ponder, B.A. An Analysis of Variation in Expression of Neurofibromatosis (NF) Type 1 (NF1): Evidence for Modifying Genes. Am. J. Hum. Genet. 1993, 53, 305–313. [Google Scholar] [PubMed]
- Szudek, J.; Joe, H.; Friedman, J.M. Analysis of Intrafamilial Phenotypic Variation in Neurofibromatosis 1 (NF1). Genet. Epidemiol. 2002, 23, 150–164. [Google Scholar] [CrossRef] [PubMed]
- Sabbagh, A.; Pasmant, E.; Laurendeau, I.; Parfait, B.; Barbarot, S.; Guillot, B.; Combemale, P.; Ferkal, S.; Vidaud, M.; Aubourg, P.; et al. Unravelling the Genetic Basis of Variable Clinical Expression in Neurofibromatosis 1. Hum. Mol. Genet. 2009, 18, 2768–2778. [Google Scholar] [CrossRef] [PubMed]
- Pinna, V.; Daniele, P.; Calcagni, G.; Mariniello, L.; Criscione, R.; Giardina, C.; Lepri, F.R.; Hozhabri, H.; Alberico, A.; Cavone, S.; et al. Prevalence, Type, and Molecular Spectrum of NF1 Mutations in Patients with Neurofibromatosis Type 1 and Congenital Heart Disease. Genes 2019, 10, 675. [Google Scholar] [CrossRef]
- Melloni, G.; Eoli, M.; Cesaretti, C.; Bianchessi, D.; Ibba, M.C.; Esposito, S.; Scuvera, G.; Morcaldi, G.; Micheli, R.; Piozzi, E.; et al. Risk of Optic Pathway Glioma in Neurofibromatosis Type 1: No Evidence of Genotype-Phenotype Correlations in A Large Independent Cohort. Cancers 2019, 11, 1838. [Google Scholar] [CrossRef] [PubMed]
- Castle, B.; Baser, M.E.; Huson, S.M.; Cooper, D.N.; Upadhyaya, M. Evaluation of Genotype-Phenotype Correlations in Neurofibromatosis Type 1. J. Med. Genet. 2003, 40, e109. [Google Scholar] [CrossRef] [PubMed]
- Kang, E.; Kim, Y.-M.; Seo, G.H.; Oh, A.; Yoon, H.M.; Ra, Y.-S.; Kim, E.K.; Kim, H.; Heo, S.-H.; Kim, G.-H.; et al. Phenotype Categorization of Neurofibromatosis Type I and Correlation to NF1 Mutation Types. J. Hum. Genet. 2020, 65, 79–89. [Google Scholar] [CrossRef]
- Upadhyaya, M.; Huson, S.M.; Davies, M.; Thomas, N.; Chuzhanova, N.; Giovannini, S.; Evans, D.G.; Howard, E.; Kerr, B.; Griffiths, S.; et al. An Absence of Cutaneous Neurofibromas Associated with a 3-Bp Inframe Deletion in Exon 17 of the NF1 Gene (c.2970-2972 DelAAT): Evidence of a Clinically Significant NF1 Genotype-Phenotype Correlation. Am. J. Hum. Genet. 2007, 80, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Koczkowska, M.; Callens, T.; Gomes, A.; Sharp, A.; Chen, Y.; Hicks, A.D.; Aylsworth, A.S.; Azizi, A.A.; Basel, D.G.; Bellus, G.; et al. Expanding the Clinical Phenotype of Individuals with a 3-Bp in-Frame Deletion of the NF1 Gene (c.2970_2972del): An Update of Genotype-Phenotype Correlation. Genet. Med. 2019, 21, 867–876. [Google Scholar] [CrossRef]
- Pinna, V.; Lanari, V.; Daniele, P.; Consoli, F.; Agolini, E.; Margiotti, K.; Bottillo, I.; Torrente, I.; Bruselles, A.; Fusilli, C.; et al. P.Arg1809Cys Substitution in Neurofibromin Is Associated with a Distinctive NF1 Phenotype without Neurofibromas. Eur. J. Hum. Genet. 2015, 23, 1068–1071. [Google Scholar] [CrossRef]
- Rojnueangnit, K.; Xie, J.; Gomes, A.; Sharp, A.; Callens, T.; Chen, Y.; Liu, Y.; Cochran, M.; Abbott, M.-A.; Atkin, J.; et al. High Incidence of Noonan Syndrome Features Including Short Stature and Pulmonic Stenosis in Patients Carrying NF1 Missense Mutations Affecting p.Arg1809: Genotype-Phenotype Correlation. Hum. Mutat. 2015, 36, 1052–1063. [Google Scholar] [CrossRef]
- Nyström, A.M.; Ekvall, S.; Allanson, J.; Edeby, C.; Elinder, M.; Holmström, G.; Bondeson, M.L.; Annerén, G. Noonan Syndrome and Neurofibromatosis Type I in a Family with a Novel Mutation in NF1. Clin. Genet. 2009, 76, 524–534. [Google Scholar] [CrossRef]
- Ekvall, S.; Sjörs, K.; Jonzon, A.; Vihinen, M.; Annerén, G.; Bondeson, M.-L. Novel Association of Neurofibromatosis Type 1-Causing Mutations in Families with Neurofibromatosis-Noonan Syndrome. Am. J. Med. Genet. A 2014, 164A, 579–587. [Google Scholar] [CrossRef]
- Santoro, C.; Maietta, A.; Giugliano, T.; Melis, D.; Perrotta, S.; Nigro, V.; Piluso, G. Arg(1809) Substitution in Neurofibromin: Further Evidence of a Genotype-Phenotype Correlation in Neurofibromatosis Type 1. Eur. J. Hum. Genet. 2015, 23, 1460–1461. [Google Scholar] [CrossRef]
- Koczkowska, M.; Callens, T.; Chen, Y.; Gomes, A.; Hicks, A.D.; Sharp, A.; Johns, E.; Uhas, K.A.; Armstrong, L.; Bosanko, K.A.; et al. Clinical Spectrum of Individuals with Pathogenic NF1 Missense Variants Affecting p.Met1149, p.Arg1276, and p.Lys1423: Genotype-Phenotype Study in Neurofibromatosis Type 1. Hum. Mutat. 2020, 41, 299–315. [Google Scholar] [CrossRef] [PubMed]
- Koczkowska, M.; Chen, Y.; Callens, T.; Gomes, A.; Sharp, A.; Johnson, S.; Hsiao, M.-C.; Chen, Z.; Balasubramanian, M.; Barnett, C.P.; et al. Genotype-Phenotype Correlation in NF1: Evidence for a More Severe Phenotype Associated with Missense Mutations Affecting NF1 Codons 844–848. Am. J. Hum. Genet. 2018, 102, 69–87. [Google Scholar] [CrossRef]
- Kehrer-Sawatzki, H.; Mautner, V.-F.; Cooper, D.N. Emerging Genotype-Phenotype Relationships in Patients with Large NF1 Deletions. Hum. Genet. 2017, 136, 349–376. [Google Scholar] [CrossRef]
- Pasmant, E.; Sabbagh, A.; Spurlock, G.; Laurendeau, I.; Grillo, E.; Hamel, M.-J.; Martin, L.; Barbarot, S.; Leheup, B.; Rodriguez, D.; et al. NF1 Microdeletions in Neurofibromatosis Type 1: From Genotype to Phenotype. Hum. Mutat. 2010, 31, E1506–E1518. [Google Scholar] [CrossRef]
- Ning, X.; Farschtschi, S.; Jones, A.; Kehrer-Sawatzki, H.; Mautner, V.-F.; Friedman, J.M. Growth in Neurofibromatosis 1 Microdeletion Patients. Clin. Genet. 2016, 89, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Pasmant, E.; de Saint-Trivier, A.; Laurendeau, I.; Dieux-Coeslier, A.; Parfait, B.; Vidaud, M.; Vidaud, D.; Bièche, I. Characterization of a 7.6-Mb Germline Deletion Encompassing the NF1 Locus and about a Hundred Genes in an NF1 Contiguous Gene Syndrome Patient. Eur. J. Hum. Genet. 2008, 16, 1459–1466. [Google Scholar] [CrossRef]
- Upadhyaya, M.; Spurlock, G.; Monem, B.; Thomas, N.; Friedrich, R.E.; Kluwe, L.; Mautner, V. Germline and Somatic NF1 Gene Mutations in Plexiform Neurofibromas. Hum. Mutat. 2008, 29, E103–E111. [Google Scholar] [CrossRef]
- Pasmant, E.; Parfait, B.; Luscan, A.; Goussard, P.; Briand-Suleau, A.; Laurendeau, I.; Fouveaut, C.; Leroy, C.; Montadert, A.; Wolkenstein, P.; et al. Neurofibromatosis Type 1 Molecular Diagnosis: What Can NGS Do for You When You Have a Large Gene with Loss of Function Mutations? Eur. J. Hum. Genet. 2015, 23, 596–601. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Imbard, A.; Pasmant, E.; Sabbagh, A.; Luscan, A.; Soares, M.; Goussard, P.; Blanché, H.; Laurendeau, I.; Ferkal, S.; Vidaud, M.; et al. NF1 Single and Multi-Exons Copy Number Variations in Neurofibromatosis Type 1. J. Hum. Genet. 2015, 60, 221–224. [Google Scholar] [CrossRef]
- Kinori, M.; Hodgson, N.; Zeid, J.L. Ophthalmic Manifestations in Neurofibromatosis Type 1. Surv. Ophthalmol. 2018, 63, 518–533. [Google Scholar] [CrossRef] [PubMed]
- Boyd, K.P.; Korf, B.R.; Theos, A. Neurofibromatosis Type 1. J. Am. Acad. Dermatol. 2009, 61, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Cambiaso, P.; Galassi, S.; Palmiero, M.; Mastronuzzi, A.; Del Bufalo, F.; Capolino, R.; Cacchione, A.; Buonuomo, P.S.; Gonfiantini, M.V.; Bartuli, A.; et al. Growth Hormone Excess in Children with Neurofibromatosis Type-1 and Optic Glioma. Am. J. Med. Genet. A 2017, 173, 2353–2358. [Google Scholar] [CrossRef] [PubMed]
- Peces, R.; Mena, R.; Martín, Y.; Hernández, C.; Peces, C.; Tellería, D.; Cuesta, E.; Selgas, R.; Lapunzina, P.; Nevado, J. Co-Occurrence of Neurofibromatosis Type 1 and Optic Nerve Gliomas with Autosomal Dominant Polycystic Kidney Disease Type 2. Mol. Genet. Genomic Med. 2020, 8, e1321. [Google Scholar] [CrossRef] [PubMed]
- Koc, F.; Guzel, A.I. Neurofibromatosis Type 1 Associated with Charcot-Marie-Tooth Type 1A. J. Dermatol. 2009, 36, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Lupski, J.R.; Pentao, L.; Williams, L.L.; Patel, P.I. Stable Inheritance of the CMT1A DNA Duplication in Two Patients with CMT1 and NF1. Am. J. Med. Genet. 1993, 45, 92–96. [Google Scholar] [CrossRef]
- Onu, D.O.; Hunn, A.W.; Peters-Willke, J. Charcot-Marie-Tooth Syndrome and Neurofibromatosis Type 1 with Multiple Neurofibromas of the Entire Spinal Nerve Roots. BMJ Case Rep. 2013, 2013. [Google Scholar] [CrossRef]
- Roos, K.L.; Pascuzzi, R.M.; Dunn, D.W. Neurofibromatosis, Charcot-Marie-Tooth Disease, or Both? Neurofibromatosis 1989, 2, 238–243. [Google Scholar]
- Sbidian, E.; Bastuji-Garin, S.; Valeyrie-Allanore, L.; Ferkal, S.; Lefaucheur, J.P.; Drouet, A.; Brugière, P.; Vialette, C.; Combemale, P.; Barbarot, S.; et al. At-Risk Phenotype of Neurofibromatose-1 Patients: A Multicentre Case-Control Study. Orphanet J. Rare Dis. 2011, 6, 51. [Google Scholar] [CrossRef]
- Akshintala, S.; Baldwin, A.; Liewehr, D.J.; Goodwin, A.; Blakeley, J.O.; Gross, A.M.; Steinberg, S.M.; Dombi, E.; Widemann, B.C. Longitudinal Evaluation of Peripheral Nerve Sheath Tumors in Neurofibromatosis Type 1: Growth Analysis of Plexiform Neurofibromas and Distinct Nodular Lesions. Neuro Oncol. 2020, 22, 1368–1378. [Google Scholar] [CrossRef] [PubMed]
- Mautner, V.-F.; Kluwe, L.; Friedrich, R.E.; Roehl, A.C.; Bammert, S.; Högel, J.; Spöri, H.; Cooper, D.N.; Kehrer-Sawatzki, H. Clinical Characterisation of 29 Neurofibromatosis Type-1 Patients with Molecularly Ascertained 1.4 Mb Type-1 NF1 Deletions. J. Med. Genet. 2010, 47, 623–630. [Google Scholar] [CrossRef]
- De Schepper, S.; Boucneau, J.; Lambert, J.; Messiaen, L.; Naeyaert, J.-M. Pigment Cell-Related Manifestations in Neurofibromatosis Type 1: An Overview. Pigment Cell Res. 2005, 18, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Allouche, J.; Bellon, N.; Saidani, M.; Stanchina-Chatrousse, L.; Masson, Y.; Patwardhan, A.; Gilles-Marsens, F.; Delevoye, C.; Domingues, S.; Nissan, X.; et al. In Vitro Modeling of Hyperpigmentation Associated to Neurofibromatosis Type 1 Using Melanocytes Derived from Human Embryonic Stem Cells. Proc. Natl. Acad. Sci. USA 2015, 112, 9034–9039. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, D.H.; Ferner, R.E.; Listernick, R.H.; Korf, B.R.; Wolters, P.L.; Johnson, K.J. Neurofibromatosis Type 1. Nat. Rev. Dis. Primers 2017, 3, 17004. [Google Scholar] [CrossRef]
- Huson, S.M.; Compston, D.A.; Harper, P.S. A Genetic Study of von Recklinghausen Neurofibromatosis in South East Wales. II. Guidelines for Genetic Counselling. J. Med. Genet. 1989, 26, 712–721. [Google Scholar] [CrossRef]
- Griffith, J.L.; Morris, S.M.; Mahdi, J.; Goyal, M.S.; Hershey, T.; Gutmann, D.H. Increased Prevalence of Brain Tumors Classified as T2 Hyperintensities in Neurofibromatosis 1. Neurol. Clin. Pract. 2018, 8, 283–291. [Google Scholar] [CrossRef]
- Baudou, E.; Nemmi, F.; Biotteau, M.; Maziero, S.; Assaiante, C.; Cignetti, F.; Vaugoyeau, M.; Audic, F.; Peran, P.; Chaix, Y. Are Morphological and Structural MRI Characteristics Related to Specific Cognitive Impairments in Neurofibromatosis Type 1 (NF1) Children? Eur. J. Paediatr. Neurol. 2020, 28, 89–100. [Google Scholar] [CrossRef]
- Jacques, C.; Dietemann, J.L. [Imaging features of neurofibromatosis type 1]. J. Neuroradiol. 2005, 32, 180–197. [Google Scholar] [CrossRef]
- Shah, K.N. The Diagnostic and Clinical Significance of Café-Au-Lait Macules. Pediatric Clin. N. Am. 2010, 57, 1131–1153. [Google Scholar] [CrossRef]
- Bernier, A.; Larbrisseau, A.; Perreault, S. Café-Au-Lait Macules and Neurofibromatosis Type 1: A Review of the Literature. Pediatric Neurol. 2016, 60, 24–29.e1. [Google Scholar] [CrossRef]
- Cannon, A.; Chen, M.-J.; Li, P.; Boyd, K.P.; Theos, A.; Redden, D.T.; Korf, B. Cutaneous Neurofibromas in Neurofibromatosis Type I: A Quantitative Natural History Study. Orphanet J. Rare Dis. 2018, 13, 31. [Google Scholar] [CrossRef] [PubMed]
- McGaughran, J.M.; Harris, D.I.; Donnai, D.; Teare, D.; MacLeod, R.; Westerbeek, R.; Kingston, H.; Super, M.; Harris, R.; Evans, D.G. A Clinical Study of Type 1 Neurofibromatosis in North West England. J. Med. Genet. 1999, 36, 197–203. [Google Scholar]
- Well, L.; Döbel, K.; Kluwe, L.; Bannas, P.; Farschtschi, S.; Adam, G.; Mautner, V.-F.; Salamon, J. Genotype-Phenotype Correlation in Neurofibromatosis Type-1: NF1 Whole Gene Deletions Lead to High Tumor-Burden and Increased Tumor-Growth. PLoS Genet. 2021, 17, e1009517. [Google Scholar] [CrossRef]
- Lobbous, M.; Bernstock, J.D.; Coffee, E.; Friedman, G.K.; Metrock, L.K.; Chagoya, G.; Elsayed, G.; Nakano, I.; Hackney, J.R.; Korf, B.R.; et al. An Update on Neurofibromatosis Type 1-Associated Gliomas. Cancers 2020, 12, 114. [Google Scholar] [CrossRef] [PubMed]
- Hernáiz Driever, P.; von Hornstein, S.; Pietsch, T.; Kortmann, R.; Warmuth-Metz, M.; Emser, A.; Gnekow, A.K. Natural History and Management of Low-Grade Glioma in NF-1 Children. J. Neurooncol. 2010, 100, 199–207. [Google Scholar] [CrossRef]
- Campen, C.J.; Gutmann, D.H. Optic Pathway Gliomas in Neurofibromatosis Type 1. J. Child Neurol. 2018, 33, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Freret, M.E.; Gutmann, D.H. Insights into Optic Pathway Glioma Vision Loss from Mouse Models of Neurofibromatosis Type 1. J. Neurosci. Res. 2019, 97, 45–56. [Google Scholar] [CrossRef]
- Frayling, I.M.; Mautner, V.-F.; van Minkelen, R.; Kallionpaa, R.A.; Aktaş, S.; Baralle, D.; Ben-Shachar, S.; Callaway, A.; Cox, H.; Eccles, D.M.; et al. Breast Cancer Risk in Neurofibromatosis Type 1 Is a Function of the Type of NF1 Gene Mutation: A New Genotype-Phenotype Correlation. J. Med. Genet. 2019, 56, 209–219. [Google Scholar] [CrossRef]
- Clementi, M.; Milani, S.; Mammi, I.; Boni, S.; Monciotti, C.; Tenconi, R. Neurofibromatosis Type 1 Growth Charts. Am. J. Med. Genet. 1999, 87, 317–323. [Google Scholar] [CrossRef]
- Szudek, J.; Birch, P.; Friedman, J.M. Growth Charts for Young Children with Neurofibromatosis 1 (NF1). Am. J. Med. Genet. 2000, 92, 224–228. [Google Scholar] [CrossRef]
- Sant, D.W.; Margraf, R.L.; Stevenson, D.A.; Grossmann, A.H.; Viskochil, D.H.; Hanson, H.; Everitt, M.D.; Rios, J.J.; Elefteriou, F.; Hennessey, T.; et al. Evaluation of Somatic Mutations in Tibial Pseudarthrosis Samples in Neurofibromatosis Type 1. J. Med. Genet. 2015, 52, 256–261. [Google Scholar] [CrossRef]
- Laycock-van Spyk, S.; Thomas, N.; Cooper, D.N.; Upadhyaya, M. Neurofibromatosis Type 1-Associated Tumours: Their Somatic Mutational Spectrum and Pathogenesis. Hum. Genom. 2011, 5, 623–690. [Google Scholar] [CrossRef]
- Serra, E.; Puig, S.; Otero, D.; Gaona, A.; Kruyer, H.; Ars, E.; Estivill, X.; Lázaro, C. Confirmation of a Double-Hit Model for the NF1 Gene in Benign Neurofibromas. Am. J. Hum. Genet. 1997, 61, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Kluwe, L.; Siebert, R.; Gesk, S.; Friedrich, R.E.; Tinschert, S.; Kehrer-Sawatzki, H.; Mautner, V.-F. Screening 500 Unselected Neurofibromatosis 1 Patients for Deletions of the NF1 Gene. Hum. Mutat. 2004, 23, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Pasmant, E.; Sabbagh, A.; Masliah-Planchon, J.; Haddad, V.; Hamel, M.-J.; Laurendeau, I.; Soulier, J.; Parfait, B.; Wolkenstein, P.; Bièche, I.; et al. Detection and Characterization of NF1 Microdeletions by Custom High Resolution Array CGH. J. Mol. Diagn. 2009, 11, 524–529. [Google Scholar] [CrossRef][Green Version]
- De Raedt, T.; Beert, E.; Pasmant, E.; Luscan, A.; Brems, H.; Ortonne, N.; Helin, K.; Hornick, J.L.; Mautner, V.; Kehrer-Sawatzki, H.; et al. PRC2 Loss Amplifies Ras-Driven Transcription and Confers Sensitivity to BRD4-Based Therapies. Nature 2014, 514, 247–251. [Google Scholar] [CrossRef]
- Oppel, F.; Ki, D.H.; Zimmerman, M.W.; Ross, K.N.; Tao, T.; Shi, H.; He, S.; Aster, J.C.; Look, A.T. Suz12 Inactivation in P53- and Nf1-Deficient Zebrafish Accelerates the Onset of Malignant Peripheral Nerve Sheath Tumors and Expands the Spectrum of Tumor Types. Dis. Models Mech. 2020, 13. [Google Scholar] [CrossRef]
- Sohier, P.; Luscan, A.; Lloyd, A.; Ashelford, K.; Laurendeau, I.; Briand-Suleau, A.; Vidaud, D.; Ortonne, N.; Pasmant, E.; Upadhyaya, M. Confirmation of Mutation Landscape of NF1-Associated Malignant Peripheral Nerve Sheath Tumors. Genes Chromosomes Cancer 2017, 56, 421–426. [Google Scholar] [CrossRef]
- Ferrari, L.; Scuvera, G.; Tucci, A.; Bianchessi, D.; Rusconi, F.; Menni, F.; Battaglioli, E.; Milani, D.; Riva, P. Identification of an Atypical Microdeletion Generating the RNF135-SUZ12 Chimeric Gene and Causing a Position Effect in an NF1 Patient with Overgrowth. Hum. Genet. 2017, 136, 1329–1339. [Google Scholar] [CrossRef]
| Clinical Features | 0–8 Years Old | 9–18 Years Old | >18 Years Old | All Patients n/N * | All Patients % |
|---|---|---|---|---|---|
| Age range | 4 months–8 yo | 9–18 yo | 18–69 yo | 4 months–69 yo | |
| Median age (years) | 5 | 13 | 32 | 21 | |
| Number of individuals (proband:relative) | 31:3 | 22:1 | 69:0 | 122:4 | |
| Male:Female | 16:18 | 12:11 | 23:46 | 51:75 | |
| Fulfilling the NIH criteria if the family history is taken into account | 28/34 | 21/23 | 67/69 | 116/126 | 92% |
| Fulfilling the NIH criteria if solely taking the physical signs into account | 27/34 | 21/23 | 67/69 | 115/126 | 91% |
| CALS a | 34/34 | 22/22 | 66/67 | 122/123 | 99% |
| 1–5 | 0/34 | 1/22 | 6/67 | 7/123 | |
| 6–100 | 27/34 | 14/22 | 42/67 | 83/123 | |
| >100 | 0/34 | 0/22 | 1/67 | 1/123 | |
| Not quantified | 7/34 | 7/22 | 17/67 | 31/123 | |
| Freckling | 23/31 | 20/20 | 55/61 | 98/112 | 88% |
| Blue–red macules | 1/22 | 0/12 | 14/40 | 15/74 | 20% |
| Lisch nodules | 3/18 | 6/15 | 31/44 | 40/77 | 52% |
| Unilateral | 0/18 | 1/15 | 2/44 | 3/77 | |
| Bilateral | 2/18 | 3/15 | 6/44 | 11/77 | |
| NS | 1/18 | 2/15 | 23/44 | 26/77 | |
| Cutaneous neurofibromas b | 2/31 | 10/19 | 59/64 | 71/114 | 62% |
| 1 | 0/31 | 0/19 | 0/64 | 0/114 | |
| 2–9 | 2/31 | 5/19 | 8/64 | 15/114 | |
| 10–99 | 0/31 | 2/19 | 20/64 | 22/114 | |
| >100 | 0/31 | 0/19 | 15/64 | 15/114 | |
| Not quantified | 0/31 | 3/19 | 16/64 | 19/114 | |
| Subcutaneous neurofibromas b | 3/27 | 11/14 | 39/53 | 53/94 | 56% |
| 1 | 2/27 | 1/14 | 5/53 | 8/94 | |
| 2–9 | 1/27 | 8/14 | 20/53 | 29/94 | |
| 10–99 | 0/27 | 2/14 | 10/53 | 12/94 | |
| >100 | 0/27 | 0/14 | 0/53 | 0/94 | |
| Not quantified | 0/27 | 0/14 | 4/53 | 4/94 | |
| Deep neurofibromas | 2/13 | 3/7 | 15/28 | 20/48 | 42% |
| Plexiform neurofibromas | 0/14 | 3/12 | 14/49 | 17/75 | 23% |
| Spinal neurofibromas | 0/5 | 3/5 | 10/25 | 13/35 | 37% |
| Symptomatic | 0/5 | 1/5 | 5/25 | 6/35 | |
| Asymptomatic | 0/5 | 1/5 | 4/25 | 5/35 | |
| NS | 0/5 | 1/5 | 1/25 | 2/35 | |
| OPGs c | 3/17 | 4/16 | 2/47 | 9/80 | 11% |
| Symptomatic OPGs | 0/17 | 2/16 | 2/47 | 4/80 | |
| Asymptomatic OPGs | 3/17 | 2/16 | 0/47 | 5/80 | |
| Malignancies d | 0/19 | 1/14 | 9/51 | 10/84 | 12% |
| MPNSTs | 0/19 | 1/14 | 3/51 | 4/84 | |
| Musculoskeletal abnormalities e | 13/26 | 14/22 | 25/59 | 52/107 | 49% |
| Scoliosis | 2/26 | 6/22 | 17/59 | 25/107 | |
| Hyperflexibilty of joints | 6/26 | 3/22 | 1/34 | 10/82 | |
| Pectus abnormalities | 5/26 | 2/22 | 2/34 | 9/82 | |
| Dysmorphism f | 14/24 | 7/14 | 8/41 | 29/79 | 37% |
| Short stature (<2 sd) | 1/22 | 2/14 | 0/41 | 3/77 | 4% |
| Tall stature (>2 sd) | 0/22 | 1/14 | 0/41 | 1/77 | 1% |
| Macrocephaly (>2 sd) | 2/17 | 0/8 | 11/33 | 13/58 | 22% |
| Overweight (BMI > 30) | 0/20 | 0/12 | 7/38 | 7/70 | 10% |
| Neurological abnormalities g | 17/23 | 14/14 | 29/41 | 60/78 | 77% |
| UBOs | 14/23 | 10/14 | 11/25 | 35/62 | |
| Cognitive impairment and/or learning disabilities | 22/28 | 19/21 | 33/53 | 74/102 | 73% |
| Cardiovascular abnormalities h | 6/16 | 1/11 | 8/37 | 15/64 | 23% |
| Pulmonic stenosis | 1/16 | 1/11 | 0/12 | 2/39 |
| Clinical Features | French Deleted Cohort a | “Classic” NF1 Phenotype a,b | p Value | Adjusted p Value |
|---|---|---|---|---|
| >5 CALS | 84/92 c (91%) | 1537/1728 (89%) | 0.61 | 0.64 |
| Skinfold freckling | 98/112 (88%) | 1403/1667 (84%) | 0.42 | 0.50 |
| Lisch nodules | 40/77 (52%) | 729/1237 (59%) | 0.24 | 0.33 |
| Cutaneous neurofibromas d | 59/64 (92%) | 656/723 (91%) | 0.82 | 0.82 |
| Subcutaneous neurofibromas d | 39/53 (74%) | 297/515 (58%) | 0.028 ↗ | 0.062 |
| Plexiform neurofibromas e,f | 17/61 (28%) | 120/648 (18%) | 0.089 | 0.13 |
| Symptomatic spinal neurofibromas | 6/35 (17%) | 36/2058 (2%) | 4.8 × 10−5 ↗ | 1.7 × 10−4 ↗ |
| Symptomatic OPGs | 4/80 (5%) | 64/1650 (4%) | 0.55 | 0.62 |
| Asymptomatic OPGs | 5/80 (6%) | 70/519 (13%) | 0.07 | 0.13 |
| Malignancies g | 10/84 (12%) | 18/523 (3%) | 0.0024 ↗ | 0.0072 ↗ |
| Skeletal abnormalities h | 42/107 (39%) | 144/948 (15%) | 2.3 × 10−8 ↗ | 2.1 × 10−7 ↗ |
| Scoliosis d | 17/59 (29%) | 51/236 (22%) | 0.30 | 0.38 |
| Dysmorphism | 29/79 (37%) | 42/389 (11%) | 1.1 × 10−7 ↗ | 6.3 × 10−7 ↗ |
| Pulmonic stenosis | 2/39 (5%) | 25/2322 (1%) | 0.072 | 0.13 |
| Short stature | 3/77 (4%) | 109/684 (16%) | 0.0033 ↘ | 0.0086 ↘ |
| Macrocephaly | 13/58 (22%) | 239/704 (34%) | 0.082 | 0.13 |
| Cognitive impairment and/or learning disabilities | 74/102 (73%) | 190/424 (45%) | 5.0 × 10−7 ↗ | 2.2 × 10−6 ↗ |
| Cardiovascular abnormalities | 15/64 (23%) | 54/2322 (2%) | 9.2 × 10−11 ↗ | 1.7 × 10−9 ↗ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pacot, L.; Vidaud, D.; Sabbagh, A.; Laurendeau, I.; Briand-Suleau, A.; Coustier, A.; Maillard, T.; Barbance, C.; Morice-Picard, F.; Sigaudy, S.; et al. Severe Phenotype in Patients with Large Deletions of NF1. Cancers 2021, 13, 2963. https://doi.org/10.3390/cancers13122963
Pacot L, Vidaud D, Sabbagh A, Laurendeau I, Briand-Suleau A, Coustier A, Maillard T, Barbance C, Morice-Picard F, Sigaudy S, et al. Severe Phenotype in Patients with Large Deletions of NF1. Cancers. 2021; 13(12):2963. https://doi.org/10.3390/cancers13122963
Chicago/Turabian StylePacot, Laurence, Dominique Vidaud, Audrey Sabbagh, Ingrid Laurendeau, Audrey Briand-Suleau, Audrey Coustier, Théodora Maillard, Cécile Barbance, Fanny Morice-Picard, Sabine Sigaudy, and et al. 2021. "Severe Phenotype in Patients with Large Deletions of NF1" Cancers 13, no. 12: 2963. https://doi.org/10.3390/cancers13122963
APA StylePacot, L., Vidaud, D., Sabbagh, A., Laurendeau, I., Briand-Suleau, A., Coustier, A., Maillard, T., Barbance, C., Morice-Picard, F., Sigaudy, S., Glazunova, O. O., Damaj, L., Layet, V., Quelin, C., Gilbert-Dussardier, B., Audic, F., Dollfus, H., Guerrot, A.-M., Lespinasse, J., ... Pasmant, E. (2021). Severe Phenotype in Patients with Large Deletions of NF1. Cancers, 13(12), 2963. https://doi.org/10.3390/cancers13122963






