MITF Promotes Cell Growth, Migration and Invasion in Clear Cell Renal Cell Carcinoma by Activating the RhoA/YAP Signal Pathway
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. Reagents and Antibodies
2.3. MITF Knockdown in RCC Cell Lines
2.4. Overexpression of MITF in RCC Cell Lines
2.5. Transient Transfection
2.6. In Vitro Cell Proliferation Assay
2.7. Anchorage-Independent Growth (Anoikis) Assay
2.8. Clonogenic Assay
2.9. Migration and Invasion Assays
2.10. Western Blot Analysis
2.11. Wound Healing Assay
2.12. Immunofluorescence
2.13. Cell Cycle Analysis
2.14. Active RhoA/Rac1/Cdc42 Pulldown Assay
2.15. Xenograft Tumors in Nude Mice
2.16. Statistical Analysis
3. Results
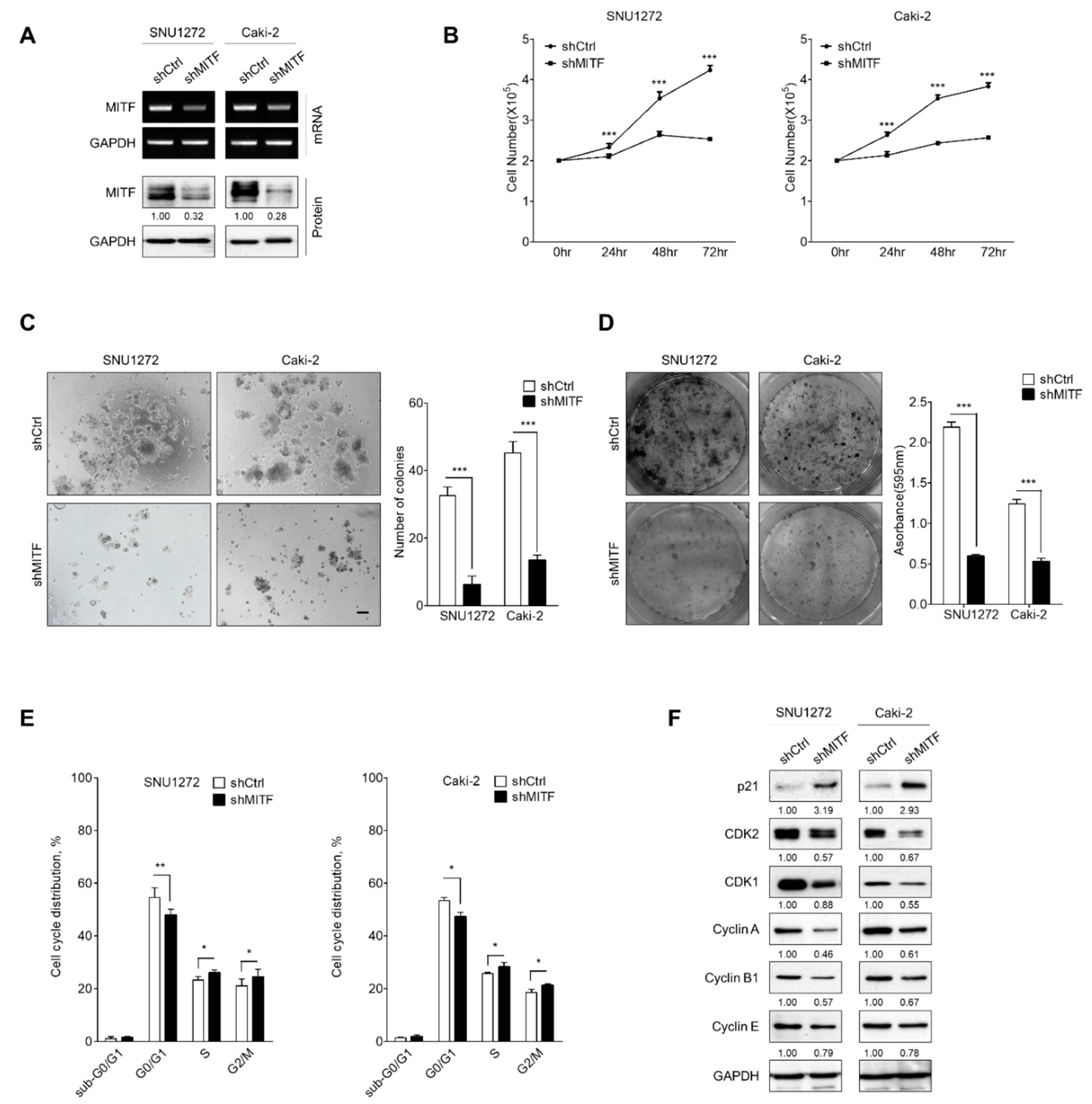
3.1. Knockdown of MITF Inhibits Cell Growth by Cell Cycle Shift
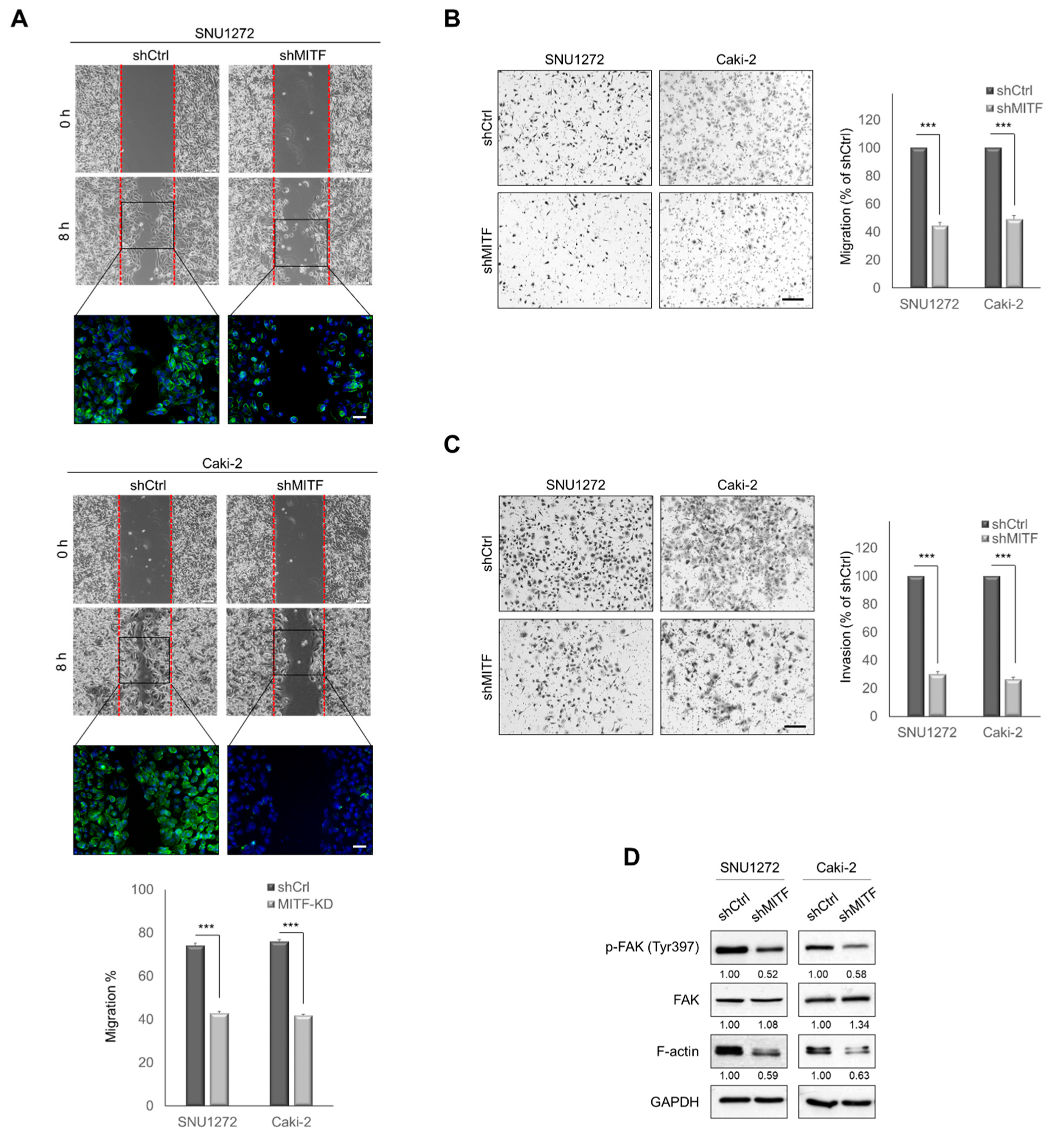
3.2. Knockdown of MITF Suppresses Cell Migration and Invasion In Vitro
3.3. Knockdown of MITF Inhibits YAP Nuclear Translocation
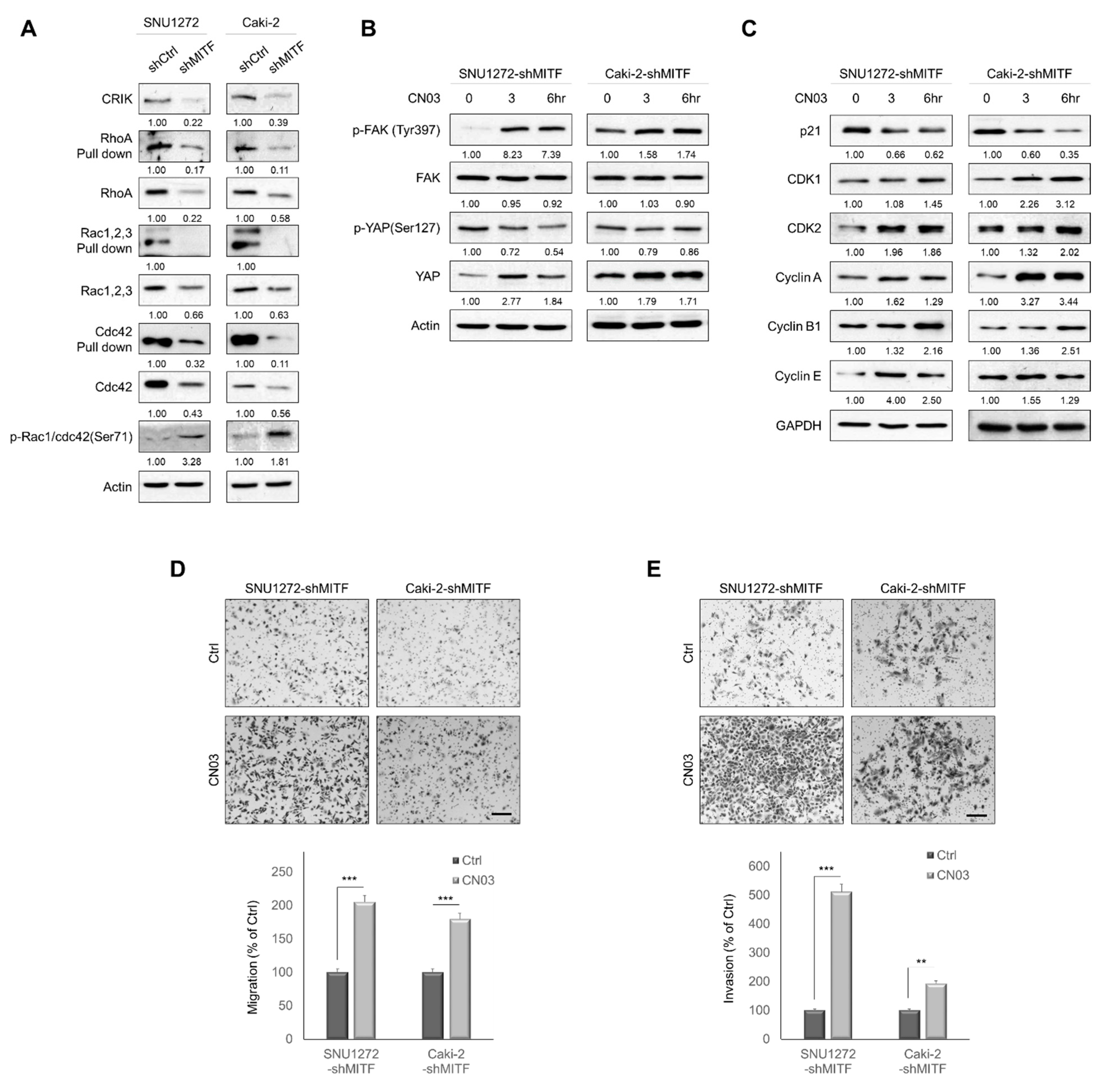
3.4. Knockdown of MITF Suppresses RhoA Activation
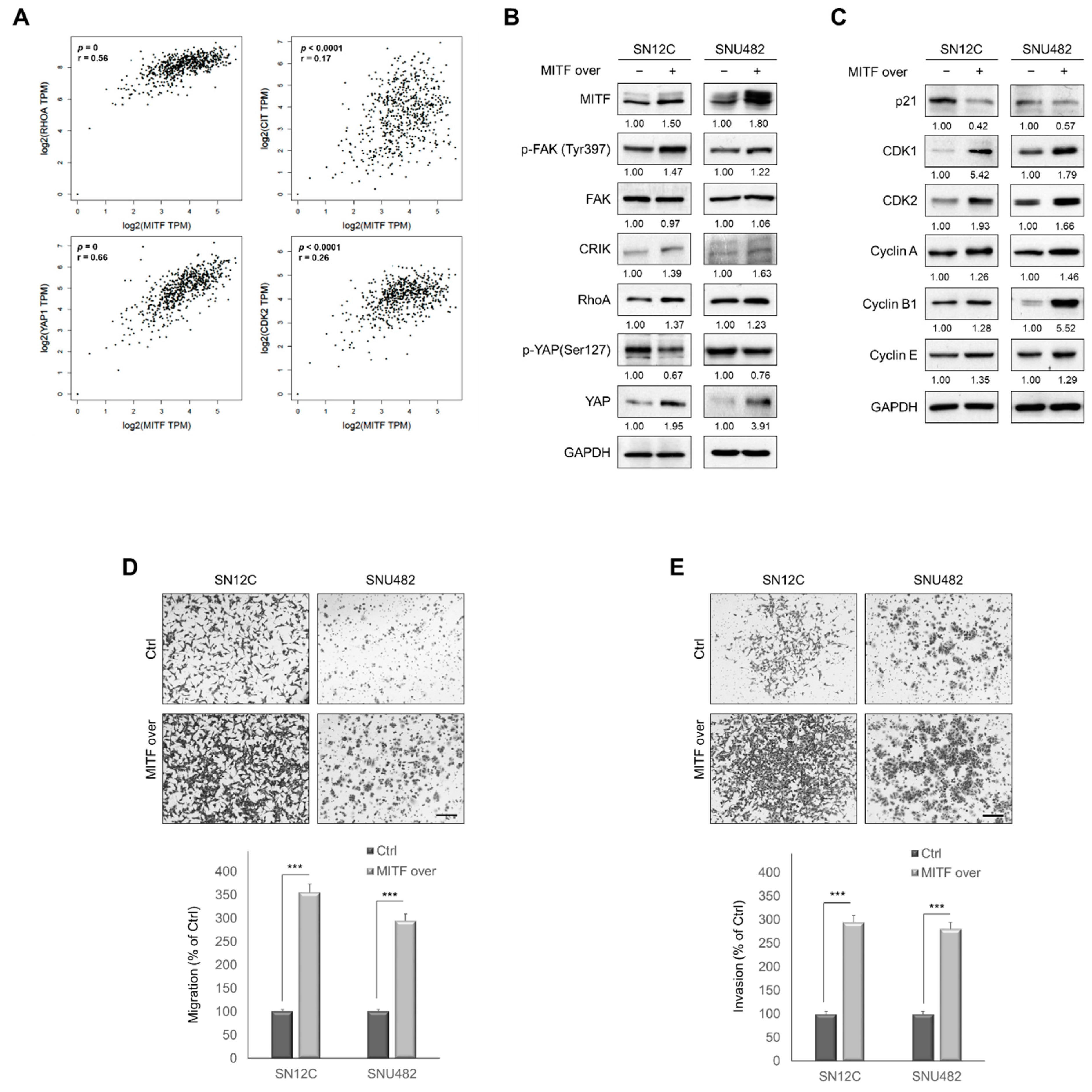
3.5. MITF Overexpression Promoted Cell Migration and Invasion by Upregulating RhoA/YAP Signaling
3.6. MITF Affects Tumorigenesis in ccRCC Cells In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kashyap, M.; Kumar, A.; Emelianenko, N.; Kashyap, A.; Kaushik, R.; Huang, R.; Khullar, M.; Sharma, S.K.; Singh, S.K.; Bhargave, A.K.; et al. Biochemical and molecular markers in renal cell carcinoma: An update and future prospects. Biomarkers 2005, 10, 258–294. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA A Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef]
- De Meerleer, G.; Khoo, V.; Escudier, B.; Joniau, S.; Bossi, A.; Ost, P.; Briganti, A.; Fonteyne, V.; Van Vulpen, M.; Lumen, N.; et al. Radiotherapy for renal-cell carcinoma. Lancet Oncol. 2014, 15, e170–e177. [Google Scholar] [CrossRef]
- Tabibu, S.; Vinod, P.K.; Jawahar, C.V. Pan-Renal Cell Carcinoma classification and survival prediction from histopathology images using deep learning. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Motzer, R.J.; Bukowski, R.M. Targeted Therapy for Metastatic Renal Cell Carcinoma. J. Clin. Oncol. 2006, 24, 5601–5608. [Google Scholar] [CrossRef] [PubMed]
- Muramatsu, T.; Imoto, I.; Matsui, T.; Kozaki, K.-I.; Haruki, S.; Sudol, M.; Shimada, Y.; Tsuda, H.; Kawano, T.; Inazawa, J. YAP is a candidate oncogene for esophageal squamous cell carcinoma. Carcinogenesis 2010, 32, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Fang, R.; Liu, B.; Shi, H.; Wang, Y.; Zhang, W.; Zhang, X.; Ye, L. Deacetylation of tumor-suppressor MST1 in Hippo pathway induces its degradation through HBXIP-elevated HDAC6 in promotion of breast cancer growth. Oncogene 2016, 35, 4048–4057. [Google Scholar] [CrossRef]
- Kim, W.; Khan, S.K.; Liu, Y.; Xu, R.; Park, O.; He, Y.; Cha, B.; Gao, B.; Yang, Y. Hepatic Hippo signaling inhibits protumoural microenvironment to suppress hepatocellular carcinoma. Gut 2017, 67, 1692–1703. [Google Scholar] [CrossRef]
- Morgan, E.L.; Patterson, M.R.; Ryder, E.L.; Lee, S.Y.; Wasson, C.W.; Harper, K.L.; Li, Y.; Griffin, S.; Blair, G.E.; Whitehouse, A.; et al. MicroRNA-18a targeting of the STK4/MST1 tumour suppressor is necessary for transformation in HPV positive cervical cancer. PLoS Pathog. 2020, 16, e1008624. [Google Scholar] [CrossRef]
- Zhao, B.; Lei, Q.-Y.; Guan, K.-L. The Hippo–YAP pathway: New connections between regulation of organ size and cancer. Curr. Opin. Cell Biol. 2008, 20, 638–646. [Google Scholar] [CrossRef]
- Seo, J.; Kim, M.H.; Hong, H.; Cho, H.; Park, S.; Kim, S.K.; Kim, J. MK5 Regulates YAP Stability and Is a Molecular Target in YAP-Driven Cancers. Cancer Res. 2019, 79, 6139–6152. [Google Scholar] [CrossRef]
- Yao, F.; Xiao, Z.; Sun, Y.; Ma, L. SKP2 and OTUD1 govern non-proteolytic ubiquitination of YAP that promotes YAP nuclear localization and activity. Cell Stress 2018, 2, 233–235. [Google Scholar] [CrossRef]
- Harvey, K.F.; Zhang, X.; Thomas, D. The Hippo pathway and human cancer. Nat. Rev. Cancer 2013, 13, 246–257. [Google Scholar] [CrossRef]
- Yoo, W.; Lee, J.; Jun, E.; Noh, K.H.; Lee, S.; Jung, D.; Jung, K.H.; Kim, J.S.; Park, Y.Y.; Kim, S.C.; et al. The YAP1–NMU Axis Is Associated with Pancreatic Cancer Progression and Poor Outcome: Identification of a Novel Diagnostic Biomarker and Therapeutic Target. Cancers 2019, 11, 1477. [Google Scholar] [CrossRef] [PubMed]
- Ehmer, U.; Sage, J. Control of Proliferation and Cancer Growth by the Hippo Signaling Pathway. Mol. Cancer Res. 2016, 14, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Yu, J.; Han, W.; Fan, X.; Qian, H.; Wei, H.; Tsai, Y.-H.S.; Zhao, J.; Zhang, W.; Liu, Q.; et al. A splicing isoform of TEAD4 attenuates the Hippo–YAP signalling to inhibit tumour proliferation. Nat. Commun. 2016, 7, 11840. [Google Scholar] [CrossRef]
- Hodgkinson, C.A.; Moore, K.J.; Nakayama, A.; Steingrímsson, E.; Copeland, N.G.; Jenkins, N.A.; Arnheiter, H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 1993, 74, 395–404. [Google Scholar] [CrossRef]
- Hughes, M.; Lingrel, J.; Krakowsky, J.; Anderson, K. A helix-loop-helix transcription factor-like gene is located at the mi locus. J. Biol. Chem. 1993, 268, 20687–20690. [Google Scholar] [CrossRef]
- Kawakami, A.; Fisher, D.E. The master role of microphthalmia-associated transcription factor in melanocyte and melanoma biology. Lab. Investig. 2017, 97, 649–656. [Google Scholar] [CrossRef]
- Nooron, N.; Ohba, K.; Takeda, K.; Shibahara, S.; Chiabchalard, A. Dysregulated Expression of MITF in Subsets of Hepatocellular Carcinoma and Cholangiocarcinoma. Tohoku J. Exp. Med. 2017, 242, 291–302. [Google Scholar] [CrossRef]
- Leachman, S.A.; Lucero, O.M.; Sampson, J.E.; Cassidy, P.; Bruno, W.; Queirolo, P.; Ghiorzo, P. Identification, genetic testing, and management of hereditary melanoma. Cancer Metastasis Rev. 2017, 36, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Qin, X.; Wang, B.; Xu, G.; Zhang, J.; Jiang, X.; Chen, C.; Qiu, F.; Zou, Z. MiTF is Associated with Chemoresistance to Cisplatin in A549 Lung Cancer Cells via Modulating Lysosomal Biogenesis and Autophagy. Cancer Manag. Res. 2020, ume 12, 6563–6573. [Google Scholar] [CrossRef]
- Lang, M.; Vocke, C.D.; Ricketts, C.J.; Metwalli, A.R.; Ball, M.W.; Schmidt, L.S.; Linehan, W.M. Clinical and Molecular Characterization of Microphthalmia-associated Transcription Factor (MITF)-related Renal Cell Carcinoma. Urology 2021, 149, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Ryu, H.; Kim, S.; Joo, M.; Jeon, H.J.; Lee, M.W.; Song, I.C.; Kim, M.N.; Kim, J.M.; Lee, H.J. CXCR7 promotes migration and invasion in head and neck squamous cell carcinoma by upregulating TGF-beta 1/Smad2/3 signaling. Sci. Rep. 2019, 9, 1. [Google Scholar]
- Yu, F.-X.; Zhao, B.; Panupinthu, N.; Jewell, J.L.; Lian, I.; Wang, L.H.; Zhao, J.; Yuan, H.; Tumaneng, K.; Li, H.; et al. Regulation of the Hippo-YAP Pathway by G-Protein-Coupled Receptor Signaling. Cell 2012, 150, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhu, X.; Xu, W.; Zhang, Y.; Chen, L.; Qiu, F.; Zhang, B.; Wu, L.; Peng, Z.; Tang, H. Up-regulation of CIT promotes the growth of colon cancer cells. Oncotarget 2017, 8, 71954–71964. [Google Scholar] [CrossRef]
- Huang, Q.B.; Ma, X.; Zhang, X.; Liu, S.W.; Ai, Q.; Shi, T.P.; Zhang, Y.; Gao, Y.; Fan, Y.; Ni, N.; et al. Down-Regulated miR-30a in Clear Cell Renal Cell Carcinoma Correlated with Tumor Hematogenous Metastasis by Targeting Angiogenesis-Specific DLL4. PLoS ONE 2013, 8, e67294. [Google Scholar] [CrossRef]
- Wei, R.; Ye, X.; Zhao, Y.; Jia, N.; Liu, T.; Lian, W.; Wei, H.; Zhang, G.; Song, L. MicroRNA-218 inhibits the cell proliferation and migration in clear cell renal cell carcinoma through targeting cancerous inhibitor of protein phosphatase 2A. Oncol. Lett. 2019, 17, 3211–3218. [Google Scholar] [CrossRef]
- Hintzy, M.C.; Camparo, P.; Vasiliu, V.; Peyromaure, M.; Vieillefond, A. Renal carcinoma associated with MiTF/TFE translocation: Report of six cases in young adults. Prog. Urol. 2008, 18, 275–280. [Google Scholar] [CrossRef]
- Fall, B.; Sow, Y.; Sarr, A.; Diao, B. Renal cell carcinoma with MiTF/TFE3 translocation in children: Report of a case at the stage of lymph node involvement. Afr. J. Paediatr. Surg. 2011, 8, 317. [Google Scholar] [CrossRef]
- Bertolotto, C.; The French Familial Melanoma Study Group; Lesueur, F.; Giuliano, S.; Strub, T.; De Lichy, M.; Bille, K.; Dessen, P.; D’Hayer, B.; Mohamdi, H.; et al. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nat. Cell Biol. 2011, 480, 94–98. [Google Scholar] [CrossRef]
- Levy, C.; Khaled, M.; Fisher, D.E. MITF: Master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 2006, 12, 406–414. [Google Scholar] [CrossRef]
- Perera, R.M.; Stoykova, S.; Nicolay, B.N.; Ross, K.N.; Fitamant, J.; Boukhali, M.; Lengrand, J.; Deshpande, V.; Selig, M.K.; Ferrone, C.R.; et al. Transcriptional control of autophagy–lysosome function drives pancreatic cancer metabolism. Nat. Cell Biol. 2015, 524, 361–365. [Google Scholar] [CrossRef]
- Carreira, S.; Goodall, J.; Denat, L.; Rodriguez-Teja, M.; Nuciforo, P.; Hoek, K.S.; Testori, A.; LaRue, L.; Goding, C.R. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 2006, 20, 3426–3439. [Google Scholar] [CrossRef]
- Hoek, K.S.; Schlegel, N.C.; Brafford, P.; Sucker, A.; Ugurel, S.; Kumar, R.; Weber, B.L.; Nathanson, K.L.; Phillips, D.J.; Herlyn, M.; et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006, 19, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Widlund, H.; Horstmann, M.A.; Ramaswamy, S.; Ross, K.; Huber, W.E.; Nishimura, E.K.; Golub, T.R.; Fisher, D.E. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell 2004, 6, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Carreira, S.; Goodall, J.; Aksan, I.; La Rocca, S.A.; Galibert, M.-D.; Denat, L.; Larue, L.; Goding, C.R. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nat. Cell Biol. 2005, 433, 764–769. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; AOCS Study Group; George, J.M.; Deb, S.; Degoutin, J.L.; Takano, E.A.; Fox, S.B.; Bowtell, D.D.L.; Harvey, K.F. The Hippo pathway transcriptional co-activator, YAP, is an ovarian cancer oncogene. Oncogene 2011, 30, 2810–2822. [Google Scholar] [CrossRef] [PubMed]
- Schütte, U.; Bisht, S.; Heukamp, L.C.; Kebschull, M.; Florin, A.; Haarmann, J.; Hoffmann, P.; Bendas, G.; Buettner, R.; Brossart, P.; et al. Hippo Signaling Mediates Proliferation, Invasiveness, and Metastatic Potential of Clear Cell Renal Cell Carcinoma. Transl. Oncol. 2014, 7, 309–321. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.W.; Lim, C.J.; Chen, L.; Chong, Y.F.; Huang, C.; Song, H.; Hong, W. The hippo pathway in biological control and cancer development. J. Cell. Physiol. 2011, 226, 928–939. [Google Scholar] [CrossRef]
- Li, H.; Wolfe, A.; Septer, S.; Edwards, G.; Zhong, X.; Abdulkarim, A.B.; Ranganathan, S.; Apte, U. Deregulation of Hippo kinase signalling in Human hepatic malignancies. Liver Int. 2011, 32, 38–47. [Google Scholar] [CrossRef]
- Steinhardt, A.A.; Gayyed, M.; Klein, A.P.; Dong, J.; Maitra, A.; Pan, D.; Montgomery, E.A.; Anders, R.A. Expression of Yes-associated protein in common solid tumors. Hum. Pathol. 2008, 39, 1582–1589. [Google Scholar] [CrossRef]
- Zanconato, F.; Forcato, M.; Battilana, G.; Azzolin, L.; Quaranta, E.; Bodega, B.; Rosato, A.; Bicciato, S.; Cordenonsi, M.; Piccolo, S. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 2015, 17, 1218–1227. [Google Scholar] [CrossRef]
- Wang, Z.; Kong, Q.; Su, P.; Duan, M.; Xue, M.; Li, X.; Tang, J.; Gao, Z.; Wang, B.; Li, Z.; et al. Regulation of Hippo signaling and triple negative breast cancer progression by an ubiquitin ligase RNF187. Oncogenesis 2020, 9, 1–11. [Google Scholar] [CrossRef]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in mechanotransduction. Nat. Cell Biol. 2011, 474, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Piccolo, S.; Dupont, S.; Cordenonsi, M. The Biology of YAP/TAZ: Hippo Signaling and Beyond. Physiol. Rev. 2014, 94, 1287–1312. [Google Scholar] [CrossRef] [PubMed]
- Etienne-Manneville, S.; Hall, A. Rho GTPases in cell biology. Nature 2002, 420, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zheng, Y. Cell type-specific functions of Rho GTPases revealed by gene targeting in mice. Trends Cell Biol. 2007, 17, 58–64. [Google Scholar] [CrossRef]
- Ridley, A.J. Rho GTPases and cell migration. J. Cell Sci. 2001, 114, 2713–2722. [Google Scholar] [CrossRef]
- Turcotte, S.; Desrosiers, R.R.; Brand, G.; Béliveau, R. von Hippel-Lindau tumor suppressor protein stimulation by thrombin involves RhoA activation. Int. J. Cancer 2004, 112, 777–786. [Google Scholar] [CrossRef]
- Bassi, Z.I.; Audusseau, M.; Riparbelli, M.G.; Callaini, G.; D’Avino, P.P. Citron kinase controls a molecular network required for midbody formation in cytokinesis. Proc. Natl. Acad. Sci. USA 2013, 110, 9782–9787. [Google Scholar] [CrossRef] [PubMed]
- Bassi, Z.I.; Verbrugghe, K.J.; Capalbo, L.; Gregory, S.; Montembault, E.; Glover, D.; D’Avino, P.P. Sticky/Citron kinase maintains proper RhoA localization at the cleavage site during cytokinesis. J. Cell Biol. 2011, 195, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Meng, D.; Yu, Q.; Feng, L.; Luo, M.; Shao, S.; Huang, S.; Wang, G.; Jing, X.; Tong, Z.; Zhao, X.; et al. Citron kinase (CIT-K) promotes aggressiveness and tumorigenesis of breast cancer cells in vitro and in vivo: Preliminary study of the underlying mechanism. Clin. Transl. Oncol. 2018, 21, 910–923. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, N.; Kim, S.; Lee, M.-W.; Jeon, H.-J.; Ryu, H.; Kim, J.-M.; Lee, H.-J. MITF Promotes Cell Growth, Migration and Invasion in Clear Cell Renal Cell Carcinoma by Activating the RhoA/YAP Signal Pathway. Cancers 2021, 13, 2920. https://doi.org/10.3390/cancers13122920
Kim N, Kim S, Lee M-W, Jeon H-J, Ryu H, Kim J-M, Lee H-J. MITF Promotes Cell Growth, Migration and Invasion in Clear Cell Renal Cell Carcinoma by Activating the RhoA/YAP Signal Pathway. Cancers. 2021; 13(12):2920. https://doi.org/10.3390/cancers13122920
Chicago/Turabian StyleKim, Nayoung, Solbi Kim, Myung-Won Lee, Heung-Jin Jeon, Hyewon Ryu, Jin-Man Kim, and Hyo-Jin Lee. 2021. "MITF Promotes Cell Growth, Migration and Invasion in Clear Cell Renal Cell Carcinoma by Activating the RhoA/YAP Signal Pathway" Cancers 13, no. 12: 2920. https://doi.org/10.3390/cancers13122920
APA StyleKim, N., Kim, S., Lee, M.-W., Jeon, H.-J., Ryu, H., Kim, J.-M., & Lee, H.-J. (2021). MITF Promotes Cell Growth, Migration and Invasion in Clear Cell Renal Cell Carcinoma by Activating the RhoA/YAP Signal Pathway. Cancers, 13(12), 2920. https://doi.org/10.3390/cancers13122920





