Exosomal microRNA in Pancreatic Cancer Diagnosis, Prognosis, and Treatment: From Bench to Bedside
Abstract
Simple Summary
Abstract
1. Introduction
2. Exosomes
2.1. Structural Composition of Exosome
2.2. Isolation and Identification of Exosomes
2.3. Role of Exosomes
3. Exosomal miRNAs and Pancreatic Cancer
4. Exosomal miRNAs as Diagnostic Biomarkers for PDAC
4.1. Exosomal miRNA in Blood Samples
4.2. Exosomal miRNA in Other Body Fluids
5. Exosomal miRNAs as a Prognostic Biomarker in PDAC
6. Exosomal miRNAs in the Treatment of PDAC
6.1. Exosomes as a Drug Delivery System
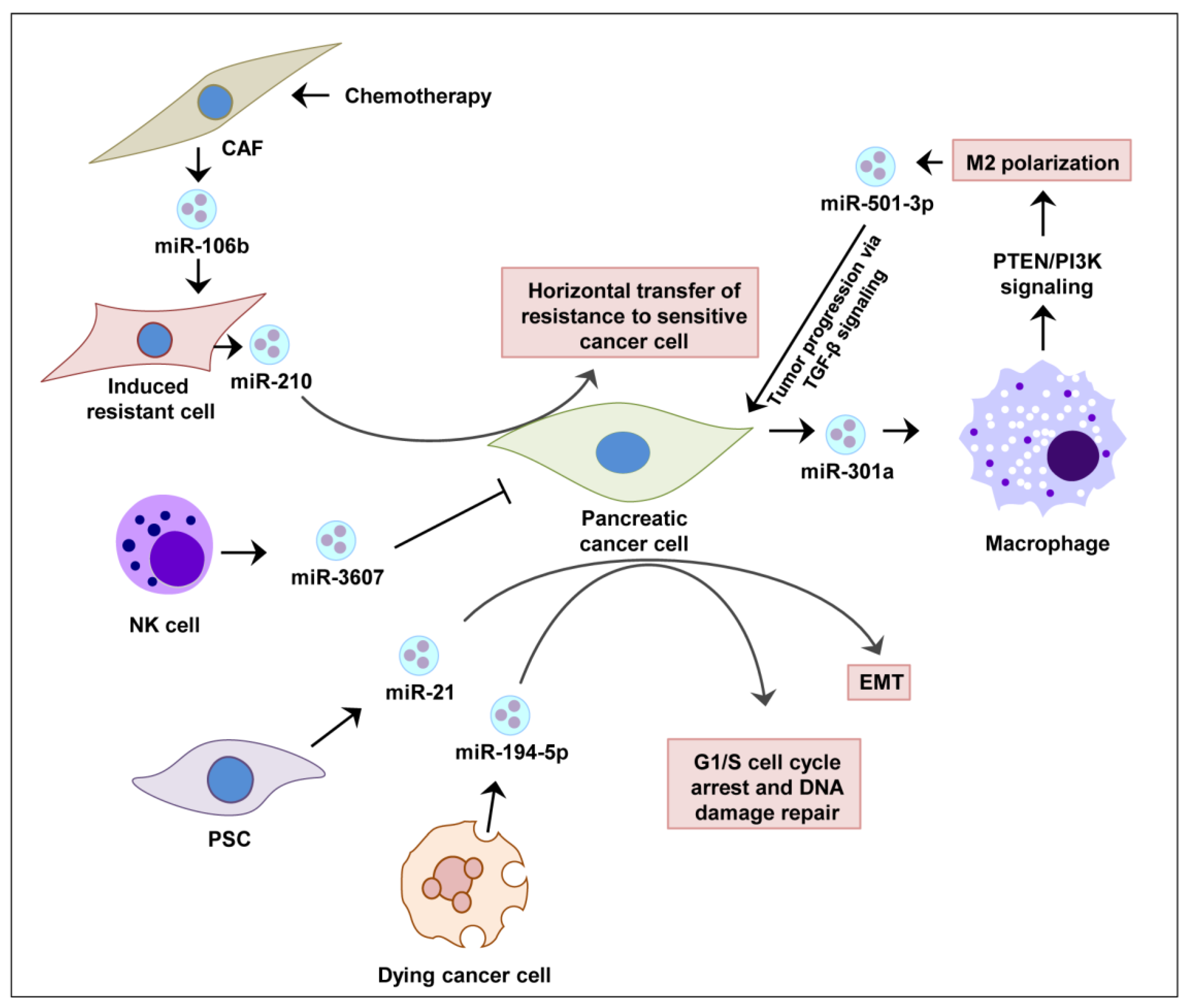
6.2. Exosomal miRNA Therapy in Pancreatic Cancer Progression and Therapy Resistance
6.3. Possibility of Exosomal miRNA to Target Key Mutations in Pancreatic Cancer
7. Conclusions and Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BM-MSC | bone marrow mesenchymal stem cell |
| CAF | cancer associated fibroblast |
| NK cell | natural killer cell |
| PCC | pancreatic cancer cells |
| PSC | pancreatic stellate cell |
| UC-MSC | umbilical cord derived mesenchymal stromal cells |
| IPMN | intraductal papillary mucinous neoplasms |
References
- Roder, P.V.; Wu, B.; Liu, Y.; Han, W. Pancreatic regulation of glucose homeostasis. Exp. Mol. Med. 2016, 48, 1–19. [Google Scholar] [CrossRef]
- Lanfredini, S.; Thapa, A.; O’Neill, E. RAS in pancreatic cancer. Biochem. Soc. Trans. 2019, 47, 961–972. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, X.; Williams, A.; Ding, W. The role of exosomal microRNAs in pancreatic cancer. Stem Cell Investig. 2020, 7. [Google Scholar] [CrossRef]
- Pishvaian, M.J.; Brody, J.R. Therapeutic Implications of Molecular Subtyping for Pancreatic Cancer. Oncology 2017, 31, 159–166, 168. [Google Scholar]
- Kleeff, J.; Korc, M.; Apte, M.; la Vecchia, C.; Johnson, C.D.; Biankin, A.V.; Neale, R.E.; Tempero, M.; Tuveson, D.A.; Hruban, R.H.; et al. Pancreatic cancer. Nat. Rev. Dis. Primers 2016, 2, 16022. [Google Scholar] [CrossRef]
- Society, A.C. Cancer Facts & Figures 2021; American Cancer Society: Atlanta, Georgia, 2021. [Google Scholar]
- Gao, X.; Wan, Z.; Wei, M.; Dong, Y.; Zhao, Y.; Chen, X.; Li, Z.; Qin, W.; Yang, G.; Liu, L. Chronic myelogenous leukemia cells remodel the bone marrow niche via exosome-mediated transfer of miR-320. Theranostics 2019, 9, 5642–5656. [Google Scholar] [CrossRef] [PubMed]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, D.; Arima, K.; Yokoyama, N.; Chikamoto, A.; Taki, K.; Inoue, R.; Kaida, T.; Higashi, T.; Nitta, H.; Ohmuraya, M.; et al. Heterogeneity of KRAS Mutations in Pancreatic Ductal Adenocarcinoma. Pancreas 2016, 45, 1111–1114. [Google Scholar] [CrossRef] [PubMed]
- Luo, J. KRAS mutation in pancreatic cancer. Semin. Oncol. 2021. [Google Scholar] [CrossRef]
- Singh, H.; Longo, D.L.; Chabner, B.A. Improving Prospects for Targeting RAS. J. Clin. Oncol. 2015, 33, 3650–3659. [Google Scholar] [CrossRef]
- Hobbs, G.A.; Baker, N.M.; Miermont, A.M.; Thurman, R.D.; Pierobon, M.; Tran, T.H.; Anderson, A.O.; Waters, A.M.; Diehl, J.N.; Papke, B.; et al. Atypical KRAS(G12R) Mutant Is Impaired in PI3K Signaling and Macropinocytosis in Pancreatic Cancer. Cancer Discov. 2020, 10, 104–123. [Google Scholar] [CrossRef]
- Waddell, N.; Pajic, M.; Patch, A.M.; Chang, D.K.; Kassahn, K.S.; Bailey, P.; Johns, A.L.; Miller, D.; Nones, K.; Quek, K.; et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015, 518, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Hingorani, S.R.; Wang, L.; Multani, A.S.; Combs, C.; Deramaudt, T.B.; Hruban, R.H.; Rustgi, A.K.; Chang, S.; Tuveson, D.A. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005, 7, 469–483. [Google Scholar] [CrossRef]
- Martinez-Useros, J.; Garcia-Foncillas, J. Can Molecular Biomarkers Change the Paradigm of Pancreatic Cancer Prognosis? BioMed Res. Int. 2016, 2016, 4873089. [Google Scholar] [CrossRef] [PubMed]
- Gopinathan, A.; Morton, J.; Jodrell, D.I.; Sansom, O.J. GEMMs as preclinical models for testing pancreatic cancer therapies. Dis. Model. Mech. 2015, 8, 1185–1200. [Google Scholar] [CrossRef] [PubMed]
- Ryan, D.P.; Hong, T.S.; Bardeesy, N. Pancreatic Adenocarcinoma. N. Engl. J. Med. 2014, 371, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.; Herman, J.; Schulick, R.; Hruban, R.H.; Goggins, M. Pancreatic cancer. Lancet 2011, 378, 607–620. [Google Scholar] [CrossRef]
- Bond-Smith, G.; Banga, N.; Hammond, T.M.; Imber, C.J. Pancreatic adenocarcinoma. BMJ 2012, 344, e2476. [Google Scholar] [CrossRef]
- Garrido-Laguna, I.; Hidalgo, M. Pancreatic cancer: From state-of-the-art treatments to promising novel therapies. Nat. Rev. Clin. Oncol. 2015, 12, 319–334. [Google Scholar] [CrossRef] [PubMed]
- Sohn, T.A.; Yeo, C.J.; Cameron, J.L.; Koniaris, L.; Kaushal, S.; Abrams, R.A.; Sauter, P.K.; Coleman, J.; Hruban, R.H.; Lillemoe, K.D. Resected adenocarcinoma of the pancreas-616 patients: Results, outcomes, and prognostic indicators. J. Gastrointest. Surg. 2000, 4, 567–579. [Google Scholar] [CrossRef]
- Winter, J.M.; Cameron, J.L.; Campbell, K.A.; Arnold, M.A.; Chang, D.C.; Coleman, J.; Hodgin, M.B.; Sauter, P.K.; Hruban, R.H.; Riall, T.S. 1423 Pancreaticoduodenectomies for Pancreatic Cancer: A Single-Institution Experience. J. Gastrointest. Surg. 2006, 10, 1199–1211. [Google Scholar] [CrossRef] [PubMed]
- van Roest, M.H.G.; Gouw, A.S.H.; Peeters, P.M.J.G.; Porte, R.J.; Slooff, M.J.H.; Fidler, V.; de Jong, K.P. Results of pancreaticoduodenectomy in patients with periampullary adenocarcinoma: Perineural growth more important prognostic factor than tumor localization. Ann. Surg. 2008, 248, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Kuhlmann, K.; De Castro, S.; Wesseling, J.; Kate, F.T.; Offerhaus, G.; Busch, O.; Van Gulik, T.; Obertop, H.; Gouma, D. Surgical treatment of pancreatic adenocarcinoma: Actual survival and prognostic factors in 343 patients. Eur. J. Cancer 2004, 40, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Verbeke, C.S.; Leitch, D.; Menon, K.V.; McMahon, M.J.; Guillou, P.J.; Anthoney, A. Redefining the R1 resection in pancreatic cancer. Br. J. Surg. 2006, 93, 1232–1237. [Google Scholar] [CrossRef]
- Khalifa, M.A.; Maksymov, V.; Rowsell, C. Retroperitoneal margin of the pancreaticoduodenectomy specimen: Anatomic mapping for the surgical pathologist. Virchows Archiv 2009, 454, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Westgaard, A.; Tafjord, S.; Farstad, I.N.; Cvancarova, M.; Eide, T.J.; Mathisen, O.; Clausen, O.P.F.; Gladhaug, I.P. Resectable adenocarcinomas in the pancreatic head: The retroperitoneal resection margin is an independent prognostic factor. BMC Cancer 2008, 8, 5. [Google Scholar] [CrossRef]
- Arnaoutakis, G.J.; Rangachari, D.; Laheru, D.A.; Iacobuzio-Donahue, C.A.; Hruban, R.H.; Herman, J.M.; Edil, B.H.; Pawlik, T.M.; Schulick, R.D.; Cameron, J.L.; et al. Pulmonary Resection for Isolated Pancreatic Adenocarcinoma Metastasis: An Analysis of Outcomes and Survival. J. Gastrointest Surg. 2011, 15, 1611–1617. [Google Scholar] [CrossRef] [PubMed]
- Ballehaninna, U.K.; Chamberlain, R.S. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: An evidence based appraisal. J. Gastrointest Oncol. 2012, 3, 105–119. [Google Scholar]
- Winter, J.M.; Yeo, C.J.; Brody, J.R. Diagnostic, prognostic, and predictive biomarkers in pancreatic cancer. J. Surg. Oncol. 2013, 107, 15–22. [Google Scholar] [CrossRef]
- Ballehaninna, U.K.; Chamberlain, R.S. Serum CA 19-9 as a Biomarker for Pancreatic Cancer—A Comprehensive Review. Indian J. Surg. Oncol. 2011, 2, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouché, O.; Guimbaud, R.; Bécouarn, Y.; Adenis, A.; Raoul, J.-L.; Gourgou-Bourgade, S.; De La Fouchardière, C.; et al. FOLFIRINOX versus Gemcitabine for Metastatic Pancreatic Cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef] [PubMed]
- Juiz, N.A.; Iovanna, J.; Dusetti, N. Pancreatic Cancer Heterogeneity Can Be Explained Beyond the Genome. Front. Oncol. 2019, 9, 246. [Google Scholar] [CrossRef]
- Seifert, L.; Werba, G.; Tiwari, S.; Ly, N.N.G.; Alothman, S.; Alqunaibit, D.; Avanzi, A.; Barilla, R.; Daley, D.; Greco, S.H.; et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature 2016, 532, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Cui, L.; Yang, L.; Wang, B.; Zhuo, Y.; Zhang, L.; Wang, X.; Zhang, Q.; Zhang, S. Pancreatic Stellate Cells Promote Tumor Progression by Promoting an Immunosuppressive Microenvironment in Murine Models of Pancreatic Cancer. Pancreas 2020, 49, 120–127. [Google Scholar] [CrossRef]
- Wu, Q.; Zhou, L.; Lv, D.; Zhu, X.; Tang, H. Exosome-mediated communication in the tumor microenvironment contributes to hepatocellular carcinoma development and progression. J. Hematol. Oncol. 2019, 12, 53. [Google Scholar] [CrossRef]
- Hannafon, B.N.; Ding, W.-Q. Intercellular Communication by Exosome-Derived microRNAs in Cancer. Int. J. Mol. Sci. 2013, 14, 14240–14269. [Google Scholar] [CrossRef]
- Chen, K.; Wang, Q.; Kornmann, M.; Tian, X.; Yang, Y. The Role of Exosomes in Pancreatic Cancer from Bench to Clinical Application: An Updated Review. Front. Oncol. 2021, 11, 644358. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-M.; Liu, H.; Liu, X.-H.; Hu, H.-B.; Liu, S.-M. Clinical significance of exosomal miRNAs and proteins in three human cancers with high mortality in China. Oncol. Lett. 2019, 17, 11–22. [Google Scholar] [CrossRef]
- Kahlert, C.; Melo, S.; Protopopov, A.; Tang, J.; Seth, S.; Koch, M.; Zhang, J.; Weitz, J.; Chin, L.; Futreal, A.; et al. Identification of Double-stranded Genomic DNA Spanning All Chromosomes with Mutated KRAS and p53 DNA in the Serum Exosomes of Patients with Pancreatic Cancer. J. Biol. Chem. 2014, 289, 3869–3875. [Google Scholar] [CrossRef]
- Kamerkar, S.; LeBleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 2017, 546, 498–503. [Google Scholar] [CrossRef]
- Mendt, M.; Kamerkar, S.; Sugimoto, H.; McAndrews, K.M.; Wu, C.-C.; Gagea, M.; Yang, S.; Blanko, E.V.R.; Peng, Q.; Ma, X.; et al. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Pan, B.T.; Teng, K.; Wu, C.; Adam, M.; Johnstone, R.M. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 1985, 101, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Harding, C.; Stahl, P. Transferrin recycling in reticulocytes: pH and iron are important determinants of ligand binding and processing. Biochem. Biophys. Res. Commun. 1983, 113, 650–658. [Google Scholar] [CrossRef]
- Pan, B.-T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef]
- Kowal, J.; Tkach, M.; Théry, C. Biogenesis and secretion of exosomes. Curr. Opin. Cell Biol. 2014, 29, 116–125. [Google Scholar] [CrossRef]
- Andaloussi, S.E.; Mäger, I.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef]
- van der Pol, E.; Böing, A.N.; Harrison, P.; Sturk, A.; Nieuwland, R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol. Rev. 2012, 64, 676–705. [Google Scholar] [CrossRef]
- Wunsch, B.H.; Smith, J.T.; Gifford, S.M.; Wang, C.; Brink, M.; Bruce, R.L.; Austin, R.H.; Stolovitzky, G.; Astier, Y. Nanoscale lateral displacement arrays for the separation of exosomes and colloids down to 20 nm. Nat. Nanotechnol. 2016, 11, 936–940. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tao, Y.; Wang, X.; Jiang, P.; Li, J.; Peng, M.; Zhang, X.; Chen, K.; Liu, H.; Zhen, P.; et al. Tumor-Secreted Exosomal miR-222 Promotes Tumor Progression via Regulating P27 Expression and Re-Localization in Pancreatic Cancer. Cell. Physiol. Biochem. 2018, 51, 610–629. [Google Scholar] [CrossRef]
- Lu, L.; Risch, H.A. Exosomes: Potential for early detection in pancreatic cancer. Future Oncol. 2016, 12, 1081–1090. [Google Scholar] [CrossRef]
- Huleihel, L.; Hussey, G.S.; Naranjo, J.D.; Zhang, L.; Dziki, J.L.; Turner, N.J.; Stolz, D.B.; Badylak, S.F. Matrix-bound nanovesicles within ECM bioscaffolds. Sci. Adv. 2016, 2, e1600502. [Google Scholar] [CrossRef]
- Sullivan, R.; Saez, F.; Girouard, J.; Frenette, G. Role of exosomes in sperm maturation during the transit along the male reproductive tract. Blood Cells Mol. Dis. 2005, 35, 1–10. [Google Scholar] [CrossRef]
- Gehrmann, U.; Näslund, T.I.; Hiltbrunner, S.; Larssen, P.; Gabrielsson, S. Harnessing the exosome-induced immune response for cancer immunotherapy. Semin. Cancer Biol. 2014, 28, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer Exosomes Perform Cell-Independent MicroRNA Biogenesis and Promote Tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef]
- Fukushima, K.; Satoh, T.; Sugihara, F.; Sato, Y.; Okamoto, T.; Mitsui, Y.; Yoshio, S.; Li, S.; Nojima, S.; Motooka, D.; et al. Dysregulated Expression of the Nuclear Exosome Targeting Complex Component Rbm7 in Nonhematopoietic Cells Licenses the Development of Fibrosis. Immunity 2020, 52, 542–556. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological Properties of Extracellular Vesicles and their Physiological Functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Keller, S.; Sanderson, M.P.; Stoeck, A.; Altevogt, P. Exosomes: From biogenesis and secretion to biological function. Immunol. Lett. 2006, 107, 102–108. [Google Scholar] [CrossRef]
- Spaull, R.; McPherson, B.; Gialeli, A.; Clayton, A.; Uney, J.; Heep, A.; Llana, O.C. Exosomes populate the cerebrospinal fluid of preterm infants with post-haemorrhagic hydrocephalus. Int. J. Dev. Neurosci. 2019, 73, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Baixauli, F.; Gutiérrez-Vázquez, C.; Sánchez-Madrid, F.; Mittelbrunn, M. Sorting it out: Regulation of exosome loading. Semin. Cancer Biol. 2014, 28, 3–13. [Google Scholar] [CrossRef]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef] [PubMed]
- Iraci, N.; Leonardi, T.; Gessler, F.; Vega, B.; Pluchino, S. Focus on Extracellular Vesicles: Physiological Role and Signalling Properties of Extracellular Membrane Vesicles. Int. J. Mol. Sci. 2016, 17, 171. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W.; Théry, C. Extracellular vesicles or exosomes? On primacy, precision, and popularity influencing a choice of nomenclature. J. Extracell. Vesicles 2019, 8, 1648167. [Google Scholar] [CrossRef] [PubMed]
- Lobb, R.J.; Becker, M.; Wen, S.W.; Wong, C.S.F.; Wiegmans, A.P.; Leimgruber, A.; Möller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef]
- Stranska, R.; Gysbrechts, L.; Wouters, J.; Vermeersch, P.; Bloch, K.; Dierickx, D.; Andrei, G.; Snoeck, R. Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J. Transl. Med. 2018, 16, 1. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Kang, M.-H.; Jeyaraj, M.; Qasim, M.; Kim, J.-H. Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef]
- Yang, D.; Zhang, W.; Zhang, H.; Zhang, F.; Chen, L.; Ma, L.; Larcher, L.; Chen, S.; Liu, N.; Zhao, Q.; et al. Progress, opportunity, and perspective on exosome isolation - efforts for efficient exosome-based theranostics. Theranostics 2020, 10, 3684–3707. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Benito-Martin, A.; Bojmar, L.; et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Niu, F.; Chen, X.; Niu, X.; Cai, Y.; Zhang, Q.; Chen, T.; Yang, H. Integrated Immunomagnetic Bead-Based Microfluidic Chip for Exosomes Isolation. Micromachines 2020, 11, 503. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Shao, H.; Weissleder, R.; Lee, H. Acoustic Purification of Extracellular Microvesicles. ACS Nano 2015, 9, 2321–2327. [Google Scholar] [CrossRef]
- Hannafon, B.N.; Carpenter, K.J.; Berry, W.L.; Janknecht, R.; Dooley, W.C.; Ding, W.-Q. Exosome-mediated microRNA signaling from breast cancer cells is altered by the anti-angiogenesis agent docosahexaenoic acid (DHA). Mol. Cancer 2015, 14, 1–13. [Google Scholar] [CrossRef]
- Tkach, M.; Théry, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef]
- Su, M.-J.; Aldawsari, H.; Amiji, M. Pancreatic Cancer Cell Exosome-Mediated Macrophage Reprogramming and the Role of MicroRNAs 155 and 125b2 Transfection using Nanoparticle Delivery Systems. Sci. Rep. 2016, 6, 30110. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef]
- Dhondt, B.; Rousseau, Q.; De Wever, O.; Hendrix, A. Function of extracellular vesicle-associated miRNAs in metastasis. Cell Tissue Res. 2016, 365, 621–641. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Pucci, F.; Garris, C.; Lai, C.P.; Newton, A.; Pfirschke, C.; Engblom, C.; Alvarez, D.; Sprachman, M.; Evavold, C.; Magnuson, A.; et al. SCS macrophages suppress melanoma by restricting tumor-derived vesicle-B cell interactions. Science 2016, 352, 242–246. [Google Scholar] [CrossRef] [PubMed]
- Skog, J.; Würdinger, T.; Van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Peinado, H.; Aleckovic, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; Garcia-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2009, 12, 19–30. [Google Scholar] [CrossRef]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Mahajan, V.; Deygen, I.; Klyachko, N.L.; Inskoe, E.; Piroyan, A.; Sokolsky, M.; Okolie, O.; et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 655–664. [Google Scholar] [CrossRef]
- Wang, T.; Gilkes, D.M.; Takano, N.; Xiang, L.; Luo, W.; Bishop, C.J.; Chaturvedi, P.; Green, J.J.; Semenza, G.L. Hypoxia-inducible factors and RAB22A mediate formation of microvesicles that stimulate breast cancer invasion and metastasis. Proc. Natl. Acad. Sci. USA 2014, 111, E3234–E3242. [Google Scholar] [CrossRef]
- Gastpar, R.; Gehrmann, M.; Bausero, M.A.; Asea, A.; Gross, C.; Schroeder, J.A.; Multhoff, G. Heat Shock Protein 70 Surface-Positive Tumor Exosomes Stimulate Migratory and Cytolytic Activity of Natural Killer Cells. Cancer Res. 2005, 65, 5238–5247. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Fu, G.; Ming, L. Role of exosomes in pancreatic cancer (Review). Oncol. Lett. 2018, 15, 7479–7488. [Google Scholar] [CrossRef]
- Lan, B.; Zeng, S.; Grützmann, R.; Pilarsky, C. The Role of Exosomes in Pancreatic Cancer. Int. J. Mol. Sci. 2019, 20, 4332. [Google Scholar] [CrossRef]
- Masamune, A.; Yoshida, N.; Hamada, S.; Takikawa, T.; Nabeshima, T.; Shimosegawa, T. Exosomes derived from pancreatic cancer cells induce activation and profibrogenic activities in pancreatic stellate cells. Biochem. Biophys. Res. Commun. 2018, 495, 71–77. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, B.; Zheng, W.; Kang, M.; Chen, Q.; Qin, W.; Li, C.; Zhang, Y.; Shao, Y.; Wu, Y. Exosomes derived from pancreatic cancer cells induce insulin resistance in C2C12 myotube cells through the PI3K/Akt/FoxO1 pathway. Sci. Rep. 2017, 7, 5384. [Google Scholar] [CrossRef]
- Javeed, N.; Sagar, G.; Dutta, S.K.; Smyrk, T.C.; Lau, J.S.; Bhattacharya, S.; Truty, M.; Petersen, G.M.; Kaufman, R.J.; Chari, S.T.; et al. Pancreatic Cancer-Derived Exosomes Cause Paraneoplastic beta-cell Dysfunction. Clin. Cancer Res. 2015, 21, 1722–1733. [Google Scholar] [CrossRef] [PubMed]
- Korc, M. Pancreatic cancer-associated diabetes is an “exosomopathy”. Clin. Cancer Res. 2015, 21, 1508–1510. [Google Scholar] [CrossRef]
- Sagar, G.; Sah, R.P.; Javeed, N.; Dutta, S.K.; Smyrk, T.C.; Lau, J.S.; Giorgadze, N.; Tchkonia, T.; Kirkland, J.L.; Chari, S.T.; et al. Pathogenesis of pancreatic cancer exosome-induced lipolysis in adipose tissue. Gut 2016, 65, 1165–1174. [Google Scholar] [CrossRef]
- Kim, J.; Reber, H.A.; Dry, S.M.; Elashoff, D.; Chen, S.L.; Umetani, N.; Kitago, M.; Hines, O.J.; Kazanjian, K.K.; Hiramatsu, S.; et al. Unfavourable prognosis associated with K-ras gene mutation in pancreatic cancer surgical margins. Gut 2006, 55, 1598–1605. [Google Scholar] [CrossRef]
- Mikamori, M.; Yamada, D.; Eguchi, H.; Hasegawa, S.; Kishimoto, T.; Tomimaru, Y.; Asaoka, T.; Noda, T.; Wada, H.; Kawamoto, K.; et al. MicroRNA-155 Controls Exosome Synthesis and Promotes Gemcitabine Resistance in Pancreatic Ductal Adenocarcinoma. Sci. Rep. 2017, 7, 42339. [Google Scholar] [CrossRef] [PubMed]
- Patel, G.K.; Khan, M.A.; Bhardwaj, A.; Srivastava, S.K.; Zubair, H.; Patton, M.C.; Singh, S.; Khushman, M.; Singh, A. Exosomes confer chemoresistance to pancreatic cancer cells by promoting ROS detoxification and miR-155-mediated suppression of key gemcitabine-metabolising enzyme, DCK. Br. J. Cancer 2017, 116, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.F.; Li, S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef]
- Wang, X.; Luo, G.; Zhang, K.; Cao, J.; Huang, C.; Jiang, T.; Liu, B.; Su, L.; Qiu, Z. Hypoxic Tumor-Derived Exosomal miR-301a Mediates M2 Macrophage Polarization via PTEN/PI3Kgamma to Promote Pancreatic Cancer Metastasis. Cancer Res. 2018, 78, 4586–4598. [Google Scholar] [CrossRef]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Chiang, H.R.; Schoenfeld, L.W.; Ruby, J.G.; Auyeung, V.C.; Spies, N.; Baek, D.; Johnston, W.K.; Russ, C.; Luo, S.; Babiarz, J.E.; et al. Mammalian microRNAs: Experimental evaluation of novel and previously annotated genes. Genes Dev. 2010, 24, 992–1009. [Google Scholar] [CrossRef]
- Sin, T.K.; Wang, F.; Meng, F.; Wong, S.C.C.; Cho, W.C.S.; Siu, P.M.; Chan, L.W.C.; Yung, B.Y.M. Implications of MicroRNAs in the Treatment of Gefitinib-Resistant Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2016, 17, 237. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuka, M.; Ling, H.; Doki, Y.; Mori, M.; Calin, G.A. MicroRNA Processing and Human Cancer. J. Clin. Med. 2015, 4, 1651–1667. [Google Scholar] [CrossRef] [PubMed]
- Hannafon, B.N.; Ding, W.-Q. miRNAs as Biomarkers for Predicting the Progression of Ductal Carcinoma in Situ. Am. J. Pathol. 2018, 188, 542–549. [Google Scholar] [CrossRef]
- Frampton, A.E.; Krell, J.; Jamieson, N.B.; Gall, T.M.; Giovannetti, E.; Funel, N.; Prado, M.M.; Krell, D.; Habib, N.A.; Castellano, L.; et al. microRNAs with prognostic significance in pancreatic ductal adenocarcinoma: A meta-analysis. Eur. J. Cancer 2015, 51, 1389–1404. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, Y.G.; Lucas, A.L. MicroRNA in pancreatic ductal adenocarcinoma and its precursor lesions. World J. Gastrointest. Oncol. 2016, 8, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Chua, C.Y.X.; Tian, W.; Zhang, Z.; Chiao, P.J.; Zhang, W. MicroRNA Signaling Pathway Network in Pancreatic Ductal Adenocarcinoma. J. Genet. Genom. 2015, 42, 563–577. [Google Scholar] [CrossRef]
- Li, H.; Xiang, H.; Ge, W.; Wang, H.; Wang, T.; Xiong, M. Expression and functional perspectives of miR-184 in pancreatic ductal adenocarcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 12313–12318. [Google Scholar]
- Le Large, T.Y.; Meijer, L.L.; Prado, M.M.; Kazemier, G.; Frampton, A.E.; Giovannetti, E. Circulating microRNAs as diagnostic biomarkers for pancreatic cancer. Expert Rev. Mol. Diagn. 2015, 15, 1525–1529. [Google Scholar] [CrossRef]
- Permuth-Wey, J.; Chen, D.-T.; Fulp, W.J.; Yoder, S.J.; Zhang, Y.; Georgeades, C.; Husain, K.; Centeno, B.A.; Magliocco, A.M.; Coppola, D.; et al. Plasma MicroRNAs as Novel Biomarkers for Patients with Intraductal Papillary Mucinous Neoplasms of the Pancreas. Cancer Prev. Res. 2015, 8, 826–834. [Google Scholar] [CrossRef] [PubMed]
- Cote, G.A.; Gore, A.J.; McElyea, S.D.; Heathers, L.E.; Xu, H.; Sherman, S.; Korc, M. A Pilot Study to Develop a Diagnostic Test for Pancreatic Ductal Adenocarcinoma Based on Differential Expression of Select miRNA in Plasma and Bile. Am. J. Gastroenterol. 2014, 109, 1942–1952. [Google Scholar] [CrossRef]
- Liu, R.; Chen, X.; Du, Y.; Yao, W.; Shen, L.; Wang, C.; Hu, Z.; Zhuang, R.; Ning, G.; Zhang, C.; et al. Serum MicroRNA Expression Profile as a Biomarker in the Diagnosis and Prognosis of Pancreatic Cancer. Clin. Chem. 2012, 58, 610–618. [Google Scholar] [CrossRef]
- Komatsu, S.; Ichikawa, D.; Takeshita, H.; Morimura, R.; Hirajima, S.; Tsujiura, M.; Kawaguchi, T.; Miyamae, M.; Nagata, H.; Konishi, H.; et al. Circulating miR-18a: A sensitive cancer screening biomarker in human cancer. In Vivo 2014, 28, 293–297. [Google Scholar] [PubMed]
- Morimura, R.; Komatsu, S.; Ichikawa, D.; Takeshita, H.; Tsujiura, M.; Nagata, H.; Konishi, H.; Shiozaki, A.; Ikoma, H.; Okamoto, K.; et al. Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. Br. J. Cancer 2011, 105, 1733–1740. [Google Scholar] [CrossRef]
- Liffers, S.-T.; Munding, J.B.; Vogt, M.; Kuhlmann, J.D.; Verdoodt, B.; Nambiar, S.; Maghnouj, A.; Mirmohammadsadegh, A.; Hahn, S.A.; Tannapfel, A. MicroRNA-148a is down-regulated in human pancreatic ductal adenocarcinomas and regulates cell survival by targeting CDC25B. Lab. Investig. 2011, 91, 1472–1479. [Google Scholar] [CrossRef]
- Munding, J.; Adai, A.T.; Maghnouj, A.; Urbanik, A.; Zöllner, H.; Liffers, S.T.; Chromik, A.M.; Uhl, W.; Szafranska-Schwarzbach, A.E.; Tannapfel, A.; et al. Global microRNA expression profiling of microdissected tissues identifies miR-135b as a novel biomarker for pancreatic ductal adenocarcinoma. Int. J. Cancer 2012, 131, E86–E95. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, C.-Y.; Zhang, S.-H.; Yu, D.-H.; Chen, Y.; Liu, Q.-H.; Shi, M.; Ni, C.-R.; Zhu, M.-H. Upregulation of miR-194 contributes to tumor growth and progression in pancreatic ductal adenocarcinoma. Oncol. Rep. 2014, 31, 1157–1164. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, J.; Zhang, S.; Yu, D.; Chen, Y.; Liu, Q.; Shi, M.; Ni, C.; Zhu, M. Diagnostic and biological significance of microRNA-192 in pancreatic ductal adenocarcinoma. Oncol. Rep. 2013, 30, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Barceló, M.; Castells, M.; Bassas, L.; Vigués, F.; Larriba, S. Semen miRNAs Contained in Exosomes as Non-Invasive Biomarkers for Prostate Cancer Diagnosis. Sci. Rep. 2019, 9, 13772. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Sai, B.; Wang, F.; Wang, L.; Wang, Y.; Zheng, L.; Li, G.; Tang, J.; Xiang, J. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol. Cancer 2019, 18, 40. [Google Scholar] [CrossRef]
- Hannafon, B.N.; Trigoso, Y.D.; Calloway, C.L.; Zhao, Y.D.; Lum, D.H.; Welm, A.L.; Zhao, Z.J.; Blick, K.E.; Dooley, W.C.; Ding, W.Q. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016, 18, 90. [Google Scholar] [CrossRef]
- Xu, Y.-F.; Hannafon, B.N.; Khatri, U.; Gin, A.; Ding, W.-Q. The origin of exosomal miR-1246 in human cancer cells. RNA Biol. 2019, 16, 770–784. [Google Scholar] [CrossRef]
- Xu, Y.-F.; Hannafon, B.N.; Zhao, Y.D.; Postier, R.G.; Ding, W.-Q. Plasma exosome miR-196a and miR-1246 are potential indicators of localized pancreatic cancer. Oncotarget 2017, 8, 77028–77040. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, Y.; Geng, C.; Zhou, H.; Gao, W.; Chen, W. Serum exosomal microRNAs as novel biomarkers for multiple myeloma. Hematol. Oncol. 2019, 37, 409–417. [Google Scholar] [CrossRef]
- Que, R.; Ding, G.; Chen, J.; Cao, L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J. Surg. Oncol. 2013, 11, 219. [Google Scholar] [CrossRef]
- Sun, H.; Shi, K.; Qi, K.; Kong, H.; Zhang, J.; Dai, S.; Ye, W.; Deng, T.; He, Q.; Zhou, M. Natural killer cell-derived exosomal miR-3607-3p inhibits pancreatic cancer progression by targeting IL-26. Front. Immunol. 2019, 10, 2819. [Google Scholar] [CrossRef]
- Ma, Q.; Wu, H.; Xiao, Y.; Liang, Z.; Liu, T. Upregulation of exosomal microRNA-21 in pancreatic stellate cells promotes pancreatic cancer cell migration and enhances Ras/ERK pathway activity. Int. J. Oncol. 2020, 56, 1025–1033. [Google Scholar] [CrossRef]
- Jiang, M.; Chen, Y.; Dai, J.; Gu, D.; Mei, Z.; Liu, F.; Huang, Q.; Tian, L. Dying tumor cell-derived exosomal miR-194-5p potentiates survival and repopulation of tumor repopulating cells upon radiotherapy in pancreatic cancer. Mol. Cancer 2020, 19, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef]
- Suetsugu, A.; Honma, K.; Saji, S.; Moriwaki, H.; Ochiya, T.; Hoffman, R.M. Imaging exosome transfer from breast cancer cells to stroma at metastatic sites in orthotopic nude-mouse models. Adv. Drug Deliv. Rev. 2013, 65, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Madhavan, B.; Yue, S.; Galli, U.; Rana, S.; Gross, W.; Müller, M.; Giese, N.A.; Kalthoff, H.; Becker, T.; Büchler, M.W.; et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int. J. Cancer 2015, 136, 2616–2627. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Wang, M.; McElyea, S.D.; Sherman, S.; House, M.; Korc, M. A microRNA signature in circulating exosomes is superior to exosomal glypican-1 levels for diagnosing pancreatic cancer. Cancer Lett. 2017, 393, 86–93. [Google Scholar] [CrossRef]
- Goto, T.; Fujiya, M.; Konishi, H.; Sasajima, J.; Fujibayashi, S.; Hayashi, A.; Utsumi, T.; Sato, H.; Iwama, T.; Ijiri, M.; et al. An elevated expression of serum exosomal microRNA-191, − 21, −451a of pancreatic neoplasm is considered to be efficient diagnostic marker. BMC Cancer 2018, 18, 116. [Google Scholar] [CrossRef] [PubMed]
- Abue, M.; Yokoyama, M.; Shibuya, R.; Tamai, K.; Yamaguchi, K.; Sato, I.; Tanaka, N.; Hamada, S.; Shimosegawa, T.; Sugamura, K.; et al. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int. J. Oncol. 2015, 46, 539–547. [Google Scholar] [CrossRef]
- Takahasi, K.; Iinuma, H.; Wada, K.; Minezaki, S.; Kawamura, S.; Kainuma, M.; Ikeda, Y.; Shibuya, M.; Miura, F.; Sano, K. Usefulness of exosome-encapsulated microRNA-451a as a minimally invasive biomarker for prediction of recurrence and prognosis in pancreatic ductal adenocarcinoma. J. Hepato-Biliary-Pancreat. Sci. 2017, 25, 155–161. [Google Scholar] [CrossRef]
- Engle, D.D.; Tiriac, H.; Rivera, K.D.; Pommier, A.; Whalen, S.; Oni, T.E.; Alagesan, B.; Lee, E.J.; Yao, M.A.; Lucito, M.S.; et al. The glycan CA19-9 promotes pancreatitis and pancreatic cancer in mice. Science 2019, 364, 1156–1162. [Google Scholar] [CrossRef]
- Kondo, N.; Murakami, Y.; Uemura, K.; Hayashidani, Y.; Sudo, T.; Hashimoto, Y.; Nakashima, A.; Sakabe, R.; Shigemoto, N.; Kato, Y.; et al. Prognostic Impact of Perioperative Serum CA 19-9 Levels in Patients with Resectable Pancreatic Cancer. Ann. Surg. Oncol. 2010, 17, 2321–2329. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Sadakari, Y.; Ohtsuka, T.; Okayama, T.; Nakashima, Y.; Gotoh, Y.; Saeki, K.; Mori, Y.; Nakata, K.; Miyasaka, Y.; et al. Pancreatic Juice Exosomal MicroRNAs as Biomarkers for Detection of Pancreatic Ductal Adenocarcinoma. Ann. Surg. Oncol. 2019, 26, 2104–2111. [Google Scholar] [CrossRef] [PubMed]
- Machida, T.; Tomofuji, T.; Maruyama, T.; Yoneda, T.; Ekuni, D.; Azuma, T.; Miyai, H.; Mizuno, H.; Kato, H.; Tsutsumi, K.; et al. miR-1246 and miR-4644 in salivary exosome as potential biomarkers for pancreatobiliary tract cancer. Oncol. Rep. 2016, 36, 2375–2381. [Google Scholar] [CrossRef]
- Kawamura, S.; Iinuma, H.; Wada, K.; Takahashi, K.; Minezaki, S.; Kainuma, M.; Shibuya, M.; Miura, F.; Sano, K. Exosome-encapsulated microRNA-4525, microRNA-451a and microRNA-21 in portal vein blood is a high-sensitive liquid biomarker for the selection of high-risk pancreatic ductal adenocarcinoma patients. J. Hepato Biliary Pancreat Sci. 2019, 26, 63–72. [Google Scholar] [CrossRef]
- Chen, D.; Wu, X.; Xia, M.; Wu, F.; Ding, J.; Jiao, Y.; Zhan, Q.; An, F. Upregulated exosomic miR-23b-3p plays regulatory roles in the progression of pancreatic cancer. Oncol. Rep. 2017, 38, 2182–2188. [Google Scholar] [CrossRef]
- Yu, Z.; Zhao, S.; Wang, L.; Wang, J.; Zhou, J. miRNA-339-5p Plays an Important Role in Invasion and Migration of Pancreatic Cancer Cells. Med. Sci. Monit. 2019, 25, 7509–7517. [Google Scholar] [CrossRef] [PubMed]
- Shang, D.; Xie, C.; Hu, J.; Tan, J.; Yuan, Y.; Liu, Z.; Yang, Z. Pancreatic cancer cell–derived exosomal microRNA-27a promotes angiogenesis of human microvascular endothelial cells in pancreatic cancer via BTG2. J. Cell. Mol. Med. 2020, 24, 588–604. [Google Scholar] [CrossRef]
- Ding, G.; Zhou, L.; Qian, Y.; Fu, M.; Chen, J.; Chen, J.; Xiang, J.; Wu, Z.; Jiang, G.; Cao, L. Pancreatic cancer-derived exosomes transfer miRNAs to dendritic cells and inhibit RFXAP expression via miR-212-3p. Oncotarget 2015, 6, 29877–29888. [Google Scholar] [CrossRef]
- Batista, I.A.; Melo, S.A. Exosomes and the Future of Immunotherapy in Pancreatic Cancer. Int. J. Mol. Sci. 2019, 20, 567. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Ma, T.; Huang, B.; Lin, L.; Zhou, Y.; Yan, J.; Zou, Y.; Chen, S. Macrophage-derived exosomal microRNA-501-3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3-mediated TGF-beta signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 310. [Google Scholar] [CrossRef]
- Yang, Z.; Zhao, N.; Cui, J.; Wu, H.; Xiong, J.; Peng, T. Exosomes derived from cancer stem cells of gemcitabine-resistant pancreatic cancer cells enhance drug resistance by delivering miR-210. Cell. Oncol. 2020, 43, 123–136. [Google Scholar] [CrossRef]
- Wu, D.-M.; Wen, X.; Han, X.-R.; Wang, S.; Wang, Y.-J.; Shen, M.; Fan, S.-H.; Zhang, Z.-F.; Shan, Q.; Li, M.-Q.; et al. Bone Marrow Mesenchymal Stem Cell-Derived Exosomal MicroRNA-126-3p Inhibits Pancreatic Cancer Development by Targeting ADAM9. Mol. Ther. Nucleic Acids 2019, 16, 229–245. [Google Scholar] [CrossRef]
- Shang, S.; Wang, J.; Chen, S.; Tian, R.; Zeng, H.; Wang, L.; Xia, M.; Zhu, H.; Zuo, C. Exosomal miRNA-1231 derived from bone marrow mesenchymal stem cells inhibits the activity of pancreatic cancer. Cancer Med. 2019, 8, 7728–7740. [Google Scholar] [CrossRef]
- Ding, Y.; Cao, F.; Sun, H.; Wang, Y.; Liu, S.; Wu, Y.; Cui, Q.; Mei, W.; Li, F. Exosomes derived from human umbilical cord mesenchymal stromal cells deliver exogenous miR-145-5p to inhibit pancreatic ductal adenocarcinoma progression. Cancer Lett. 2019, 442, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Richards, K.E.; Zeleniak, A.E.; Fishel, M.L.; Wu, J.; Littlepage, L.E.; Hill, R. Cancer-associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene 2017, 36, 1770–1778. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Zhou, W.; Rong, Y.; Kuang, T.; Xu, X.; Wu, W.; Wang, D.; Lou, W. Exosomal miRNA-106b from cancer-associated fibroblast promotes gemcitabine resistance in pancreatic cancer. Exp. Cell Res. 2019, 383, 111543. [Google Scholar] [CrossRef]
- Liu, J.; Gao, J.; Du, Y.; Li, Z.; Ren, Y.; Gu, J.; Wang, X.; Gong, Y.; Wang, W.; Kong, X. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int. J. Cancer 2012, 131, 683–691. [Google Scholar] [CrossRef]
- He, L.; Ping, F.; Fan, Z.; Zhang, C.; Deng, M.; Cheng, B.; Xia, J. Salivary exosomal miR-24-3p serves as a potential detective biomarker for oral squamous cell carcinoma screening. Biomed. Pharmacother. 2020, 121, 109553. [Google Scholar] [CrossRef]
- Ye, H.; Wang, H.; Wang, P.; Song, C.-H.; Wang, K.-J.; Dai, L.-P.; Shi, J.-X.; Liu, X.-X.; Sun, C.-Q.; Wang, X.; et al. Systematic review: Exosomal microRNAs associated with pancreatic cancer for early detection and prognosis. Eur. Rev. Med Pharmacol. Sci. 2019, 23, 9351–9361. [Google Scholar]
- Thind, A.; Wilson, C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J. Extracell. Vesicles 2016, 5, 31292. [Google Scholar] [CrossRef] [PubMed]
- Pitt, J.M.; Kroemer, G.; Zitvogel, L. Extracellular vesicles: Masters of intercellular communication and potential clinical interventions. J. Clin. Investig. 2016, 126, 1139–1143. [Google Scholar] [CrossRef]
- Mirzaei, H.; Sahebkar, A.; Jaafari, M.R.; Goodarzi, M.; Mirzaei, H.R. Diagnostic and Therapeutic Potential of Exosomes in Cancer: The Beginning of a New Tale? J. Cell. Physiol. 2017, 232, 3251–3260. [Google Scholar] [CrossRef]
- Pigati, L.; Yaddanapudi, S.C.S.; Iyengar, R.; Kim, D.-J.; Hearn, S.A.; Danforth, D.; Hastings, M.; Duelli, D.M. Selective Release of MicroRNA Species from Normal and Malignant Mammary Epithelial Cells. PLoS ONE 2010, 5, e13515. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Suresh, R.; Banerjee, S.; Bao, B.; Xu, Z.; Wilson, J.; Philip, P.A.; Apte, M.; Sarkar, F.H. Contribution of microRNAs in understanding the pancreatic tumor microenvironment involving cancer associated stellate and fibroblast cells. Am. J. Cancer Res. 2015, 5, 1251–1264. [Google Scholar]
- Song, M.; Sun, M.; Xia, L.; Chen, W.; Yang, C. miR-19b-3p promotes human pancreatic cancer Capan-2 cells proliferation by targeting phosphatase and tension homolog. Ann. Transl. Med. 2019, 7, 236. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Shivapurkar, N.; Wu, Z.; Hwang, J.J.; Pishvaian, M.J.; Weiner, L.M.; Ley, L.; Zhou, D.; Zhi, X.; Wellstein, A.; et al. Circulating microRNA profile predicts disease progression in patients receiving second-line treatment of lapatinib and capecitabine for metastatic pancreatic cancer. Oncol. Lett. 2016, 11, 1645–1650. [Google Scholar] [CrossRef]
- Jabbari, N.; Akbariazar, E.; Feqhhi, M.; Rahbarghazi, R.; Rezaie, J. Breast cancer-derived exosomes: Tumor progression and therapeutic agents. J. Cell. Physiol. 2020, 235, 6345–6356. [Google Scholar] [CrossRef]
- Jabbari, N.; Karimipour, M.; Khaksar, M.; Akbariazar, E.; Heidarzadeh, M.; Mojarad, B.; Aftab, H.; Rahbarghazi, R.; Rezaie, J. Tumor-derived extracellular vesicles: Insights into bystander effects of exosomes after irradiation. Lasers Med Sci. 2019, 35, 531–545. [Google Scholar] [CrossRef] [PubMed]
- Akbari, A.; Jabbari, N.; Sharifi, R.; Ahmadi, M.; Vahhabi, A.; Seyedzadeh, S.J.; Nawaz, M.; Szafert, S.; Mahmoodi, M.; Jabbari, E.; et al. Free and hydrogel encapsulated exosome-based therapies in regenerative medicine. Life Sci. 2020, 249, 117447. [Google Scholar] [CrossRef] [PubMed]
- Fitts, C.A.; Ji, N.; Li, Y.; Tan, C. Exploiting Exosomes in Cancer Liquid Biopsies and Drug Delivery. Adv. Healthc. Mater. 2019, 8, 1801268. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Badawi, M.; Pomeroy, S.; Sutaria, D.S.; Xie, Z.; Baek, A.; Jiang, J.; Elgamal, O.A.; Mohamed, B.; La Perle, K.; et al. Comprehensive toxicity and immunogenicity studies reveal minimal effects in mice following sustained dosing of extracellular vesicles derived from HEK293T cells. J. Extracell. Vesicles 2017, 6, 1324730. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J.A. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, S.W.; Nguyen, J. Exosomes as therapeutics: The implications of molecular composition and exosomal heterogeneity. J. Control. Release 2016, 228, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Kordelas, L.; Rebmann, V.; Ludwig, A.-K.; Radtke, S.; Ruesing, J.; Doeppner, T.R.; Epple, M.; Horn, P.A.; Beelen, D.W.; Giebel, B. MSC-derived exosomes: A novel tool to treat therapy-refractory graft-versus-host disease. Leukemia 2014, 28, 970–973. [Google Scholar] [CrossRef]
- Gabriel, A.N.A.; Wang, F.; Jiao, Q.; Yvette, U.; Yang, X.; Al-Ameri, S.A.; Du, L.; Wang, Y.-S.; Wang, C. The involvement of exosomes in the diagnosis and treatment of pancreatic cancer. Mol. Cancer 2020, 19, 132. [Google Scholar] [CrossRef]
- Kamisawa, T.; Wood, L.D.; Itoi, T.; Takaori, K. Pancreatic cancer. Lancet 2016, 388, 73–85. [Google Scholar] [CrossRef]
- Cheng, H.; Fan, K.; Luo, G.; Fan, Z.; Yang, C.; Huang, Q.; Jin, K.; Xu, J.; Yu, X.; Liu, C. KrasG12D mutation contributes to regulatory T cell conversion through activation of the MEK/ERK pathway in pancreatic cancer. Cancer Lett. 2019, 446, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Sharples, R.A.; Scicluna, B.J.; Hill, A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 2014, 3, 23743. [Google Scholar] [CrossRef] [PubMed]
- Mittal, A.; Chitkara, D.; Behrman, S.W.; Mahato, R.I. Efficacy of gemcitabine conjugated and miRNA-205 complexed micelles for treatment of advanced pancreatic cancer. Biomaterials 2014, 35, 7077–7087. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Liu, J.; Xiang, X.; Liu, S.; Zhong, P.; Xie, F.; Mou, T.; Lai, L. Expression of miRNA-143 in Pancreatic Cancer and Its Clinical Significance. Cancer Biother. Radiopharm. 2018, 33, 373–379. [Google Scholar] [CrossRef]
- Tesfaye, A.A.; Azmi, A.S.; Philip, P.A. miRNA and Gene Expression in Pancreatic Ductal Adenocarcinoma. Am. J. Pathol. 2019, 189, 58–70. [Google Scholar] [CrossRef]
- Qadir, M.I.; Faheem, A. miRNA: A Diagnostic and Therapeutic Tool for Pancreatic Cancer. Crit. Rev. Eukaryot. Gene Expr. 2017, 27, 197–204. [Google Scholar] [CrossRef]
- Daoud, A.Z.; Mulholland, E.J.; Cole, G.; McCarthy, H.O. MicroRNAs in Pancreatic Cancer: Biomarkers, prognostic, and therapeutic modulators. BMC Cancer 2019, 19, 1130. [Google Scholar] [CrossRef] [PubMed]
- Rawat, M.; Kadian, K.; Gupta, Y.; Kumar, S.; Chain, P.S.; Kovbasnjuk, O.; Parasher, G. MicroRNA in Pancreatic Cancer: From Biology to Therapeutic Potential. Genes 2019, 10, 752. [Google Scholar] [CrossRef]
- Xia, J.; Cao, T.; Ma, C.; Shi, Y.; Sun, Y.; Wang, Z.P.; Ma, J. miR-7 Suppresses Tumor Progression by Directly Targeting MAP3K9 in Pancreatic Cancer. Mol. Ther. Nucleic Acids 2018, 13, 121–132. [Google Scholar] [CrossRef]
- Wang, S.; Ji, J.; Song, J.; Li, X.; Han, S.; Lian, W.; Cao, C.; Zhang, X.; Li, M. MicroRNA-182 promotes pancreatic cancer cell proliferation and migration by targeting beta-TrCP2. Acta Biochim. Biophys. Sin. 2016, 48, 1085–1093. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, P.; Li, J.; Peng, M.; Zhao, X.; Zhang, X.; Chen, K.; Zhang, Y.; Liu, H.; Gan, L.; et al. Tumor-derived exosomal lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in pancreatic ductal adenocarcinoma. Oncogene 2018, 37, 3822–3838. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Boussac, M.; Véron, P.; Ricciardi-Castagnoli, P.; Raposo, G.; Garin, J.; Amigorena, S. Proteomic Analysis of Dendritic Cell-Derived Exosomes: A Secreted Subcellular Compartment Distinct from Apoptotic Vesicles. J. Immunol. 2001, 166, 7309–7318. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef] [PubMed]
- Ferraiuolo, R.-M.; Manthey, K.C.; Stanton, M.J.; Triplett, A.A.; Wagner, K.-U. The Multifaceted Roles of the Tumor Susceptibility Gene 101 (TSG101) in Normal Development and Disease. Cancers 2020, 12, 450. [Google Scholar] [CrossRef] [PubMed]
| Body Fluids (Source) | Exosomal miRNA | Biomarker or Other Use | References |
|---|---|---|---|
| Serum | miR-17-5p | Diagnostic | [133] |
| Serum | miR-1246, miR-4644, miR-3976 and miR-4306 | Diagnostic | [140] |
| Serum | miR-10b, miR-21, miR-30c, and miR-181a (up) and miR-let7a (down) | Diagnostic | [141] |
| Serum | miR-191, miR-21 and miR-451a | Diagnostic | [142] |
| Serum | miR-21 | Early diagnostic; prognostic; recurrence | [133] |
| Plasma | miR-1246 and miR-196a | Diagnostic | [130,131] |
| Plasma | miR-483-3p | Diagnostic (differentiation of pancreatic cancer with IPMN) | [143] |
| Plasma | miR-451a | Prognostic and recurrence | [144] |
| Blood | miRNA-16a and miRNA-196a | Diagnostic | [145,146] |
| Pancreatic juice | miR-21 and miR-155 | Diagnostic or prognostic | [147] |
| Saliva | miR-1246 and miR-4644 | Diagnostic | [148] |
| Portal vein blood | miR-4525, miR-451a and miR-21 | Prognostic and recurrence | [149] |
| PCC | miR-23b-3p | Prognostic | [150] |
| PCC | miR-339-5p | Prognostic | [151] |
| PCC | miR-222 | Prognostic | [52] |
| PCC | miR-222 | Prognostic | [52] |
| PCC | miR-155 (in vitro and in vivo study) | Prognostic (gemcitabine resistance) | [101] |
| PCC | miR-27a | Prognostic (promote angiogenesis of human microvascular endothelial cells) | [152] |
| PCC | miR-212-3p | Prognostic immune suppression (MHC II downregulation) | [153,154] |
| PCC (under hypoxia) | miR-301a | Prognostic (PTEN/PI3K signaling) | [7] |
| Dying PCC | miR-194-5p | Prognostic | [114] |
| M2 macrophage | miR-501-3p | Prognostic (TGF-beta signaling) | [155] |
| NK cells | miR-3607-3p | Prognostic | [134] |
| PSC | miR-210 | Prognostic (gemcitabine resistance) | [156] |
| BM-MSC | miR-126-3p | Prognostic (suppress development via ADAM9 down regulation) | [157] |
| BM-MSC | miR-1231 | Prognostic (suppress development) | [158] |
| UC-MSC | miR-145-5p | Prognostic (suppress progression) | [159] |
| CAF | miR-146a | Prognostic (gemcitabine resistance) | [160] |
| CAF | miR-106b | Prognostic (gemcitabine resistance) | [161] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uddin, M.H.; Al-Hallak, M.N.; Philip, P.A.; Mohammad, R.M.; Viola, N.; Wagner, K.-U.; Azmi, A.S. Exosomal microRNA in Pancreatic Cancer Diagnosis, Prognosis, and Treatment: From Bench to Bedside. Cancers 2021, 13, 2777. https://doi.org/10.3390/cancers13112777
Uddin MH, Al-Hallak MN, Philip PA, Mohammad RM, Viola N, Wagner K-U, Azmi AS. Exosomal microRNA in Pancreatic Cancer Diagnosis, Prognosis, and Treatment: From Bench to Bedside. Cancers. 2021; 13(11):2777. https://doi.org/10.3390/cancers13112777
Chicago/Turabian StyleUddin, Md. Hafiz, Mohammed Najeeb Al-Hallak, Philip A. Philip, Ramzi M. Mohammad, Nerissa Viola, Kay-Uwe Wagner, and Asfar S. Azmi. 2021. "Exosomal microRNA in Pancreatic Cancer Diagnosis, Prognosis, and Treatment: From Bench to Bedside" Cancers 13, no. 11: 2777. https://doi.org/10.3390/cancers13112777
APA StyleUddin, M. H., Al-Hallak, M. N., Philip, P. A., Mohammad, R. M., Viola, N., Wagner, K.-U., & Azmi, A. S. (2021). Exosomal microRNA in Pancreatic Cancer Diagnosis, Prognosis, and Treatment: From Bench to Bedside. Cancers, 13(11), 2777. https://doi.org/10.3390/cancers13112777







