Advances in the Pathogenesis of EBV-Associated Diffuse Large B Cell Lymphoma
Abstract
Simple Summary
Abstract
1. Introduction
2. EBV Association with Lymphoma
3. Epidemiology of EBV + DLBCL in Different Populations
4. EBV Latent and Lytic Antigen Expression
5. Cellular Gene Expression
6. Genetic Alterations
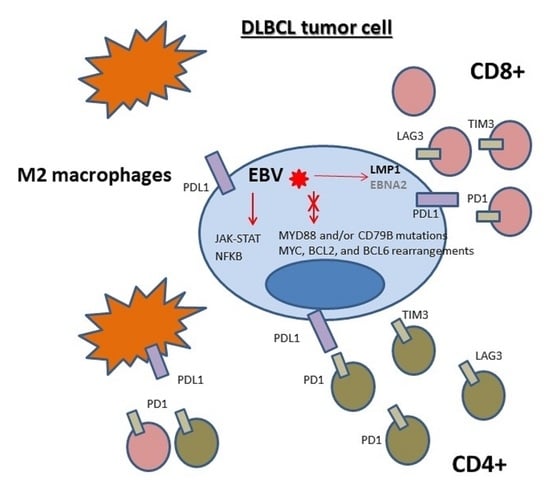
7. Microenvironment Composition
8. Survival
9. DLBCL Associated with Chronic Inflammation
10. Conclusions
Funding
Conflicts of Interest
References
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J. International Agency for Research on Cancer (IARC). WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th ed.; WHO Press: Geneva, Switzerland, 2008; Volume 2. [Google Scholar]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef]
- Nakamura, S.; Jaffe, E.S.; Swerdlow, S.H. EBV-positive diffuse large B-cell lymphoma, not otherwise specified (NOS). In IARC. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th ed.; Swerdlow, S.H., Campo, E., Harris, N.L., Jaffe, E.S., Pileri, S.A., Stein, H., Thiele, J., Eds.; WHO Press: Geneva, Switzerland, 2017; Volume 2. [Google Scholar]
- Zapatka, M.; Pathogens, P.; Borozan, I.; Brewer, D.S.; Iskar, M.; Grundhoff, A.; Alawi, M.; Desai, N.; Sültmann, H.; Moch, H.; et al. The landscape of viral associations in human cancers. Nat. Genet. 2020, 52, 320–330. [Google Scholar] [CrossRef]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Epstein-Barr Virus and Kaposi’s Sarcoma Herpesvirus/Human Herpesvirus 8. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, No. 70. Available online: https://www.ncbi.nlm.nih.gov/books/NBK385507/ (accessed on 20 December 2020).
- Young, L.S.; Rickinson, A.B. Epstein–Barr virus: 40 years on. Nat. Rev. Cancer 2004, 4, 757–768. [Google Scholar] [CrossRef]
- Chesnokova, L.S.; Jiang, R.; Hutt-Fletcher, L.M. Viral Entry. Curr. Top. Microbiol. Immunol. 2015, 391, 221–235. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.A. EBV Persistence—Introducing the Virus. Curr. Top. Microbiol. Immunol. 2015, 390, 151–209. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.A.; Hawkins, J.B.; Tracy, S.I.; Shapiro, M. The pathogenesis of Epstein–Barr virus persistent infection. Curr. Opin. Virol. 2013, 3, 227–232. [Google Scholar] [CrossRef]
- Rickinson, A.B.; Long, H.M.; Palendira, U.; Münz, C.; Hislop, A.D. Cellular immune controls over Epstein–Barr virus infection: New lessons from the clinic and the laboratory. Trends Immunol. 2014, 35, 159–169. [Google Scholar] [CrossRef]
- Smith, N.A.; Coleman, C.B.; Gewurz, B.E.; Rochford, R. CD21 (Complement Receptor 2) Is the Receptor for Epstein-Barr Virus Entry into T Cells. J. Virol. 2020, 94, e00428-20. [Google Scholar] [CrossRef] [PubMed]
- Coleman, C.B.; Lang, J.; Sweet, L.A.; Smith, N.A.; Freed, B.M.; Pan, Z.; Haverkos, B.; Pelanda, R.; Rochford, R. Epstein-Barr Virus Type 2 Infects T Cells and Induces B Cell Lymphomagenesis in Humanized Mice. J. Virol. 2018, 92, e00813-18. [Google Scholar] [CrossRef] [PubMed]
- Chijioke, O.; Müller, A.; Feederle, R.; Barros, M.H.M.; Krieg, C.; Emmel, V.; Marcenaro, E.; Leung, C.S.; Antsiferova, O.; Landtwing, V.; et al. Human Natural Killer Cells Prevent Infectious Mononucleosis Features by Targeting Lytic Epstein-Barr Virus Infection. Cell Rep. 2013, 5, 1489–1498. [Google Scholar] [CrossRef] [PubMed]
- Azzi, T.; Lünemann, A.; Murer, A.; Ueda, S.; Béziat, V.; Malmberg, K.-J.; Staubli, G.; Gysin, C.; Berger, C.; Münz, C.; et al. Role for early-differentiated natural killer cells in infectious mononucleosis. Blood 2014, 124, 2533–2543. [Google Scholar] [CrossRef]
- Taylor, G.S.; Long, H.M.; Brooks, J.M.; Rickinson, A.B.; Hislop, A.D. The Immunology of Epstein-Barr Virus–Induced Disease. Annu. Rev. Immunol. 2015, 33, 787–821. [Google Scholar] [CrossRef]
- Kempkes, B.; Robertson, E.S. Epstein-Barr virus latency: Current and future perspectives. Curr. Opin. Virol. 2015, 14, 138–144. [Google Scholar] [CrossRef]
- Shannon-Lowe, C.; Rickinson, A. The Global Landscape of EBV-Associated Tumors. Front. Oncol. 2019, 9, 713. [Google Scholar] [CrossRef]
- Oyama, T.; Ichimura, K.; Suzuki, R.; Suzumiya, J.; Ohshima, K.; Yatabe, Y.; Yokoi, T.; Kojima, M.; Kamiya, Y.; Taji, H.; et al. Senile EBV+ B-cell lymphoproliferative disorders: A clinicopathologic study of 22 patients. Am. J. Surg. Pathol. 2003, 27, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Oyama, T.; Yamamoto, K.; Asano, N.; Oshiro, A.; Suzuki, R.; Kagami, Y.; Morishima, Y.; Takeuchi, K.; Izumo, T.; Mori, S.; et al. Age-Related EBV-Associated B-Cell Lymphoproliferative Disorders Constitute a Distinct Clinicopathologic Group: A Study of 96 Patients. Clin. Cancer Res. 2007, 13, 5124–5132. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lee, J.; Ko, Y.H.; Han, A.; Jun, H.J.; Lee, S.C.; Hwang, I.G.; Park, Y.H.; Ahn, J.S.; Jung, C.W.; et al. The impact of Epstein-Barr virus status on clinical outcome in diffuse large B-cell lymphoma. Blood 2007, 110, 972–978. [Google Scholar] [CrossRef] [PubMed]
- Kuze, T.; Nakamura, N.; Hashimoto, Y.; Sasaki, Y.; Abe, M. The Characteristics of Epstein-Barr Virus (EBV)-positive Diffuse Large B-Cell Lymphoma: Comparison between EBV+ and EBV−Cases in Japanese Population. Jpn. J. Cancer Res. 2000, 91, 1233–1240. [Google Scholar] [CrossRef]
- Sato, A.; Nakamura, N.; Kojima, M.; Ohmachi, K.; Carreras, J.; Kikuti, Y.Y.; Numata, H.; Ohgiya, D.; Tazume, K.; Amaki, J.; et al. Clinical outcome of Epstein–Barr virus-positive diffuse large B-cell lymphoma of the elderly in the rituximab era. Cancer Sci. 2014, 105, 1170–1175. [Google Scholar] [CrossRef] [PubMed]
- Wada, N.; Ikeda, J.; Hori, Y.; Fujita, S.; Ogawa, H.; Soma, T.; Sugiyama, H.; Fukuhara, S.; Kanamaru, A.; Hino, M.; et al. Epstein-barr virus in diffuse large B-Cell lymphoma in immunocompetent patients in Japan is as low as in Western Countries. J. Med. Virol. 2011, 83, 317–321. [Google Scholar] [CrossRef]
- Yamauchi, A.; Fujita, S.; Ikeda, J.; Nakamichi, I.; Fukuhara, S.; Hino, M.; Kanakura, Y.; Ogawa, H.; Sugiyama, H.; Kanamaru, A.; et al. Diffuse large B-cell lymphoma in the young in Japan: A study by the Osaka Lymphoma Study Group. Am. J. Hematol. 2007, 82, 893–897. [Google Scholar] [CrossRef]
- Pan, Y.; Meng, B.; Zhang, H.; Cao, W.; Wang, H.; Bi, C.; Liu, F.; Sun, B.; Hao, X.; Ai, W.Z.; et al. Low incidence of Epstein–Barr virus-positive diffuse large B-cell lymphoma of the elderly in Tianjin, Northern China. Leuk. Lymphoma 2012, 54, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Park, M.; Lee, M.H.; Woo, H.J.; Kim, H.-W.; Yang, J.Y.; Eom, Y.-B.; Kim, S.-H.; Yoo, C.; Kim, J.-B. Development of EBV-encoded small RNA targeted PCR to classify EBV positive diffuse large B-cell lymphoma (DLBCL) of the elderly. Int. J. Clin. Exp. Pathol. 2015, 8, 7859–7868. [Google Scholar]
- Lu, T.-X.; Liang, J.-H.; Miao, Y.; Fan, L.; Wang, L.; Qu, X.-Y.; Cao, L.; Gong, Q.-X.; Wang, Z.; Zhang, Z.-H.; et al. Epstein-Barr virus positive diffuse large B-cell lymphoma predict poor outcome, regardless of the age. Sci. Rep. 2015, 5, 12168. [Google Scholar] [CrossRef]
- Chang, S.-T.; Lu, Y.-H.; Lu, C.-L.; Weng, S.-F.; Lin, S.-H.; Kuo, S.-Y.; Chuang, Y.-T.; Takeuchi, K.; Ohshima, K.; Chuang, S.-S. Epstein–Barr virus is rarely associated with diffuse large B cell lymphoma in Taiwan and carries a trend for a shorter median survival time. J. Clin. Pathol. 2013, 67, 326–332. [Google Scholar] [CrossRef]
- Hong, J.Y.; Yoon, D.H.; Suh, C.; Huh, J.; Do, I.-G.; Sohn, I.; Jo, J.; Jung, S.-H.; Hong, M.E.; Yoon, H.; et al. EBV-positive diffuse large B-cell lymphoma in young adults: Is this a distinct disease entity? Ann. Oncol. 2015, 26, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Gibson, S.E.; Hsi, E.D. Epstein-Barr virus–positive B-cell lymphoma of the elderly at a United States tertiary medical center: An uncommon aggressive lymphoma with a nongerminal center B-cell phenotype. Hum. Pathol. 2009, 40, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Hoeller, S.; Tzankov, A.; Pileri, S.A.; Went, P.; Dirnhofer, S. Epstein-Barr virus–positive diffuse large B-cell lymphoma in elderly patients is rare in Western populations. Hum. Pathol. 2010, 41, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Hofscheier, A.; Ponciano, A.; Bonzheim, I.; Adam, P.; Lome-Maldonado, C.; Vela, T.; Cortes, E.; Ortiz-Hidalgo, C.; Fend, F.; Quintanilla-Martinez, L. Geographic variation in the prevalence of Epstein–Barr virus-positive diffuse large B-cell lymphoma of the elderly: A comparative analysis of a Mexican and a German population. Mod. Pathol. 2011, 24, 1046–1054. [Google Scholar] [CrossRef]
- Tracy, S.I.; Habermann, T.M.; Feldman, A.L.; Maurer, M.J.; Dogan, A.; Perepu, U.S.; Syrbu, S.; Ansell, S.M.; Thompson, C.A.; Weiner, G.J.; et al. Outcomes among North American patients with diffuse large B-cell lymphoma are independent of tumor Epstein-Barr virus positivity or immunosuppression. Haematologica 2018, 103, 297–303. [Google Scholar] [CrossRef]
- Beltran, B.E.; Castillo, J.J.; Morales, D.; de Mendoza, F.H.; Quinones, P.; Miranda, R.N.; Gallo, A.; Lopez-Ilasaca, M.; Butera, J.N.; Sotomayor, E.M. EBV-positive diffuse large B-cell lymphoma of the elderly: A case series from Peru. Am. J. Hematol. 2011, 86, 663–667. [Google Scholar] [CrossRef]
- Beltran, B.E.; Morales, D.; Quinones, P.; Medeiros, L.J.; Miranda, R.N.; Castillo, J.J. EBV-Positive Diffuse Large B-Cell Lymphoma in Young Immunocompetent Individuals. Clin. Lymphoma Myeloma Leuk. 2011, 11, 512–516. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.; Narbaitz, M.; Metrebian, F.; De Matteo, E.; Preciado, M.V.; Chabay, P.A. Epstein-Barr virus-positive diffuse large B-cell lymphoma association is not only restricted to elderly patients. Int. J. Cancer 2014, 135, 2816–2824. [Google Scholar] [CrossRef] [PubMed]
- Cohen, M.; De Matteo, E.; Narbaitz, M.; Carreño, F.A.; Preciado, M.V.; Chabay, P.A. Epstein-Barr virus presence in pediatric diffuse large B-cell lymphoma reveals a particular association and latency patterns: Analysis of viral role in tumor microenvironment. Int. J. Cancer 2012, 132, 1572–1580. [Google Scholar] [CrossRef] [PubMed]
- Uccini, S.; Al-Jadiry, M.F.; Scarpino, S.; Ferraro, D.; Alsaadawi, A.R.; Al-Darraji, A.F.; Moleti, M.L.; Testi, A.M.; Al-Hadad, S.A.; Ruco, L. Epstein-Barr virus–positive diffuse large B-cell lymphoma in children: A disease reminiscent of Epstein-Barr virus–positive diffuse large B-cell lymphoma of the elderly. Hum. Pathol. 2015, 46, 716–724. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Karube, K.; Yamamoto, K.; Takizawa, J.; Tsuzuki, S.; Yatabe, Y.; Kanda, T.; Katayama, M.; Ozawa, Y.; Ishitsuka, K.; et al. Gene expression profiling of Epstein–Barr virus-positive diffuse large B-cell lymphoma of the elderly reveals alterations of characteristic oncogenetic pathways. Cancer Sci. 2014, 105, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, A.; Kato, S.; Okamoto, A.; Inaguma, Y.; Satou, A.; Tsuzuki, T.; Emi, N.; Okamoto, M.; Nakamura, S. Reappraisal of Epstein-Barr virus (EBV) in diffuse large B-cell lymphoma (DLBCL): Comparative analysis between EBV-positive and EBV-negative DLBCL with EBV-positive bystander cells. Histopathology 2017, 71, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Ok, C.Y.; Li, L.; Xu-Monette, Z.Y.; Visco, C.; Tzankov, A.; Manyam, G.C.; Montes-Moreno, S.; Dybaer, K.; Chiu, A.; Orazi, A.; et al. Prevalence and Clinical Implications of Epstein–Barr Virus Infection in De Novo Diffuse Large B-Cell Lymphoma in Western Countries. Clin. Cancer Res. 2014, 20, 2338–2349. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-H.; Lee, K.-F.; Chen, C.-C.; Chen, Y.-Y.; Huang, C.-E.; Tsai, P.-S.; Tsou, H.-Y.; Chou, H.-J.; Chen, M.-F.; Chen, P.-T.; et al. Clinical Characteristics and Treatment Outcome in a Taiwanese Population of Patients with Epstein-Barr Virus-positive Diffuse Large B-cell Lymphoma. Jpn. J. Clin. Oncol. 2014, 44, 1164–1171. [Google Scholar] [CrossRef]
- Ahn, J.-S.; Yang, D.-H.; Choi, Y.D.; Jung, S.-H.; Yhim, H.-Y.; Kwak, J.-Y.; Park, H.S.; Shin, M.-G.; Kim, Y.-K.; Kim, H.-J.; et al. Clinical outcome of elderly patients with Epstein-Barr virus positive diffuse large B-cell lymphoma treated with a combination of rituximab and CHOP chemotherapy. Am. J. Hematol. 2013, 88, 774–779. [Google Scholar] [CrossRef]
- Beltran, B.E.; Quiñones, P.; Morales, D.; Malaga, J.M.; Chavez, J.C.; Sotomayor, E.M.; Castillo, J.J. Response and survival benefit with chemoimmunotherapy in Epstein-Barr virus-positive diffuse large B-cell lymphoma. Hematol. Oncol. 2018, 36, 93–97. [Google Scholar] [CrossRef]
- Balfour, H.H.; Dunmire, S.K.; Hogquist, K.A. Infectious mononucleosis. Clin. Transl. Immunol. 2015, 4, e33. [Google Scholar] [CrossRef] [PubMed]
- Montes-Moreno, S.; Odqvist, L.; Diaz-Perez, J.A.; Lopez, A.B.; De Villambrosía, S.G.; Mazorra, F.; Castillo, M.E.; Lopez, M.; Pajares, R.; García, J.F.; et al. EBV-positive diffuse large B-cell lymphoma of the elderly is an aggressive post-germinal center B-cell neoplasm characterized by prominent nuclear factor-kB activation. Mod. Pathol. 2012, 25, 968–982. [Google Scholar] [CrossRef] [PubMed]
- Mundo, L.; Del Porro, L.; Granai, M.; Siciliano, M.C.; Mancini, V.; Santi, R.; Marcar, L.; Vrzalikova, K.; Vergoni, F.; Di Stefano, G.; et al. Frequent traces of EBV infection in Hodgkin and non-Hodgkin lymphomas classified as EBV-negative by routine methods: Expanding the landscape of EBV-related lymphomas. Mod. Pathol. 2020, 33, 2407–2421. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Robertson, E.S. Mechanisms of B-Cell Oncogenesis Induced by Epstein-Barr Virus. J. Virol. 2019, 93, e00238-19. [Google Scholar] [CrossRef] [PubMed]
- Shindiapina, P.; Ahmed, E.H.; Mozhenkova, A.; Abebe, T.; Baiocchi, R.A. Immunology of EBV-Related Lymphoproliferative Disease in HIV-Positive Individuals. Front. Oncol. 2020, 10, 1723. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.-S.; Kieff, E. Epstein–Barr virus latent genes. Exp. Mol. Med. 2015, 47, e131. [Google Scholar] [CrossRef]
- Münz, C. Latency and lytic replication in Epstein–Barr virus-associated oncogenesis. Nat. Rev. Genet. 2019, 17, 691–700. [Google Scholar] [CrossRef]
- Mancao, C.; Hammerschmidt, W. Epstein-Barr virus latent membrane protein 2A is a B-cell receptor mimic and essential for B-cell survival. Blood 2007, 110, 3715–3721. [Google Scholar] [CrossRef] [PubMed]
- Nguyen-Van, D.; Keane, C.; Han, E.; Jones, K.; Nourse, J.P.; Vari, F.; Ross, N.; Crooks, P.; Ramuz, O.; Green, M.; et al. Epstein-Barr virus-positive diffuse large B-cell lymphoma of the elderly expresses EBNA3A with conserved CD8+ T-cell epitopes. Am. J. Blood Res. 2011, 1, 146–159. [Google Scholar]
- Nicolae, A.; Pittaluga, S.; Abdullah, S.; Steinberg, S.M.; Pham, T.A.; Davies-Hill, T.; Xi, L.; Raffeld, M.; Jaffe, E.S. EBV-positive large B-cell lymphomas in young patients: A nodal lymphoma with evidence for a tolerogenic immune environment. Blood 2015, 126, 863–872. [Google Scholar] [CrossRef]
- Cohen, M.; Vistarop, A.G.; Huaman, F.; Narbaitz, M.; Metrebian, F.; De Matteo, E.; Preciado, M.V.; Chabay, P.A. Epstein-Barr virus lytic cycle involvement in diffuse large B cell lymphoma. Hematol. Oncol. 2018, 36, 98–103. [Google Scholar] [CrossRef]
- Adam, P.; Bonzheim, I.; Fend, F.; Quintanilla-Martínez, L. Epstein-Barr Virus-positive Diffuse Large B-cell Lymphomas of the Elderly. Adv. Anat. Pathol. 2011, 18, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Marques-Piubelli, M.L.; Salas, Y.I.; Pachas, C.; Becker-Hecker, R.; Vega, F.; Miranda, R.N. Epstein–Barr virus-associated B-cell lymphoproliferative disorders and lymphomas: A review. Pathology 2020, 52, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Basso, K.; Dalla-Favera, R. Germinal centres and B cell lymphomagenesis. Nat. Rev. Immunol. 2015, 15, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.B.; Corinaldesi, C.; Shen, Q.; Kumar, R.; Compagno, N.; Wang, Z.; Nitzan, M.; Grunstein, E.; Pasqualucci, L.; Dalla-Favera, R.; et al. Single-cell analysis of germinal-center B cells informs on lymphoma cell of origin and outcome. J. Exp. Med. 2020, 217, 20200483. [Google Scholar] [CrossRef] [PubMed]
- Lenz, G.; Wright, G.; Dave, S.; Xiao, W.; Powell, J.; Zhao, H.; Xu, W.; Tan, B.; Goldschmidt, N.; Iqbal, J.; et al. Stromal Gene Signatures in Large-B-Cell Lymphomas. N. Engl. J. Med. 2008, 359, 2313–2323. [Google Scholar] [CrossRef]
- Scott, D.W.; Mottok, A.; Ennishi, D.; Wright, G.W.; Farinha, P.; Ben-Neriah, S.; Kridel, R.; Barry, G.S.; Hother, C.; Abrisqueta, P.; et al. Prognostic Significance of Diffuse Large B-Cell Lymphoma Cell of Origin Determined by Digital Gene Expression in Formalin-Fixed Paraffin-Embedded Tissue Biopsies. J. Clin. Oncol. 2015, 33, 2848–2856. [Google Scholar] [CrossRef] [PubMed]
- Ennishi, D.; Mottok, A.; Ben-Neriah, S.; Shulha, H.P.; Farinha, P.; Chan, F.C.; Meissner, B.; Boyle, M.; Hother, C.; Kridel, R.; et al. Genetic profiling of MYC and BCL2 in diffuse large B-cell lymphoma determines cell-of-origin–specific clinical impact. Blood 2017, 129, 2760–2770. [Google Scholar] [CrossRef]
- Hans, C.P.; Weisenburger, D.D.; Greiner, T.C.; Gascoyne, R.D.; Delabie, J.; Ott, G.; Müller-Hermelink, H.K.; Campo, E.; Braziel, R.M.; Jaffe, E.S.; et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004, 103, 275–282. [Google Scholar] [CrossRef]
- Abdulla, M.; Hollander, P.; Pandzic, T.; Mansouri, L.; Ednersson, S.B.; Andersson, P.; Hultdin, M.; Fors, M.; Erlanson, M.; Degerman, S.; et al. Cell-of-origin determined by both gene expression profiling and immunohistochemistry is the strongest predictor of survival in patients with diffuse large B-cell lymphoma. Am. J. Hematol. 2020, 95, 57–67. [Google Scholar] [CrossRef]
- Healy, J.A.; Dave, S.S. The Role of EBV in the Pathogenesis of Diffuse Large B Cell Lymphoma. Curr. Top. Microbiol. Immunol. 2015, 390, 315–337. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Li, J.; Wang, Y.; Liu, S.; Yue, B. Clinical characteristics and prognostic significance of EBER positivity in diffuse large B-cell lymphoma: A meta-analysis. PLoS ONE 2018, 13, e0199398. [Google Scholar] [CrossRef]
- Cassim, S.; Antel, K.; Chetty, D.R.; Oosthuizen, J.; Opie, J.; Mohamed, Z.; Verburgh, E. Diffuse large B-cell lymphoma in a South African cohort with a high HIV prevalence: An analysis by cell-of-origin, Epstein–Barr virus infection and survival. Pathology 2020, 52, 453–459. [Google Scholar] [CrossRef]
- Cohen, M.; Vistarop, A.G.; Huaman, F.; Narbaitz, M.; Metrebian, F.; De Matteo, E.; Preciado, M.V.; Chabay, P.A. Cytotoxic response against Epstein Barr virus coexists with diffuse large B-cell lymphoma tolerogenic microenvironment: Clinical features and survival impact. Sci. Rep. 2017, 7, 10813. [Google Scholar] [CrossRef] [PubMed]
- Battle-Lopez, A.; De Villambrosia, S.G.; Nuñez, J.; Cagigal, M.-L.; Montes-Moreno, S.; Conde, E.; Piris, M.A. Epstein-Barr virus-associated diffuse large B-cell lymphoma: Diagnosis, difficulties and therapeutic options. Expert Rev. Anticancer. Ther. 2016, 16, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.; Park, S.; Ju, H.; Ha, S.Y.; Sohn, I.; Jo, J.; Do, I.; Min, S.; Kim, S.J.; Kim, W.S.; et al. Integrated copy number and gene expression profiling analysis of epstein–barr virus-positive diffuse large b-cell lymphoma. Genes Chromosom. Cancer 2015, 54, 383–396. [Google Scholar] [CrossRef]
- Sehn, L.H.; Gascoyne, R.D. Diffuse large B-cell lymphoma: Optimizing outcome in the context of clinical and biologic heterogeneity. Blood 2015, 125, 22–32. [Google Scholar] [CrossRef]
- El Hussein, S.; Shaw, K.R.M.; Vega, F. Evolving insights into the genomic complexity and immune landscape of diffuse large B-cell lymphoma: Opportunities for novel biomarkers. Mod. Pathol. 2020, 33, 2422–2436. [Google Scholar] [CrossRef]
- Pasqualucci, L.; Dalla-Favera, R. Genetics of diffuse large B-cell lymphoma. Blood 2018, 131, 2307–2319. [Google Scholar] [CrossRef]
- Vermaat, J.S.; Somers, S.F.; De Wreede, L.C.; Kraan, W.; De Groen, R.A.L.; Schrader, A.M.R.; Kerver, E.D.; Scheepstra, C.G.; Beerenschot, H.; Deenik, W.; et al. MYD88 mutations identify a molecular subgroup of diffuse large B-cell lymphoma with an unfavorable prognosis. Haematolongica 2019, 105, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Gebauer, N.; Gebauer, J.; Hardel, T.T.; Bernard, V.; Biersack, H.; Lehnert, H.; Rades, D.; Feller, A.C.; Thorns, C. Prevalence of targetable oncogenic mutations and genomic alterations in Epstein–Barr virus-associated diffuse large B-cell lymphoma of the elderly. Leuk. Lymphoma 2015, 56, 1100–1106. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, K.; Miyoshi, H.; Sakata, S.; Dobashi, A.; Couronné, L.; Kogure, Y.; Sato, Y.; Nishida, K.; Gion, Y.; Shiraishi, Y.; et al. Frequent structural variations involving programmed death ligands in Epstein-Barr virus-associated lymphomas. Leukemia 2019, 33, 1687–1699. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xu, Z.; Lin, W.; Duan, Y.; Lu, C.; Liu, W.; Su, W.; Yan, Y.; Liu, H.; Liu, L.; et al. Comprehensive Genomic Profiling of EBV-Positive Diffuse Large B-cell Lymphoma and the Expression and Clinicopathological Correlations of Some Related Genes. Front. Oncol. 2019, 9, 683. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, Z.; Zhou, X.; Liu, Q.; Chen, G.; Xiao, H.; Yin, W.; Nakamura, S.; Rao, H. Genetic heterogeneity and mutational signature in Chinese Epstein-Barr virus-positive diffuse large B-cell lymphoma. PLoS ONE 2018, 13, e0201546. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Rodig, S.; Juszczynski, P.; Ouyang, J.; Sinha, P.; O’Donnell, E.; Neuberg, D.; Shipp, M.A. Constitutive AP-1 Activity and EBV Infection Induce PD-L1 in Hodgkin Lymphomas and Posttransplant Lymphoproliferative Disorders: Implications for Targeted Therapy. Clin. Cancer Res. 2012, 18, 1611–1618. [Google Scholar] [CrossRef]
- Georgiou, K.; Chen, L.; Berglund, M.; Ren, W.; de Miranda, N.; Lisboa, S.; Fangazio, M.; Zhu, S.; Hou, Y.; Wu, K.; et al. Genetic basis of PD-L1 overexpression in diffuse large B-cell lymphomas. Blood 2016, 127, 3026–3034. [Google Scholar] [CrossRef]
- Takahara, T.; Satou, A.; Ishikawa, E.; Kohno, K.; Kato, S.; Suzuki, Y.; Takahashi, E.; Ohashi, A.; Asano, N.; Tsuzuki, T.; et al. Clinicopathological analysis of neoplastic PD-L1-positive EBV+ diffuse large B cell lymphoma, not otherwise specified, in a Japanese cohort. Virchows Arch. 2021, 478, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.; Kim, S.; Kim, P.-J.; Go, H.; Nam, S.J.; Paik, J.H.; Kim, Y.A.; Kim, T.M.; Heo, D.S.; Kim, C.W.; et al. Clinicopathological analysis of programmed cell death 1 and programmed cell death ligand 1 expression in the tumour microenvironments of diffuse large B cell lymphomas. Histopathology 2015, 68, 1079–1089. [Google Scholar] [CrossRef]
- Barzyk, G.A.; Sheriff, V. EBV Positivity and Programmed Death-ligand 1 Expression in Diffuse Large B-cell Lymphoma: A Systematic Review. Anticancer Res. 2020, 40, 5951–5968. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.W.; Visser, L.; Tan, L.P.; Berg, A.V.D.; Diepstra, A. The Microenvironment in Epstein–Barr Virus-Associated Malignancies. Pathogens 2018, 7, 40. [Google Scholar] [CrossRef] [PubMed]
- Kamper, P.; Bendix, K.; Hamilton-Dutoit, S.; Honoré, B.; Nyengaard, J.R.; D’Amore, F. Tumor-infiltrating macrophages correlate with adverse prognosis and Epstein-Barr virus status in classical Hodgkin’s lymphoma. Haematologica 2010, 96, 269–276. [Google Scholar] [CrossRef]
- Barros, M.H.M.; Hassan, R.; Niedobitek, G. Tumor-Associated Macrophages in Pediatric Classical Hodgkin Lymphoma: Association with Epstein-Barr Virus, Lymphocyte Subsets, and Prognostic Impact. Clin. Cancer Res. 2012, 18, 3762–3771. [Google Scholar] [CrossRef] [PubMed]
- Derks, S.; Liao, X.; Chiaravalli, A.M.; Xu, X.; Camargo, M.C.; Solcia, E.; Sessa, F.; Fleitas, T.; Freeman, G.J.; Rodig, S.J.; et al. Abundant PD-L1 expression in Epstein-Barr Virus-infected gastric cancers. Oncotarget 2016, 7, 32925–32932. [Google Scholar] [CrossRef] [PubMed]
- Saito, R.; Abe, H.; Kunita, A.; Yamashita, H.; Seto, Y.; Fukayama, M. Overexpression and gene amplification of PD-L1 in cancer cells and PD-L1+ immune cells in Epstein–Barr virus-associated gastric cancer: The prognostic implications. Mod. Pathol. 2017, 30, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.-Y.; Guo, S.-S.; Zhang, Y.; Lu, J.-B.; Chen, Q.-Y.; Tang, L.-Q.; Zhang, L.; Liu, L.-T.; Zhang, L.; Mai, H.-Q. Tumor CTLA-4 overexpression predicts poor survival in patients with nasopharyngeal carcinoma. Oncotarget 2016, 7, 13060–13068. [Google Scholar] [CrossRef] [PubMed]
- Kiyasu, J.; Miyoshi, H.; Hirata, A.; Arakawa, F.; Ichikawa, A.; Niino, D.; Sugita, Y.; Yufu, Y.; Choi, I.; Abe, Y.; et al. Expression of programmed cell death ligand 1 is associated with poor overall survival in patients with diffuse large B-cell lymphoma. Blood 2015, 126, 2193–2201. [Google Scholar] [CrossRef]
- Chen, B.J.; Chapuy, B.; Ouyang, J.; Sun, H.H.; Roemer, M.G.; Xu, M.L.; Yu, H.; Fletcher, C.D.; Freeman, G.J.; Shipp, M.A.; et al. PD-L1 Expression Is Characteristic of a Subset of Aggressive B-cell Lymphomas and Virus-Associated Malignancies. Clin. Cancer Res. 2013, 19, 3462–3473. [Google Scholar] [CrossRef]
- Veloza, L.; Teixido, C.; Castrejon, N.; Climent, F.; Carrió, A.; Marginet, M.; Soldini, D.; Gonzalez-Farre, B.; Ribera-Cortada, I.; Lopez-Guillermo, A.; et al. Clinicopathological evaluation of the programmed cell death 1 (PD1)/programmed cell death-ligand 1 (PD-L1) axis in post-transplant lymphoproliferative disorders: Association with Epstein–Barr virus, PD-L1copy number alterations, and outcome. Histopathology 2019, 75, 799–812. [Google Scholar] [CrossRef] [PubMed]
- Quan, L.; Chen, X.; Liu, A.; Zhang, Y.; Guo, X.; Yan, S.; Liu, Y. PD-1 Blockade Can Restore Functions of T-Cells in Epstein-Barr Virus-Positive Diffuse Large B-Cell Lymphoma In Vitro. PLoS ONE 2015, 10, e0136476. [Google Scholar] [CrossRef]
- Auclair, H.; Ouk-Martin, C.; Roland, L.; Santa, P.; Al Mohamad, H.; Faumont, N.; Feuillard, J.; Jayat-Vignoles, C. EBV Latency III–Transformed B Cells Are Inducers of Conventional and Unconventional Regulatory T Cells in a PD-L1–Dependent Manner. J. Immunol. 2019, 203, 1665–1674. [Google Scholar] [CrossRef] [PubMed]
- Cárdenas, D.; Velez, G.; Orfao, A.; Herrera, M.V.; Solano, J.; Olaya, M.; Uribe, A.M.; Saavedra, C.; Duarte, M.; Rodríguez, M.; et al. Epstein–Barr virus-specific CD8+ T lymphocytes from diffuse large B cell lymphoma patients are functionally impaired. Clin. Exp. Immunol. 2015, 182, 173–183. [Google Scholar] [CrossRef]
- Keane, C.; Tobin, J.; Gunawardana, J.; Francis, S.; Gifford, G.; Gabrielli, S.; Gill, A.; Stevenson, W.; Talaulikar, D.; Gould, C.; et al. The tumour microenvironment is immuno-tolerogenic and a principal determinant of patient outcome in EBV-positive diffuse large B-cell lymphoma. Eur. J. Haematol. 2019, 103, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Dolcetti, R. Cross-talk between Epstein-Barr virus and microenvironment in the pathogenesis of lymphomas. Semin. Cancer Biol. 2015, 34, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, K.; Urano, E.; Yoshiyama, H.; Komano, J. Cytokine signatures of transformed B cells with distinct Epstein-Barr virus latencies as a potential diagnostic tool for B cell lymphoma. Cancer Sci. 2011, 102, 1236–1241. [Google Scholar] [CrossRef] [PubMed]
- Vaysberg, M.; Lambert, S.L.; Krams, S.M.; Martinez, O.M. Activation of the JAK/STAT Pathway in Epstein Barr Virus+-Associated Posttransplant Lymphoproliferative Disease: Role of Interferon-γ. Arab. Archaeol. Epigr. 2009, 9, 2292–2302. [Google Scholar] [CrossRef][Green Version]
- Incrocci, R.; McCormack, M.; Swanson-Mungerson, M. Epstein–Barr virus LMP2A increases IL-10 production in mitogen-stimulated primary B-cells and B-cell lymphomas. J. Gen. Virol. 2013, 94, 1127–1133. [Google Scholar] [CrossRef]
- Mahot, S.; Sergeant, A.; Drouet, E.; Gruffat, H. A novel function for the Epstein–Barr virus transcription factor EB1/Zta: Induction of transcription of the hIL-10 gene. J. Gen. Virol. 2003, 84, 965–974. [Google Scholar] [CrossRef] [PubMed]
- Skinnider, B.F.; Mak, T.W. The role of cytokines in classical Hodgkin lymphoma. Blood 2002, 99, 4283–4297. [Google Scholar] [CrossRef]
- Iwakiri, D.; Takada, K. Role of EBERs in the Pathogenesis of EBV Infection. Adv. Cancer Res. 2010, 107, 119–136. [Google Scholar] [CrossRef]
- Beltran, B.E.; Castro, D.; Paredes, S.; Miranda, R.N.; Castillo, J.J. EBV-positive diffuse large B-cell lymphoma, not otherwise specified: 2020 update on diagnosis, risk-stratification and management. Am. J. Hematol. 2020, 95, 435–445. [Google Scholar] [CrossRef]
- Song, C.-G.; Huang, J.-J.; Li, Y.-J.; Xia, Y.; Wang, Y.; Bi, X.-W.; Jiang, W.-Q.; Huang, H.-Q.; Lin, T.-Y.; Li, Z.-M. Epstein-Barr Virus-Positive Diffuse Large B-Cell Lymphoma in the Elderly: A Matched Case-Control Analysis. PLoS ONE 2015, 10, e0133973. [Google Scholar] [CrossRef]
- Hong, J.Y.; Ryu, K.J.; Park, C.; Hong, M.; Ko, Y.H.; Kim, W.S.; Kim, S.J. Clinical impact of serum survivin positivity and tissue expression of EBV-encoded RNA in diffuse large B-cell lymphoma patients treated with rituximab-CHOP. Oncotarget 2017, 8, 13782–13791. [Google Scholar] [CrossRef][Green Version]
- Okamoto, A.; Yanada, M.; Inaguma, Y.; Tokuda, M.; Morishima, S.; Kanie, T.; Yamamoto, Y.; Mizuta, S.; Akatsuka, Y.; Yoshikawa, T.; et al. The prognostic significance of EBV DNA load and EBER status in diagnostic specimens from diffuse large B-cell lymphoma patients. Hematol. Oncol. 2015, 35, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Witte, H.M.; Merz, H.; Biersack, H.; Bernard, V.; Riecke, A.; Gebauer, J.; Lehnert, H.; Von Bubnoff, N.; Feller, A.C.; Gebauer, N. Impact of treatment variability and clinicopathological characteristics on survival in patients with Epstein-Barr-Virus positive diffuse large B cell lymphoma. Br. J. Haematol. 2020, 189, 257–268. [Google Scholar] [CrossRef]
- Lee, H.; Shin, H.; Kim, N.Y.; Park, H.S.; Park, J. Diffuse Large B-Cell Lymphoma Arising within Ileal Neobladder: An Expanding Spectrum of Diffuse Large B-Cell Lymphoma Associated with Chronic Inflammation. Cancer Res. Treat. 2019, 51, 1666–1670. [Google Scholar] [CrossRef] [PubMed]
- Mescam, L.; Camus, V.; Schiano, J.-M.; Adélaïde, J.; Picquenot, J.-M.; Guille, A.; Bannier, M.; Ruminy, P.; Viailly, P.-J.; Jardin, F.; et al. EBV+ diffuse large B-cell lymphoma associated with chronic inflammation expands the spectrum of breast implant-related lymphomas. Blood 2020, 135, 2004–2009. [Google Scholar] [CrossRef]
- Khoo, C.; McTigue, C.; Hunter-Smith, D.J.; Walker, P. EBV positive fibrin/chronic inflammation associated diffuse large B-cell lymphoma: An incidental finding associated with a breast implant. Pathology 2020, 31. in press. [Google Scholar]
- Sukswai, N.; Lyapichev, K.; Khoury, J.D.; Medeiros, L.J. Diffuse large B-cell lymphoma variants: An update. Pathology 2020, 52, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, T.; Matsuo, K.; Hashida, Y.; Kitahata, K.; Ujihara, T.; Taniguchi, A.; Yoshie, O.; Nakayama, T.; Daibata, M. Epstein–Barr virus-positive pyothorax-associated lymphoma expresses CCL17 and CCL22 chemokines that attract CCR4-expressing regulatory T cells. Cancer Lett. 2019, 453, 184–192. [Google Scholar] [CrossRef]
- Grimm, K.E.; O’Malley, D.P. Aggressive B cell lymphomas in the 2017 revised WHO classification of tumors of hematopoietic and lymphoid tissues. Ann. Diagn. Pathol. 2019, 38, 6–10. [Google Scholar] [CrossRef] [PubMed]

| Study | EBV + DLBCL Incidence | EBV + Cells Cut Off | Patients’ Origin | Latency Type | EBV Effect on Survival |
|---|---|---|---|---|---|
| Oyama et al., 2007 [18] | 5.5% | >50% | Japan | II/III | 60% (17 months) # |
| Oyama et al., 2007 [19] | 14% | >50% | Japan | II/III | EBV + inferior survival |
| Park et al., 2007 [20] | 9% | >20% | Korea | NA | EBV + inferior survival |
| Sato et al., 2014 [22] | 6.9% | >30% | Japan | II/III | EBV + inferior survival |
| Wada et al., 2011 [23] | 5.2% | >20% | Japan | NA | No difference |
| Yamauchi et al., 2007 [24] | 2% | NS | Japan | NA | NS |
| Pan et al., 2013 [25] | 3.8% | >50% | China | NA | EBV + inferior survival |
| Lu et al., 2015 [27] | 14% | >20% | China | NA | EBV + inferior survival |
| Chang et al., 2014 [28] | 4.5% | >10% | Taiwan | II/III | EBV + inferior survival (trend) |
| Hong et al., 2015 [29] | 8.4% | >20% | Korea | NA | EBV + inferior survival |
| Gibson et al., 2009 [30] | 5.3% | 80% | USA | NA | NS |
| Hoeller et al., 2010 [31] | 3.1% | >10% | Switzerland, Italy, Austria | II/III | EBV + inferior survival |
| Hofscheier et al., 2011 [32] | 8.1% | 90% | Mexico | II/III | NS |
| Tracy et al., 2018 [33] | 4.4% | >30% | USA | NA | No difference |
| Beltran et al., 2011 [34] | 19.6% | >20% | Peru | 40% (3 years) # | |
| Cohen et al., 2014 [36] and 2017 [39] | 9.3% | >20% | Argentina | II/III | EBV + inferior survival |
| Uccini et al., 2015 [38] | 54% | NS | Irak | II | 2/6 CR # |
| Ohashi et al., 2017 [40] | 4.5% | >80% | Japan | NA | EBV + inferior survival |
| Ok et al., 2014 [41] | 4% | >10% | USA | NA | No difference |
| Lu et al., 2014 [42] | 16.9% | >20% | Taiwan | NA | No difference |
| Ahn et al., 2013 [43] | 8.1% | >50% | Korea | NA | EBV + inferior survival |
| Beltran et al., 2018 [44] | 28% | >20% | Peru | NA | 54% (5 years) # |
| EBV-Associated Neoplasia | Latency Type | Viral Antigen Expression |
|---|---|---|
| Burkitt lymphoma (sporadic and endemic) | I | EBERs,miRNAs, EBNA1 |
| Gastric carcinoma | I | EBERs,miRNAs, EBNA1 |
| NK/T cell lymphoma | I | EBERs,miRNAs, EBNA1 |
| Plasmablastic lymphoma | I | EBERs,miRNAs, EBNA1 |
| Primary effusion lymphoma | I | EBERs,miRNAs, EBNA1 |
| Hodgkin lymphoma | II | EBERs,miRNAs, EBNA1, LMP1, 2A, 2B |
| Nasopharyngeal carcinoma | II | EBERs,miRNAs, EBNA1, LMP1, 2A, 2B |
| Diffuse large B cell lymphoma | II | EBERs,miRNAs, EBNA1, LMP1, 2A, 2B |
| Diffuse large B cell lymphoma | III | EBERs,miRNAs, EBNA1, LMP1, 2A, 2B, EBNA2, EBNA3A, 3B, 3C, LP |
| Postransplant lymphoproliferative disorder | III | EBERs,miRNAs, EBNA1, LMP1, 2A, 2B, EBNA2, EBNA3A, 3B, 3C, LP |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chabay, P. Advances in the Pathogenesis of EBV-Associated Diffuse Large B Cell Lymphoma. Cancers 2021, 13, 2717. https://doi.org/10.3390/cancers13112717
Chabay P. Advances in the Pathogenesis of EBV-Associated Diffuse Large B Cell Lymphoma. Cancers. 2021; 13(11):2717. https://doi.org/10.3390/cancers13112717
Chicago/Turabian StyleChabay, Paola. 2021. "Advances in the Pathogenesis of EBV-Associated Diffuse Large B Cell Lymphoma" Cancers 13, no. 11: 2717. https://doi.org/10.3390/cancers13112717
APA StyleChabay, P. (2021). Advances in the Pathogenesis of EBV-Associated Diffuse Large B Cell Lymphoma. Cancers, 13(11), 2717. https://doi.org/10.3390/cancers13112717